ABSTRACT
Receptor-like kinases (RLKs) play key roles in disease resistance, in particular basal immunity. They recognize patterns produced by the pathogen invasion and often work as complexes in the plasma membrane. Among these RLKs, there is increasing evidence in several plant species of the key role of Wall-associated kinases (WAKs) in disease resistance. We recently showed using rice (Oryza sativa) loss-of-function mutants of three transcriptionaly co-regulated OsWAK genes that individual OsWAKs are positively required for quantitative resistance to the rice blast fungus, Magnaporthe oryzae. This finding was unexpected since WAK genes belong to large gene families where functional redundancy is expected. Here we provide evidence that this may be due to complex physical interaction between OsWAK proteins.
Plants have evolved the ability to detect microorganisms via pattern-recognition receptors (PRRs) localized on the surface of plant cells.Citation1 Many of the PRRs encode Receptor-Like Kinase (RLK) proteins. PRR proteins recognize PAMPs (Pathogen Associated Molecular Patterns) that are conserved motifs from pathogens and DAMPs (Damage Associated Molecular Patterns) that derive from the damages caused by pathogen development.Citation2 For instance in rice the CEBiP PRR protein recognizes chitin from fungal cell-wall.Citation3 In contrast to PAMP, our knowledge on DAMP detection is much less advanced.Citation4 One example is the PRR/DAMP pair between the Arabidopsis WAK1 and oligogalacturonides.Citation5 Wall-Associated Kinases (WAK) contain an extracellular EGF (Epidermal Growth Factor) domain known in animals to bind small peptidesCitation6 and to form homo and heterodimers.Citation7 It is not known whether WAKs associate with other RLKs to ensure appropriate function like several other RLKs do.Citation8
Several studies provided evidence that WAK genes participate to resistance to pathogens in many plant species against fungi and bacteria (for an updated list of reports seeCitation9). In a recent study, we reported that three rice WAKs, OsWAK14, OsWAK91 and OsWAK92, act as positive regulators of quantitative resistance.Citation9 Since the WAK genes belong to large gene families (e.g. 125 OsWAKs were identified in riceCitation10), functional redundancy was expected and it was surprising that the loss of individual genes led to reduced resistance for three OsWAKs. A possible explanation for the non-redundant function of the rice OsWAKs could be the formation hetero-complexes of OsWAKs that would then act in a cooperative manner.
Indeed receptor-like kinases are often found in protein complexes.Citation8 We thus hypothesized that these OsWAK proteins could form complexes with each other. Since co-localization is a pre-requisite for such complex formation, the cellular localization OsWAKs was first determined. Only a few WAKs have been localized to the plasma membrane in ArabidopsisCitation11 or rice (OsWAK1Citation12; OsDEES1/OsWAK91Citation13). More recently, the Maize ZmWAK was shown to be localized to the plasma membrane.Citation14 Translational fusions between GFP and OsWAK14 or OsWAK92 were transiently expressed in Nicotiana benthamiana by agrobacterium-mediated transient transformation and visualized by confocal fluorescence microscopy. The OsWAK92:GFP and OsWAK14:GFP fusion proteins, that were correctly expressed (data not shown), were strongly expressed and localized to the plasma membrane (). Thus in addition to OsWAK91 which was previously shown to be membrane localized,Citation13 the three OsWAKs demonstrated to be required for disease resistance are all localized to the membrane.
Figure 1. Cellular localization of WAKs and interactions between WAKs. The C-terminal fusions of OsWAK proteins with either GFP or HA tags were transiently expressed in N. benthamiana by Agrobacterium infiltration. GFP fusions were localized using confocal microscopy (A) and protein-protein interactions between WAKs were evaluated (B). WAK14, WAK91 and WAK92 coding sequences were amplified and cloned in frame with the HA tag in the expression vector pBIN19-P35S-GTW-HACitation18. Protein extracts of N. benthamiana leaves and related immune-blotting and co-IP experiments were performed as reported.Citation18 Protein extracts were analyzed by immune-blotting with anti-GFP and anti-HA antibodies (Input). In addition, immune-precipitation was performed with anti-GFP beads (IP) and analyzed by immune-blotting with anti-GFP antibodies for the detection of immune-precipitated OsWAK proteins and with anti-HA antibodies for the detection of co-precipitated OsWAK proteins. (C) Protein-protein interactions between EGF domains of OsWAK proteins in Yeast. The EGF domains WAKs were amplified and ligated into pGBKT7 and pGADT7 vectors (Clontech). The corresponding vectors were transformed into the GOLD and Y187 yeast strains (Clontech). Yeast Two Hybrid Assay was performed as previously reported.Citation18 Interaction of OsWAK14, OsWAK91 and OsWAK92 EGF domains (cloned in AD vector) with other EGF OsWAK domains (cloned in BD vector) was assayed. Empty-BD vectors were used as controls. Cultures of diploid yeast clones and serial dilutions were spotted on synthetic medium (-Trp/-Leu/-His supplemented with 0.5 mM 3AT) to assay for interactions and on synthetic double dropout medium (-Trp/-Leu) to monitor proper growth.
