ABSTRACT
Cytochrome b5 (CB5) proteins are small heme-binding proteins, that influence cytochrome P450 activity. While only one CB5 isoform is found in mammals, higher plants have several isoforms of these proteins. The roles of the many CB5 isoforms in plants remain unknown. We hypothesized that CB5 proteins support the cytochrome P450 enzymes of plant specialized metabolism and found CB5C from Arabidopsis thaliana to co-express with glucosinolate biosynthetic genes. We characterized the glucosinolate profiles of 2 T-DNA insertion mutants of CB5C, and found that long-chained aliphatic glucosinolates were reduced in one of the mutant lines – a phenotype that was exaggerated upon methyl-jasmonate treatment. These results support the hypothesis, that CB5C influences glucosinolate biosynthesis, however, the mode of action remains unknown. Furthermore, the mutants differed in their biomass response to methyl jasmonate treatment. Thereby, our results highlight the varying effects of T-DNA insertion sites, as the 2 analyzed alleles show different phenotypes.
Abbreviations
| CB5 | = | cytochrome b5 |
| ER | = | endoplasmic reticulum |
| GLS | = | glucosinolates |
| LC | = | long-chained aliphatic |
| MeJA | = | methyl-jasmonate |
| P450 | = | cytochrome P450 |
| SC | = | short-chained aliphatic |
| 4MOI3M | = | 4-methoxyindole-3-methyl glucosinolate |
Introduction
Cytochrome b5 (CB5) proteins are small (∼15 kD) heme-binding proteins, anchored to the endoplasmic reticulum (ER). These proteins have been studied extensively in animal systems due to their role in cellular detoxification and drug metabolism.Citation1,2,3,4 CB5 proteins interact with ER-anchored cytochrome P450 (P450) enzymes; a large family of versatile enzymes able to catalyze the transformation of otherwise inert molecules.Citation5 CB5 proteins influence P450 enzymes either by supplying electrons – similar to the NADPH-dependent P450 reductases - or by modulating their activity via physical interaction independent of electron donation.Citation6,7 The majority of studies have focused on mammalian CB5 proteins, whereas CB5 proteins from other organisms have been examined to a lesser extent. In contrast to mammals, which have a single copy of the CB5 gene,Citation8 the genomes of higher plants encode multiple CB5 isoforms,Citation9 e.g. 6 in the model plant Arabidopsis thalianaCitation10 and 11 in Glycine max (soybean).Citation11 The purpose of the increased number of CB5 isoforms remains elusive.
As sessile organisms, plants excel in their capability to synthesize diverse specialized chemicals - e.g., toxins, fragrances and pigments – which enable them to interact with, and adapt to, their ever-changing environment.Citation12 Essential for this specialized metabolism are P450 enzymes that carry out central catalytic steps in several of these specialized biosynthetic pathways.Citation13,14 The number of these enzymes are widely expanded in higher plants, with A. thaliana having more than 240.Citation15 Plants may therefore have evolved a larger number of CB5 isoforms to keep up with the increased number of P450s.Citation16,17 A connection between CB5 proteins and specialized plant metabolism was found in petunia, where discoloration of the flower petals was observed in a CB5 knock-out mutant.Citation18 The change in flower color coincided with a decrease in anthocyanins and a decrease in the activity of the biosynthetic P450, flavonoid 3´, 5´-hydroxylase, indicating that this enzyme requires CB5 for full enzymatic activity. Further evidence for a role of CB5 proteins in specialized metabolism comes from the biosynthesis of artemisinic acid – the precursor for the anti-malarial drug, artemisinin. Upon heterologous expression of the artemisinic acid biosynthetic pathway (including a P450) from Artimisia annua, co-expression of a CB5 resulted in higher levels of artemisinic acid.Citation19 These findings suggest that CB5 proteins can be critical for metabolic efficiency, possibly through interactions with the P450 enzymes.
