Abstract
Cryptochromes are blue-light absorbing flavoproteins with many important signaling roles in plants, including in de-etiolation, development, and stress response. They interact with downstream signaling partners such as transcription factors and components of the proteasome, and thereby alter regulation of nuclear gene expression in a light dependent manner. In a prior study, it has also been shown that Arabidopsis cry1 activation by blue light results in direct enzymatic conversion of molecular oxygen (O2) to ROS (reactive oxygen species) in vivo leading to cell death in overexpressing lines. Here we extend these observations to show that Atcry2 is translocated from the cytosol to the nucleus in response to blue light illumination, resulting in nuclear accumulation of ROS in expressing insect cell cultures. These observations suggest that ROS formation may represent a novel means of signaling by Atcry2 distinct from, and perhaps complementary to, the currently known mechanism of light-mediated conformational change.
Introduction
Cryptochromes are flavoprotein blue light receptors with many signaling roles in plants including elongation growth, entrainment of the circadian clock, de-etiolation, and resistance to many different forms of stress.Citation1 There are 2 highly similar cryptochrome genes in Arabidopsis with overlapping functions but somewhat different light sensitivity; cry1 is stable at high light intensity whereas cry2 is rapidly degraded in the presence of blue light due to ubiquitination and targeting to the proteasome.Citation2 Until the present time, attempts to understand the mechanism of cry activation have focused on light-driven conformational change in the receptor that lead to interaction with transcription factors and/or elements of the proteasome. Certain changes in gene expression, for instance in the induction of flowering time and in de-etiolation responses have indeed been convincingly linked to cryptochrome interaction with these biological partners (reviewed inCitation1).
Recently, we have shown that Arabidopsis cryptochrome-1 (Atcry1) activation by blue light leads directly to the formation of ROS (Reactive Oxygen Species) and hydrogen peroxide in living insect cells that overexpress this protein.Citation3 We show the accumulation of ROS using a fluorescence-based assay for the detection of H2O2 in cell culture media (Amplex Ultra Red) and by direct visualization of ROS in living cells by immunofluorescence imaging techniques. Of specific interest, ROS were shown to accumulate in the nucleus in Atcry1 expressing cells, and therefore are in potential contact with transcriptional regulators and elements of the transcriptional machinery. ROS are known to act as secondary messengers, which can modify elements of signaling pathways in numerous organisms.Citation4 In plants, reactive oxygen species are formed primarily as byproducts of photosynthesis, respiration, and metabolic processes and therefore are abundant in compartments such as the chloroplast and mitochondrion.Citation5 The cell has furthermore developed many protective systems for rapid removal of potentially damaging ROS, as a result of which there is little diffusion of ROS between cellular compartments.Citation5 Therefore, a mechanism whereby the cell has the possibility to generate a localized, light-dependent accumulation of ROS in the nucleus itself may be of potential importance for signaling.
Here, we provide new data showing that ROS accumulation in the nucleus also results from illumination of cryptochrome. We show by direct immunostaining with anti-cry2 antibodies that cry2 protein is initially localized exclusively in the cytoplasm in dark-adapted cell cultures, and then travels rapidly to the nucleus upon illumination. We further show the accumulation of ROS in the nucleus occurs as a direct consequence of cry2 illumination by blue light.
Atcry2 is translocated into the nucleus in a blue-light dependent manner
Insect cell cultures expressing high levels of Atcry2 maintained in darkness show staining surrounding the plasma membrane .() Permeabilization of cells with 0.1% Triton X100 (see reference 3) results in detection of the Atcry2 protein within the cytosol as well, but does not detect the protein in the nucleus .() This indicates that all cry2 protein in insect cells is cytosolic in the dark. However, after a 15-minute pulse of high intensity blue light (100μmol/m−2/sec−1), a number of differences are observed. Firstly, there is a significant increase in staining observed at the plasma membrane () and in the cytosol .() This more intense signal is not due to an increase in the concentration of the protein, but rather likely to a conformational change in the C-terminal domain of Atcry2 which ‘flips outwards’ from the protein as a result of illumination. In this way, there is improved accessibility of the antibody to its antigenic sites in the C-terminal domain. Such effects have been observed for cryptochromes in other systems as well.Citation7
Figure 1. Subcellular localization of Atcry2 in insect cells by immunofluorescence staining and confocal microscopy.Sf21 cells stably expressing Atcry2 were fixed with paraformaldehyde, permeabilized with Triton X100 (A) to (F) or not (G) to (L), incubated with a rabbit polyclonal anti-Atcry2 antibody and an Alexa 488-conjugated anti-rabbit secondary antibody, DNA were stained with Dapi. Cells were observed with an inverted Leica TCS SP5 microscope. Images (A) to (F) show projection of optical sections, scale bar 10 μm. Images (G) to (L) show single confocal z-section that cross the nucleus, scale bar 10 μm.