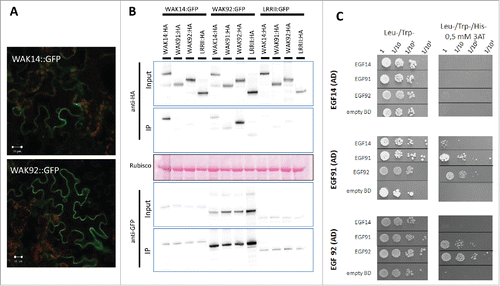
Using co-immuno precipitation with full length tagged OsWAK proteins, we next tested the possible interactions between the three OsWAK proteins studied. The GFP fusions with OsWAK14 and OsWAK92 were expressed in infiltrated leaves of N. benthamiana in combination with different OsWAK fusions with the HA tag (; anti-HA in input protein extract). The OsWAK91:GFP could not be tested as the fusion protein was cleaved (data not shown). Proteins were immuno-precipitated using GFP antibodies and an antibody against the HA tag was used to detect potential OsWAK partners (; anti-HA in IP). The OsWAK14:GFP and OsWAK92:GFP proteins co-immuno precipitated with their respective HA tagged versions, indicating that these proteins formed homo dimers. Some signals were also detected when the couples tested were OsWAK14-OsWAK92 and OsWAK91-OsWAK92. However, those signals were similar than with the negative control used and coding for another trans-membrane protein (LRRIICitation15). It is thus difficult to conclude for these specific interactions giving weak signals. Nevertheless, co-immuno precipitation data indicate that OsWAK14 and OsWAK92 full length proteins form homodimers in planta.
Using a complementary protein-protein interaction assay (Yeast-two-hybrid), we further tested the possible direct interactions between the extracellular EGF domains of OsWAK14, OsWAK91 and OsWAK92 (). The EGF domains were cloned into the Yeast-two-hybrid vectors without the adjacent trans-membrane domain to avoid targeting to the membrane and ensure nuclear localization required for the assay. The EGF domain of OsWAK92 could interact with itself, further confirming protein co-immuno precipitation experiments. Moreover, the EGF domains of OsWAK91 and OsWAK92 interacted together while the EGF domain of OsWAK91 and OsWAK92 and also weakly interacted with the EGF domain of OsWAK14. Thus the EGF domains of OsWAK14, OsWAK91 and OsWAK92 showed combinatorial interactions indicating that both homo and hetero dimers are likely formed by these proteins.
Plasma membrane PAMP receptors are often multi-protein complexes involving homo and hetero protein-protein interactions. For instance, FLS2 forms heterodimers and interacts with the BAK1 co-receptor to induce defense signaling after flg22 recognition.Citation8 The mode of action of WAKs is badly defined. Our co-immuno precipitation experiments and Yeast-two-hybrid data () indicate that OsWAK14, OsWAK91 and OsWAK92 can form homo and hetero complexes. This hypothesis is consistent with the observation that AtWAK1 resides in a large protein complexCitation16 and from experiments in rice showing that the OsWAK112 protein is found in a complex containing several proteins among which was found the OsWAK35a protein.Citation17 We propose that the absence of functional redundancy between OsWAK14, OsWAK91 and OsWAK92Citation9 may be the consequence of the existence of protein complexes comprising these three proteins. The loss of function of any of these proteins may destabilized these complexes and thus affect their functioning. Whether these protein complexes are required or not for appropriate targeting of defense responses remains now to be demonstrated.
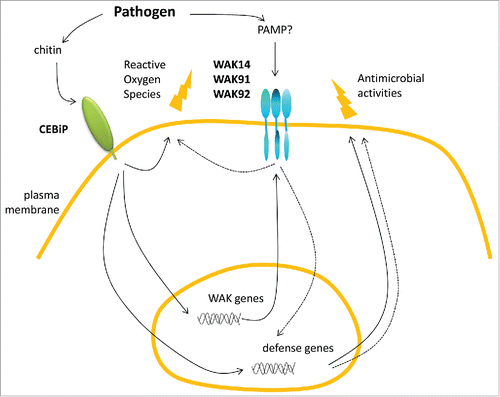
Figure 2. Hypothetical model of defense controlled by WAKs in rice. Upon infection by a fungal pathogen, chitin is recognized by the CEBiP receptor. This triggers production of reactive oxygen species and other antimicrobial defense molecules. The chitin receptor CEBiP also activates the transcription of OsWAK genes which in turn participate (dashed arrows) to the production of antimicrobial defense.Citation9 Based on Yeast Two Hybrid and co-immuno precipitation experiments, three Wall-Associated Kinases are proposed to form complexes through their EGF extracellular domains. The signal activating this complex is still unknown.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
AD's work was supported by a PhD grant from CIRAD and Région Languedoc-Roussillon. EG was supported by a INRA post-doc fellowship from INRA-BAP division.This work was supported by the IRMA grant (ANR-07-GPLA-007) from the Génoplante program and the Cerealdefense grant (ANR-09-GENM-106) from the ANR-Génomique program.