A. thaliana produces glucosinolates (GLS) as their major class of specialized metabolites involved in plant defense.Citation20,21 P450s carry out essential enzymatic steps in the GLS biosynthetic pathway. Based on the idea that CB5 proteins might be involved in plant specialized metabolism through interactions with P450s, we hypothesized that a CB5 protein plays a role for the efficiency of this pathway.Citation22 In this study, we investigated the role of the CB5 isoform C, which co-expresses with the GLS pathway in A. thaliana. We characterized GLS profiles of 2 T-DNA insertional mutants of CB5C in the presence and absence of the GLS-inducing hormone methyl-jasmonate (MeJA). Our results show that CB5C affects GLS profiles by influencing especially long-chained aliphatic (LC) GLS.
Results
Co-expression analysis revealed that only one of the 6 A. thaliana CB5 genes – CB5C (also referred to as CYB5-1) Citation23 – co-expresses with the GLS biosynthesis genes (). We obtained 2 T-DNA insertion lines, cb5c-1 and cb5c-2 () and analyzed the expression levels of CB5C (). As previously reported, cb5c-2 is a knock-down mutant,Citation23 and also cb5c-1 shows reduced transcript levels compared to wildtype (WT).
Figure 1. Sequence structure of CB5C and mutants. a) Gene structure of A. thaliana CB5C, showing the T-DNA insertion sites for the 2 mutants (cb5c-1 and cb5c-2). Black boxes depict exons; white boxes indicate introns and untranslated regions. b) Sequence comparison between the protein products of WT and cb5c-1. The N-terminal heme-binding domain (gray), the C-terminal transmembrane helix (black) and the additional domain of the cb5c-1 mutant.
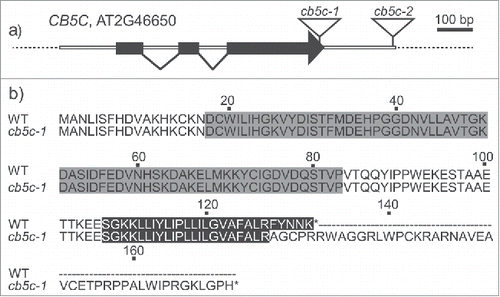
Figure 2. Expression levels of CB5C. The three genotypes (WT, cb5c-1 and cb5c-2) were analyzed for CB5C expression. Bar graphs display the mean expression (normalized to WT levels) and error-bars display the standard deviation of 3 independent biological replicates for each genotype.
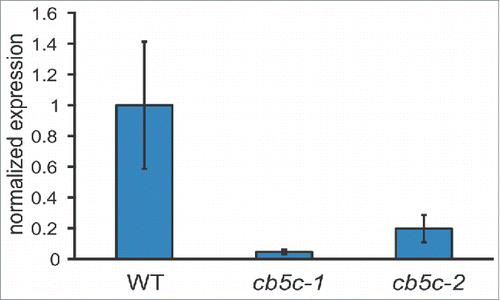
Table 1. Genes co-expressing with A. thaliana CB5C
Analysis by mass-spectrometry enabled quantification of 15 different GLS in individual seedlings. Total levels of GLS were similar in mutants and WT plants (, Table S1). However, the mutants showed differences in the levels of individual GLS. cb5c-1 showed significantly reduced levels of the LC GLS 7-methylsulfinylheptyl and 8-methylsulfinyloctyl GLS. By contrast, cb5c-2 showed reduced levels of the seed-derived 3-benzoyloxypropyl GLS. Common for both mutants was an increased accumulation of the short-chained aliphatic (SC) 4-methylthiobutyl GLS (p = 0.072 and 0.012 for cb5c-1 and cb5c-2, respectively).
Table 2. Glucosinolate levels in seedlings of the cb5c mutants (nmol/g FW).
