ABSTRACT
Salinity stress is one of the major environmental challenges which adversely affects plant growth and productivity. The acquisition of salinity stress tolerance has been an interesting area of investigation for plant abiotic stress management. Recently, we investigated the interdependency of reactive oxygen species (ROS) generating and scavenging system for offering salt stress adaptation in rice. In continuation to our earlier findings, in the present study we analyzed the transcript level expression of different respiratory burst oxidase homologs (Rbohs) genes in salt sensitive and salt tolerant cultivars of rice to corroborate this result with their activities. Brassinosteroid (BR) is known to confer abiotic stress tolerance by modulating ROS machinery, and hence in the present study, the expression of key genes associated in brassinosteroid biosynthesis and signaling in salt sensitive and tolerant cultivar of rice was also conducted. In the present investigation, the other stress markers involving proline catabolism and anabolism along with chlorophyllase has been analyzed to get a better insights to our understanding of salt stress adaptation mechanisms in rice.
Abbreviations
| BRs | = | Brassinosteroids |
| NADPH | = | Nicotinamide Adenine Dinucleotide Phosphate Hydrogen |
| P5CS | = | Δ-1- pyrroline-5-carboxylate synthase |
| PDH | = | Proline Dehydrogenase |
| Rboh | = | Respiratory Burst Oxidase Homologs |
| ROS | = | Reactive Oxygen Species |
Rice is an important food crop and provides food security to large population of the world.Citation1 The rise in population and decline in rice productivity has been a major concern for many rice growing nations.Citation2 Salinity stress has been identified as a key factor that adversely affects rice cultivation and hence a continuous effort in understanding the mechanism and deciphering strategies for salt tolerance has been the forefront of scientific interest.Citation3 Salinity stress tolerance is a complex process governed by various intrinsic factors. Recently, we conducted a comparative study of salt sensitive (IR64 and Pusa Basmati-1) and salt tolerant cultivars (Luna Sankhi and Luna Suvarna) of rice and found that cellular ROS generating and ROS scavenging pathways work in a synchronized manner for achieving salt tolerance in rice.Citation1 In this work, the enzymatic activity of Nicotinamide Adenine Dinucleotide Phosphate Hydrogen (NADPH) oxidase and superoxide dismutase (SOD) was found to be more in the tolerant cultivars that leads to increased level of hydrogen peroxide in these cultivars. The enzymatic activity and transcripts of different antioxidant enzymes were also observed to be higher in the tolerant varieties than the sensitive. Further, the level of proline concentration was found to be more in the tolerant varieties while the amount of chlorophyll was more in the sensitive varieties.Citation1
In continuation to the above studies and to further substantiate this information, in the present paper, expression analysis of critical genes involved in cellular ROS machinery were analyzed. NADPH oxidase which is involved in the generation of ROS is implicated in several critical functions including abiotic stress tolerance in plants.Citation4 In rice there are many members of Rbohs and the functions of most of them are still unknown. Moreover, to best of our knowledge information on comparative expression analysis of Rboh genes in both salt sensitive and tolerant rice cultivars is lacking. Similarly, although it is well understood that brassinosteroids (BRs) plays a vital role in abiotic stress mitigation through ROS system,Citation5 yet the dynamics of expression of BR biosynthesis and signaling genes in rice cultivars with contrasting salt tolerance level is missing. Further, the analysis of key genes involved in proline metabolism and chlorophyll degradation is worth investigating as they are known as stress markers and have probable link to ROS machinery.Citation1,6
Methodology
14 days old seedlings of 4 rice cultivars viz. IR64 and Pusa Basmati-1 (salt sensitive) and Luna Sankhi and Luna Suvarna (salt tolerant) were used for the present work. Total RNA was isolated using the trizol method as per the manufacturer's instructions and cDNA was prepared using commercial cDNA synthesis kit. Gene expression was studied using qRT-PCR in StepOne Plus Realtime PCR machine (Applied Biosystems, USA).