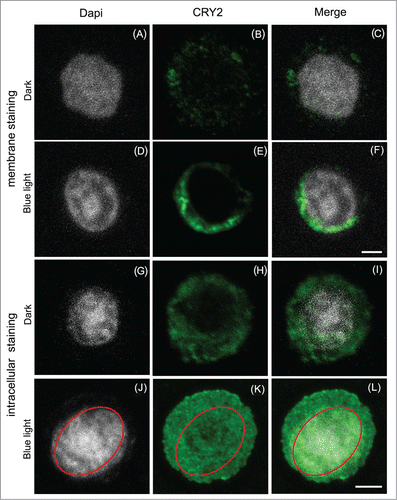
Secondly, a new localization for cry2 can also be clearly observed inside the nucleus of permeabilized cells (). The position of the nucleus stained with DAPI has been delineated with a red ellipse (). Atcry2 staining co-localizes with DAPI in the merged image () and can be clearly seen inside the red ellipse (). Atcry2 nuclear localization has also been confirmed by nuclear 3D analysis as described previouslyCitation3 (data not shown). It is therefore evident that cry2 undergoes light-dependent translocation into the nucleus in this heterologous insect cell culture system, somewhat in contrast to what has been reported in plants.Citation8
Since the nuclear localization signal of cry2 is located within the C-terminal domain, a likely explanation for this translocation is that a conformational change in the C-terminal domain could also provide greater accessibility of the nuclear localization signal to the cellular machinery. In this way, we provide evidence that significant structural rearrangement of the Atcry2 C-terminus follows from light activation, similarly to what has been previously reported for Arabidopsis cry1.Citation9
Blue-light induced accumulation of ROS in response to light activation of cry2
ROS accumulation in vivo in cell cultures can be detected by fluorescence staining with hydroxyphenylfluorescein.Citation3 Living insect cells were treated with fluorescent substrate and then either kept in the dark or illuminated with blue light (). Under these conditions, no ROS was detected in Atcry2 –expressing insect cell culture kept in darkness ( top panels). In a strong contrast, 5 or 15 min blue light illumination induced an accumulation of ROS staining that perfectly matched the nuclear compartment observed by D.I.C. .(). Thus accumulation of ROS in blue light matches the blue-light dependent Atcry2 nuclear immunolocalization (). From these data it is evident that Atcry2 induces blue-light induced accumulation of ROS in the nucleus.