As GLS levels are inducible upon application of exogenous jasmonate,Citation22,24 we treated the plants with methyl-jasmonate (MeJA) to assess whether this would exaggerate the subtle chemotypes of cb5c-1 and cb5c-2, thus revealing a potential condition-specific role of CB5C. MeJA treatment led to an increase in almost all measured GLS, and indeed resulted in more pronounced differences in GLS between genotypes (). Interestingly, MeJA-treated WT plants showed an increase in 3-benzoyloxypropyl and 4-benzoyloxybutyl GLS. These GLS originate from the seed, but are not de novo synthesized in the seedlings.Citation25 A similar – but less pronounced – increase upon MeJA, was observed for the cb5c-2 mutant, but no significant increase was found in cb5c-1.
In the MeJA-treated cb5c-1 mutant LC GLS continued to be lower compared to WT, but the decrease now involved all LC GLS. In addition, one SC GLS (4-benzoyloxybutyl GLS) was significantly lower in cb5c-1 upon MeJA treatment compared to the WT. Similarly, in cb5c-2, we found the levels of 3-benzoyloxypropyl GLS to be lower than WT when treated with MeJA. In this mutant line, the 4-methoxyindol-3-ylmethyl GLS (4MOI3M) also showed a decrease ().
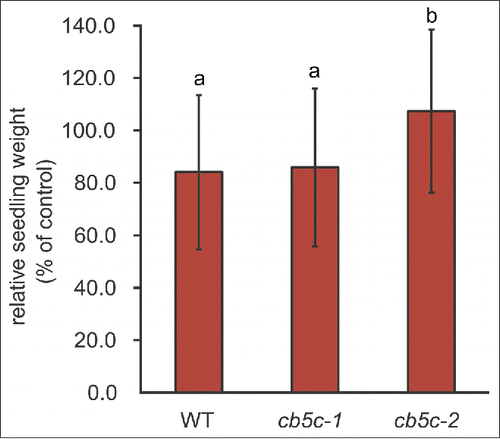
By visual inspection of the plates, we noticed a difference in growth between the genotypes in response to MeJA treatment. Quantification of this phenotype revealed a difference in growth between genotypes upon hormone treatment, whereas no significant difference between untreated plants was observed. WT and cb5c-1 plants showed decreased weight upon treatment, whereas the weight of cb5c-2 was unaffected ().
Figure 3. Biomass response to MeJA. Biomass responses of 10-day-old seedlings of the 3 genotypes (WT, cb5c-1 and cb5c-2). The weight of seedlings treated with methyl-jasmonate (MeJA) was normalized to the weight of untreated seedlings (ctrl). Different letters refer to statistical groupings (pairwise t-test as post-hoc, p < 0.05, n = 30). See Table S1 for the ANOVA table.
Discussion
We analyzed GLS levels of 2 T-DNA mutants of the CB5C gene. The two mutants have different T-DNA insertion sites. The cb5c-1 knock-down mutant has an insertion within the coding sequence of the CB5C gene, causing a disruption of the reading frame that is likely to result in a recombinant protein with 39 new C-terminal residues (). As the residues C-terminal to the transmembrane helix determine the localization of CB5 proteins in A. thaliana,Citation10 an altered amino acid sequence at the C-terminus could result in mislocalization of the CB5C protein. The cb5c-2 mutant carries a T-DNA insertion in the 3′-untranslated region of the gene, classifying it as a “knock-about” mutant ().Citation26 Although the 2 mutants show comparable decreases in transcript levels (), the corresponding proteins might differ fundamentally in their C-terminal amino acid sequence. The recombinant CB5C-1 mutant protein likely mislocalizes, whereas the cb5c-2 mutant encodes the native CB5C protein, with full functionality. This may render the phenotypic effects of the T-DNA insertions very different, although CB5C transcript levels are comparable in the 2 mutant lines.