References
- Macho AP, Zipfel C. Plant PRRs and the activation of innate immune signaling. Mol Cell 2014; 54:263-72; PMID:24766890; http://dx.doi.org/10.1016/j.molcel.2014.03.028
- Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Ann Rev Plant Biol 2009; 60:379-406; PMID:19400727; http://dx.doi.org/10.1146/annurev.arplant.57.032905.105346
- Kaku H, Nishizawa Y, Ishii-Minami N, Akimoto-Tomiyama C, Dohmae N, Takio K, Minami E, Shibuya N. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci USA 2006; 103:11086-91; PMID:16829581; http://dx.doi.org/10.1073/pnas.0508882103
- Zipfel C. Plant pattern-recognition receptors. Trends Immunol 2014; 35:345-51; PMID:24946686; http://dx.doi.org/10.1016/j.it.2014.05.004
- Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci USA 2010; 107:9452-7; PMID:20439716; http://dx.doi.org/10.1073/pnas.1000675107
- Hynes NE, MacDonald G. ErbB receptors and signaling pathways in cancer. Curr Opin Cell Biol 2009; 21:177-84; PMID:19208461; http://dx.doi.org/10.1016/j.ceb.2008.12.010
- Schlessinger J. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell 2002; 110:669-672; PMID:12297041; http://dx.doi.org/10.1016/S0092-8674(02)00966-2
- Böhm H, Albert I, Fan L, Reinhard A, Nürnberger T. Immune receptor complexes at the plant cell surface. Curr Opin Plant Biol 2014; 20:47-54; PMID:24835204; http://dx.doi.org/10.1016/j.pbi.2014.04.007
- Delteil A, Gobbato E, Cayrol B, Estevan J, Michel-Romiti C, Dievart A, Kroj T, Morel JB. Several wall-associated kinases participate positively and negatively in basal defense against rice blast fungus. BMC Plant Biol 2016; 16:17; PMID:26772971; http://dx.doi.org/10.1186/s12870-016-0711-x
- Zhang S, Chen C, Li L, Meng L, Singh J, Jiang N, Deng XW, He ZH, Lemaux PG. Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family. Plant Physiol 2005; 139:1107-24; PMID:16286450; http://dx.doi.org/10.1104/pp.105.069005
- He ZH, He DZ, Kohorn BD. Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J 1998; 14:55-63; PMID:9681026; http://dx.doi.org/10.1046/j.1365-313X.1998.00092.x
- Li H, Zhou SY, Zhao WS, Su SC, Peng YL. A novel wall-associated receptor-like protein kinase gene, OsWAK1, plays important roles in rice blast disease resistance. Plant Mol Biol 2009; 69:337-46; PMID:19039666; http://dx.doi.org/10.1007/s11103-008-9430-5
- Wang N, Huang HJ, Ren ST, Li JJ, Sun Y, Sun DY, Zhang SQ. The rice wall-associated receptor-like kinase gene OsDEES1 plays a role in female gametophyte development. Plant Physiol 2012; 160:696-707; PMID:22885936; http://dx.doi.org/10.1104/pp.112.203943
- Zuo W, Chao Q, Zhang N, Ye J, Tan G, Li B, Xing Y, Zhang B, Liu H, Fengler KA, et al. A maize wall-associated kinase confers quantitative resistance to head smut. Nat Genet 2015; 47:151-157; PMID:25531751; http://dx.doi.org/10.1038/ng.3170
- Lefebvre B, Timmers T, Mbengue M, Moreau S, Hervé C, Tóth K, Bittencourt-Silvestre J, Klaus D, Deslandes L, Godiard L, et al. A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc Natl Acad Sci USA 2010; 107:2343-8; PMID:20133878; http://dx.doi.org/10.1073/pnas.0913320107
- Park AR, Cho SK, Yun UJ, Jin MY, Lee SH, Sachetto-Martins G, Park OK. Interaction of the arabidopsis receptor protein kinase Wak1 with a glycine-rich protein, AtGRP-3. J Biol Chem 2001; 276:26688-93; PMID:11335717; http://dx.doi.org/10.1074/jbc.M101283200
- Rohila JS, Chen M, Chen S, Chen J, Cerny R, Dardick C, Canlas P, Xu X. Gribskov M, Kanrar S, Zhu JK, Ronald P, Fromm ME. Protein-protein interactions of tandem affinity purification-tagged protein kinases in rice. Plant J 2006; 46:1-13; PMID:16553892; http://dx.doi.org/10.1111/j.1365-313X.2006.02671.x
- Cesari S, Thilliez G, Ribot C, Chalvon V, Michel C, Jauneau A, Rivas S, Alaux L, Kanzaki H, Okuyama Y, et al. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 2013; 25:1463-81; PMID:23548743; http://dx.doi.org/10.1105/tpc.112.107201