Figure 2. Production and subcellular localization of ROS by Sf21 CRY2 exposed to blue light.Living Sf21 stably expressing CRY2 were exposed to dark or blue light, treated with DCFH-DA [5-(and-6)-chloromethyl-2′,7′-dichlorofluorecein diacetate] and viewed by an inverted Leica TCS SP5 microscope. Images show single confocal z section that cross the nucleus. Intense ROS staining can be seen inside the nucleus (N) whose membrane can be clearly observed by differential interference contrast (D. I. C.). Scale bars 10 μm. Methods for AtCry2 expression, immunohistochemical staining, and fluorescence staining for ROS are described in our original paper.Citation3 Polyclonal antibody used for cry2 detection has been raised to the C-terminal domain as used previously. Citation6
![Figure 2. Production and subcellular localization of ROS by Sf21 CRY2 exposed to blue light.Living Sf21 stably expressing CRY2 were exposed to dark or blue light, treated with DCFH-DA [5-(and-6)-chloromethyl-2′,7′-dichlorofluorecein diacetate] and viewed by an inverted Leica TCS SP5 microscope. Images show single confocal z section that cross the nucleus. Intense ROS staining can be seen inside the nucleus (N) whose membrane can be clearly observed by differential interference contrast (D. I. C.). Scale bars 10 μm. Methods for AtCry2 expression, immunohistochemical staining, and fluorescence staining for ROS are described in our original paper.Citation3 Polyclonal antibody used for cry2 detection has been raised to the C-terminal domain as used previously. Citation6](/cms/asset/ceeaa652-8245-4361-8663-43a939a40d5f/kpsb_a_1042647_f0002_oc.gif)
Conclusions
In our original study Citation3 and in this accompanying manuscript, we have shown a potentially novel way in which cryptochromes may participate in signaling. Whereas there is convincing evidence that conformational changes induced by primary photochemical events lead to interaction with cry signaling partners, this does not exclude that formation of ROS may also play a role in cryptochrome signaling. Moreover, this novel signaling mechanism may be of general relevance, since the primary photochemical events that lead to ROS formation are conserved in cryptochromes through a wide variety of organisms. It will be interesting to re-examine known cryptochrome signaling pathways for the presence of proteins or signaling intermediates that may be responsive to ROS.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the “Imagerie Paris Seine” imaging platform for confocal microscopy analyses and Alfred Batschauer for the baculovirus construct for Atcry2 expression.
Funding
We are grateful to AFOSR (FA9550-14-0-0409) for funding. M. Elesawi is recipient of a postdoctoral fellowship from the Ministère de l'éducation Nationale, France.
References
- Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GT, Batschauer A, Ahmad M. The cryptochromes: blue light photoreceptors in plants and animals. Ann Rev Plant Biol 2011; 62:335–64; PMID:21526969; http://dx.doi.org/10.1146/annurev-arplant-042110-103759
- Yu X, Sayegh, R, Maymon M, Warpeha K, Klejnot J, Yang H, Huang J, Lee J, Kaufman L, Lin C. Formation of nuclear bodies of arabidopsis CRY2 in response to blue light is associated with its blue light-dependent degradation. Plant Cell 2009, 21:118–30; PMID:19141709
- Consentino L, Lambert S, Martino C, Jourdan N, Bouchet PE, Witczak J, Castello P, El-Esawi M, Corbineau F, d'Harlingue A, et al. Blue-light dependent reactive oxygen species formation by arabidopsis cryptochrome may define a novel evolutionarily conserved signaling mechanism. New Phytol 2015; 206(4):1450–62; PMID:25728686
- Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F. ROS signaling: the new wave? Trends Plant Sci 2011; 16:300–9; PMID:21482172; http://dx.doi.org/10.1016/j.tplants.2011.03.007
- Schmidt R, Schippers JH. ROS-mediated redox signaling during cell differentiation in plants. Biochim Biophys Acta 2014 Dec 24. pii: S0304-4165(14)00428-0; http://dx.doi.org/10.1016/j.bbagen.2014.12.020
- Bouly JP, Schleicher E, Dionisio-Sese M, Vandenbussche F, Van Der Straeten D, Bakrim N, Meier S, Batschauer A, Galland P, et al. Cryptochrome blue light photoreceptors are activated through interconversion of flavin redox states. J Biol Chem 2007; 282:9383–91; PMID:17237227; http://dx.doi.org/10.1074/jbc.M609842200
- Nießner C, Denzau S, Stapput K, Ahmad M, Peichl L, Wiltschko W, Wiltschko R. Magnetoreception: activated cryptochrome 1a concurs with magnetic orientation in birds. J R Soc Interface 2013; 10:20130638; PMID:23966619
- Guo H, Duong H, Ma N, Lin C. The Arabidopsis blue light receptor cryptochrome 2 is a nuclear protein regulated by a blue light-dependent post-transcriptional mechanism. Plant J 1999; 19:279–87; PMID:10476075; http://dx.doi.org/10.1046/j.1365-313X.1999.00525.x
- Kondoh M, Shiraishi C, Müller P, Ahmad M, Hitomi K, Getzoff ED, Terazima M. Light-induced conformational changes in full-length Arabidopsis thaliana cryptochrome. J Mol Biol 2011; 413:128–37; PMID:21875594; http://dx.doi.org/10.1016/j.jmb.2011.08.031