Both mutants show subtle but distinct alterations in the levels of individual GLS. Treatment with MeJA exaggerates the differences between WT and mutants. Prolonged treatment of A. thaliana seedlings with exogenous MeJA – as done in these experiments - does likely not reflect physiological conditions, but allows us to induce GLS levels, Citation27 and provoke a stronger phenotype. These findings support our hypothesis that CB5C influences GLS biosynthesis, in a condition-specific manner. The function of CB5C does not seem to be essential for GLS biosynthesis, but it affects accumulation of particularly LC GLS, especially when induced by MeJA treatment. The decrease in almost exclusively LC GLS seen for the cb5c-1 mutant suggests specificity in the function of CB5C and potentially links the gene to the CYP79F enzymes that catalyze a key step in the biosynthetic step of specifically LC GLS.Citation28,29 We observed no differences in LC GLS in the cb5c-2 mutant, where we anticipate that CB5C is properly localized. Here, the residual expression might be able to sustain its role in supporting GLS biosynthesis, whereas the cb5c-1 mutant protein may be mislocalized and therefore non-functional. Another discrepancy between the 2 mutants is their relatively low levels of the seed-specific 3-benzoyloxypropyl and 4-benzoyloxybutyl GLS. Upon MeJA treatment WT plants accumulate more of these GLS, indicating less GLS turnover. The cb5c-1 did not show a significant change in these 2 GLS upon MeJA treatment, whereas cb5c-2 showed an increase comparable to - but less pronounced than - WT plants. This indicates that the effect of CB5C on GLS profiles results from increase in turnover, as is seen for these seed-specific glucosinolates.
The different phenotypic consequences of the 2 T-DNA insertions are further illustrated by the differences in biomass response to MeJA treatment (). Exogenously applied MeJA reduces seedling biomass,Citation30-32 but strikingly, seedlings of the cb5c-2 mutants did not show this response. In comparison, WT and cb5c-1 mutants showed reduced growth upon MeJA treatment. Noticeably, A. thaliana CB5 proteins interact with a series of other proteins than P450s such as 1) a plasma membrane-localized sucrose transporter,Citation23 2) several enzymes involved in fatty acid metabolismCitation11,33 and 3) the ethylene response regulator RTE1.Citation34 This – together with our findings – suggests that CB5C could play a broader physiological role, in addition to directly influencing GLS metabolism.
Methods
Co-expression analysis
Genes co-expressing with CB5C were identified using the ATTED-II database (http://atted.jp/).Citation35
Plant lines and growth conditions
Two T-DNA insertion lines targeting CB5C (AT2G46650) – cb5c-1 (SALK_027748C) and cb5c-2 (cyb5-1; SALK_065187) - were acquired from the Nottingham Arabidopsis Stock Center (NASC). The cb5c-1 line was homozygous. The cb5c-2 line was heterozygous; a homozygous line and a segregating wildtype (referred to as WT) were obtained from this heterozygote. Plants were germinated and grown on MS (Murashige and Skoog medium including vitamins; M0222, Duchefa) agar plates with 1% sucrose. For hormone treatment, the medium was supplemented with 10 µM MeJA (392707–5ML,Sigma-Aldrich). Seeds were sterilized by incubating in 70% (v/v) ethanol, 0.05 % (v/v) Triton X-100 for 5 min and washing in 70% (v/v) ethanol. The air-dried seeds were plated and cold-stratified for 24 hours at 4°C in the dark. Seedlings were grown for 10 d at 20°C under 16 hours of light with an intensity of 110–140 µE.
Quantitative RT-PCR
RNA was extracted using TRIzol® reagent (Invitrogen 15596–018). For each biological repetition, 3 seedlings were pooled and placed in a tube with 0.5 mL TRIzol and a chrome ball. Samples were homogenized in a Retch Mixer Mill 303 at 30 Hz for 30 sec. Samples were then incubated 5 min at room temperature. 0.2 mL chloroform was added and the samples mixed before being centrifuged for 15 min at 12.000x g at 4°C. The upper phase was collected and mixed with an equal volume of cold isopropanol. The samples were kept on ice for 30 min before being centrifuged for 10 min at 15.000x g at 4°C. The resulting pellet was washed with 75 % ethanol and centrifuged for 5 min at 7.500x g at 4°C. Supernatant was removed and the resulting pellet was air-dried before resuspension in 20 µL nuclease free water. The extracted RNA was DNase treated using DNaseI (AMPDK1-1KT, Sigma-Aldrich) according to manufactures instructions. 1 µg of RNA was reverse transcribed into cDNA with the iScript cDNA synthesis kit (#1708891, Bio-Rad) following the manufactures instructions.
qRT-PCR was performed using DyNAmo Flash SYBR Green qPCR Kit (F-415L, Thermo Scientific) and the following primers to quantify CB5C expression: 5′-AGCACTTTCATGGACGAACA-‘3 and 5′-TTGGCATCTTTGCTATGGTTC-‘3. PEX4/UBC21 (AT5G25760)Citation36 was used as reference gene using the following primers: 5′- CTGAGCCGGACAGTCCTCTTAACTG-‘3 and 5′-CGGCGAGGCGTGTATACATTTGTG-‘3. Two rounds of technical replicates were run, each including all 3 biological replicates. The CT values for CB5C were normalized to PEX4/UBC21 – taking primer efficiencies into account (close to 1 for both primer sets). Finally, the CB5C expression values of each mutant were further normalized to WT.
GLS and biomass analysis
GLS were extracted from single 10-day-old seedlings. The extraction was performed according to Andersen et al.Citation37 using 1 nmol p-hydroxybenzyl GLS as internal standard. Quantification was performed via liquid chromatography coupled to mass-spectrometry according to Jensen et al.Citation38 Two independent rounds of experiments were performed with 15 seedlings for each combination of genotype and treatment per round – seedlings of these experiments were weighed individually on a scale with sub-milligram sensitivity. Prior to any statistical analysis, the weight of MeJA-treated seedlings were normalized to the mean weight of the corresponding untreated seedlings – yielding the biomass response values. Statistical analysis of biomass response and GLS levels was performed across experimental rounds via ANOVA, using the following linear models: GLS = TREATMENT + GENOTYPE + EXPERIMENT + EXPERIMENT:GENOTYPE + TREATMENT:GENOTYPE + EXPERIMENT:TREATMENT and Biomass response = genotype + experiment + genotype:experiment. Holm-corrected pairwise t-tests were used for post hoc testing.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplementary_Table_1.xlsx
Download MS Excel (18.7 KB)Funding
Financial support for the work was provided by the Danish National Research Foundation DNRF grant 99 (DV, CC, TGA, MB, BAH).
References
- Porter TD. The roles of cytochrome b 5 in cytochrome P450 reactions. J Biochem Mol Toxicol 2002; 16:311-6; PMID:12481306; http://dx.doi.org/10.1002/jbt.10052
- Zhang H, Myshkin E, Waskell L. Role of cytochrome b 5 in catalysis by cytochrome P450 2B4. Biochem Biophys Res Commun 2005; 338:499-506; PMID:16182240; http://dx.doi.org/10.1016/j.bbrc.2005.09.022
- Masters BS, Marohnic CC. Cytochromes P450 — A family of proteins and scientists – understanding their relationships scientists – understanding their relationships. 2015; 2532; http://dx.doi.org/10.1080/03602530600570065
- Kandel SE, Lampe JN. The role of protein-protein interactions in cytochrome P450-mediated drug metabolism and Toxicity. Chem Res Toxicol [Internet] 2014; Available from: http://www.ncbi.nlm.nih.gov/pubmed/25133307; 27:1474-86; PMID:25133307; http://dx.doi.org/10.1021/tx500203s
- Meunier B, de Visser SP, Shaik S. Mechanism of oxidation reactions catalyzed by cytochrome P450 enzymes. Chem Rev 2004; 104:3947-80; PMID:15352783; http://dx.doi.org/10.1021/cr020443g
- Schenkman JB, Jansson I. The many roles of cytochrome b5. Pharmacol Ther 2003; 97:139-52; PMID:12559387; http://dx.doi.org/10.1016/S0163-7258(02)00327-3
- Im S-C, Waskell L. The interaction of microsomal cytochrome P450 2B4 with its redox partners, cytochrome P450 reductase and cytochrome b(5). Arch Biochem Biophys [Internet] 2011 [cited 2011 Aug 31]; 507:144-53. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid = 3073529&tool = pmcentrez&rendertype=abstract; PMID:21055385; http://dx.doi.org/10.1016/j.abb.2010.10.023
- Altuve A, Wang L, Benson DR, Rivera M. Mammalian mitochondrial and microsomal cytochromes. 2004; 314:602-9; PMID:14733950; http;//dx.doi.org/10.1016/j.bbrc.2003.12.138
- Fan R-C, Peng C-C, Xu Y-H, Wang X-F, Li Y, Shang Y, Du S-Y, Zhao R, Zhang X-Y, Zhang L-Y, et al. Apple sucrose transporter SUT1 and sorbitol transporter SOT6 interact with cytochrome b5 to regulate their affinity for substrate sugars. Plant Physiol [Internet] 2009 [cited 2011 Jul 21]; 150:1880-901. Available from: http: //www.pubmedcentral.nih.gov /articlerender.fcgi?artid = 2719124&tool = pmcentrez&rendertype =abstract; PMID:19502355; http://dx.doi.org/10.1104/pp.109.141374
- Maggio C, Barbante A, Ferro F, Frigerio L, Pedrazzini E. Intracellular sorting of the tail-anchored protein cytochrome b5 in plants: a comparative study using different isoforms from rabbit and Arabidopsis. J Exp Bot [Internet] 2007 [cited 2011 Nov 7]; 58:1365-79. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17322552; PMID:17322552; http://dx.doi.org/10.1093/jxb/erl303
- Kumar R, Tran L-SP, Neelakandan AK, Nguyen HT. Higher plant cytochrome b5 polypeptides modulate fatty acid desaturation. PLoS One [Internet] 2012 [cited 2012 Feb 28]; 7:e31370. Available from: http://dx.plos.org/10.1371/journal.pone.0031370; PMID:22384013; http://dx.doi.org/10.1371/journal.pone.0031370
- Nishida R. Chemical ecology of insect–plant interactions: ecological significance of plant secondary metabolites. Biosci Biotechnol Biochem [Internet] 2014; 78:1-13. Available from: http://www.tandfonline.com/doi/abs/10.1080/09168451.2014.877836; PMID:25036477; http://dx.doi.org/10.1080/09168451.2014.877836
- Hamberger B, Bak S. Plant P450s as versatile drivers for evolution of species-specific chemical diversity. Philos Trans R Soc Lond B Biol Sci [Internet] 2013; 368:20120426. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3538417&tool=pmcentrez&rendertype=ab stract; PMID:23297350; http://dx.doi.org/10.1098/rstb.2012.0426
- Omura T. Contribution of cytochrome P450 to the diversification of eukaryotic organisms. Biotechnol Appl Biochem 2013; 60:4-8; PMID:23586987; http://dx.doi.org/10.1002/bab.1099
- Bak S, Beisson F, Bishop G, Hamberger B, Höfer R, Paquette S, Werck-Reichhart D. Cytochromes P450. Arab B [Internet] 2011; 9:e0144. Available from: http://www.bioone.org/doi/abs/10.1199/tab.0144; PMID:22303269; http://dx.doi.org/10.1199/tab.0144
- Renault H, Bassard J, Hamberger B, Werck-Reichhart D. Cytochrome P450-mediated metabolic engineering: current progress and future challenges. Curr Opin Plant Biol [Internet] 2014; 19C:27-34. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24709279; PMID:24709279; http://dx.doi.org/10.1016/j.pbi.2014.03.004
- Laursen T, Møller BL, Bassard J-E. Plasticity of specialized metabolism as mediated by dynamic metabolons. Trends Plant Sci [Internet] 2014 [cited 2015 Jan 2]; 1-13. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25435320
- de Vetten N, ter Horst J, van Schaik HP, de Boer A, Mol J, Koes R. A cytochrome b5 is required for full activity of flavonoid 3′, 5′-hydroxylase, a cytochrome P450 involved in the formation of blue flower colors. Proc Natl Acad Sci U S A 1999; 96:778-83; PMID:9892710; http://dx.doi.org/10.1073/pnas.96.2.778
- Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, Leavell MD, Tai A, Main A, Eng D, et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature [Internet] 2013; 496:528-32. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23575629; PMID:23575629; http://dx.doi.org/10.1038/nature12051
- Halkier BA, Gershenzon J. Biology and biochemistry of glucosinolates. Annu Rev Plant Biol [Internet] 2006 [cited 2011 Jul 20]; 57:303-33. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16669764; PMID:16669764; http://dx.doi.org/10.1146/annurev.arplant.57.032905.105228
- Sønderby IE, Geu-Flores F, Halkier BA. Biosynthesis of glucosinolates–gene discovery and beyond. Trends Plant Sci [Internet] 2010 [cited 2011 Jul 22]; 15:283-90. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20303821; PMID:20303821; http://dx.doi.org/10.1016/j.tplants.2010.02.005
- Schweizer F, Fernández-Calvo P, Zander M, Diez-Diaz M, Fonseca S, Glauser G, Lewsey MG, Ecker JR, Solano R, Reymond P. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell [Internet] 2013 [cited 2014 Jul 10]; 25:3117-32. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3784603&tool=pmcentrez&rendertype=abstrac t; PMID:23943862; http://dx.doi.org/10.1105/tpc.113.115139
- Li Y, Li L-L, Fan R-C, Peng C-C, Sun H-L, Zhu S-Y, Wang X-F, Zhang L-Y, Zhang D-P. Arabidopsis sucrose transporter SUT4 interacts with cytochrome b5-2 to regulate seed germination in response to sucrose and glucose. Mol Plant [Internet] 2012 [cited 2012 Feb 26]; 1-13. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22311778; PMID:21772031; http://dx.doi.org/10.1093/mp/sss001
- Alvarez S, He Y, Chen S. Comparative investigations of the glucosinolate-myrosinase system in Arabidopsis suspension cells and hypocotyls. Plant Cell Physiol 2008; 49:324-33; PMID:18202003; http://dx.doi.org/10.1093/pcp/pcn007
- Petersen BL, Chen S, Hansen CH, Olsen CE, Halkier BA. Composition and content of glucosinolates in developing Arabidopsis thaliana. Planta 2002; 214:562-71; PMID:11925040; http://dx.doi.org/10.1007/s004250100659
- Krysan PJ. T-DNA as an Insertional Mutagen in Arabidopsis. Plant Cell Online [Internet] 1999 [cited 2015 May 18]; 11:2283-90. Available from: http://www.plantcell.org/content/11/12/2283.full; http://dx.doi.org/10.1105/tpc.11.12.2283; PMID:10590158; http;//dx.doi.org/10.1105/tpc.11.12.2283
- Frerigmann H, Gigolashvili T. MYB34, MYB51, and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Mol Plant [Internet] 2014 [cited 2014 Jul 24]; 7:814-28. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24431192; PMID:24431192; http://dx.doi.org/10.1093/mp/ssu004
- Hansen CH, Wittstock U, Olsen CE, Hick AJ, Pickett JA, Halkier BA. Cytochrome p450 CYP79F1 from arabidopsis catalyzes the conversion of dihomomethionine and trihomomethionine to the corresponding aldoximes in the biosynthesis of aliphatic glucosinolates. J Biol Chem [Internet] 2001 [cited 2015 Nov 16]; 276:11078-85. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11133994; PMID:11133994; http://dx.doi.org/10.1074/jbc.M010123200
- Chen S, Glawischnig E, Jørgensen K, Naur P, Jørgensen B, Olsen C-E, Hansen CH, Rasmussen H, Pickett JA, Halkier BA. CYP79F1 and CYP79F2 have distinct functions in the biosynthesis of aliphatic glucosinolates in Arabidopsis. Plant J [Internet] 2003 [cited 2015 Nov 12]; 33:923-37. Available from: http://doi.wiley.com/10.1046/j.1365-313X.2003.01679.x; PMID:12609033; http://dx.doi.org/10.1046/j.1365-313X.2003.01679.x
- Yang D, Yao J, Mei C, Tong X, Zeng L, Li Q, Xiao L, Sun T, Li J, Deng X-W, et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci U S A [Internet] 2012; 109:E1192-200. Available from: http://www.pnas.org/content/109/19/E1192.full; PMID:22529386; http://dx.doi.org/10.1073/pnas.1201616109
- Wasternack C, Hause B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot [Internet] 2013 [cited 2014 May 23]; 111:1021-58. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23558912; PMID:23558912; http://dx.doi.org/10.1093/aob/mct067
- Tiryaki I, Staswick PE. An Arabidopsis mutant defective in jasmonate response is allelic to the auxin-signaling mutant axr1. Plant Physiol 2002; 130:887-94; PMID:12376653; http://dx.doi.org/10.1104/pp.005272
- Nagano M, Ihara-Ohori Y, Imai H, Inada N, Fujimoto M, Tsutsumi N, Uchimiya H, Kawai-Yamada M. Functional association of cell death suppressor, Arabidopsis Bax inhibitor-1, with fatty acid 2-hydroxylation through cytochrome b(5). Plant J [Internet] 2009; 58:122-34. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19054355\nhttp://onlinelibrary.wiley.com/store/10.1111/j.1365-313X.2008.03765.x/asset/j.1365-313X.2008.03765.x.pdf?v=1&t=hrvnp91f&s=fdb36bf490c5cd401a654ace5df2fc2a5bd00105;PMID:19054355; http://dx.doi.org/10.1111/j.1365-313X.2008.03765.x
- Chang J, Clay JM, Chang C. Association of cytochrome b5 with ETR1 ethylene receptor signaling through RTE1 in Arabidopsis. Plant J 2014; 77:558-67; PMID:24635651; http://dx.doi.org/10.1111/tpj.12401
- Aoki Y, Okamura Y, Tadaka S, Kinoshita K, Obayashi T. ATTED-II in 2016: a plant coexpression database towards lineage-specific coexpression. Plant Cell Physiol [Internet] 2015 [cited 2015 Dec 11]; pcv165 – .Available from: http://pcp.oxfordjournals.org/content/early/2015/11/05/pcp.pcv165.abstract; PMID:26546318; PMID:26546318 ; http://dx.doi.org/10.1093/pcp/pcv165
- Czechowski T, Stitt M, Altmann T, Udvardi MK. Genome-wide identification and testing of superior reference genes for transcript normalization. Society 2005; 139:5-17; PMID:16166256; http://dx.doi.org/10.1104/pp.105.063743.1
- Andersen TG, Nour-Eldin HH, Fuller VL, Olsen CE, Burow M, Halkier BA. Integration of biosynthesis and long-distance transport establish organ-specific glucosinolate profiles in vegetative arabidopsis. Plant Cell [Internet] 2013; 25:3133-45. Available from: http://www.plantcell.org/cgi/doi/10.1105/tpc.113.110890; PMID:23995084; http://dx.doi.org/10.1105/tpc.113.110890
- Jensen LM, Jepsen HSK, Halkier BA, Kliebenstein DJ, Burow M. Natural variation in cross-talk between glucosinolates and onset of flowering in Arabidopsis. Front Plant Sci [Internet] 2015 [cited 2015 Sep 14]; 6:697. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4561820&tool=pmcentrez&rendertype=abstract; PMID:26442014; http://dx.doi.org/10.3389/fpls.2015.00697