Differential regulation of Rboh genes in salt sensitive and salt tolerant cultivars of rice
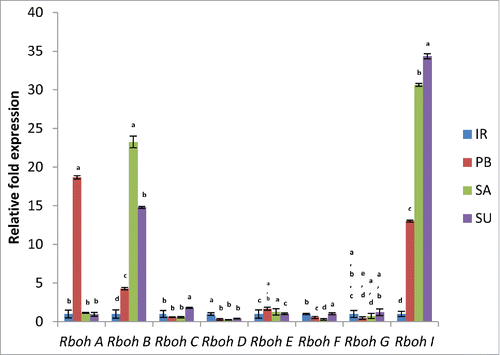
NADPH oxidases are the key ROS generating systems and are implicated in versatile functions in plants.Citation4 In our earlier study, we observed that the NADPH oxidase enzyme activity was more in the salt tolerant rice cultivars as compared to the sensitive ones.Citation1 To validate this information at gene level, we analyzed the expression of various Rboh genes in salt sensitive and salt tolerant cultivars of rice. Among various Rbohs, OsRboh I (Os12g0541300) showed 30.65 and 34.33 fold increase in tolerant cultivars Luna Sankhi and Luna Suvarna, respectively as compared to sensitive cultivar IR64. On the other hand, the expression level in another sensitive cultivar Pusa Basmati-1 was just 13.03 fold higher with respect to IR64 (). The expression level of OsRboh B (Os01g0360200) was also found to 23.26 and 14.79 fold higher in tolerant cultivars Luna Sankhi and Luna Suvarna, respectively as compared to the sensitive cultivar IR64 whereas its expression in Pusa Basmati-1 was only 4.28 fold higher than IR64 (). However, other Rboh genes either did not show much variation or were downregulated in some cases like OsRboh D. Earlier, it was observed that OsRboh I significantly up regulates under the effect of NaCl.Citation7 Together these results suggest clear involvement of Rboh B and Rboh I in providing salt tolerance in rice.
Figure 1. Real-time PCR analysis of Rboh genes in 2 salt sensitive (IR64 and Pusa Basmati-1 ) and 2 salt tolerant (Luna Sankhi and Luna Suvarna) cultivars of rice. (IR: IR64, PB: Pusa Basmati-1, SA: Luna Sankhi, SU: Luna Suvarna). Bars represent mean ± SE (n = 3). Different letters (a, b, c, d, e) within cultivars are significantly different (Fisher LSD, p ≤ 0.05).
Dynamics of BR biosynthesis and signaling genes in salt sensitive and tolerant cultivars of rice
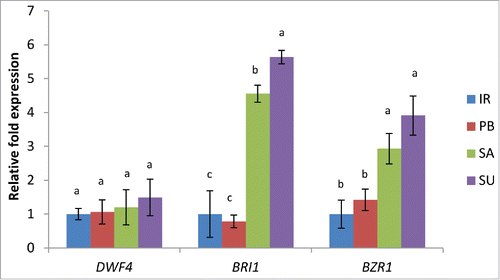
Salt stress adaptation in plants through the regulation of ROS pathway has also been linked to the plant growth regulator BRs.Citation5,8 Hence in the present study, an attempt was made to analyze the key genes involved in BR biosynthesis and signaling pathway in salt sensitive and tolerant cultivars of rice. OsDWF4 (Os03g40540) encodes for a cytochrome P450 protein and is a key gene involved in BR biosynthesis. The expression level of OsDWF4 was observed to be 1.20 and 1.49 fold high in tolerant cultivars Luna Sankhi and Luna Suvarna, respectively as compared to IR64, although this increase was found to be non significant (). Besides the biosynthetic gene, the signaling genes OsBRI1 (Os01g52050.1) which is a receptor that perceives BR signal also showed 4.55 fold increase in Luna Sankhi and 5.63 fold increase in Luna Suvarna when observed against IR64, whereas the expression in Pusa Basmati-1 was found to be downregulated as compared to IR64 (). Another gene, OsBZR1 (Os06g35900.1) that encodes for transcription factor regulating the activity of BR regulated genes also showed 2.92 and 3.90 fold increase in Luna Sankhi and Luna Suvarna, respectively than IR64, but the expression in Pusa Basmati was almost 0.4 fold less than IR64 ().These results indicate that the endogenous BR level might be playing a key role in conferring salt stress tolerance in rice plant. However, the nonsignificant increase in the expression of OsDWF4 could be either due to the tight regulation of BR biosynthesis by expression level of OsBZR1 transcription factor in a feedback inhibition manner or due to the crosstalk of BRs with other PGRs.Citation9,10 In one of the previous reports from our lab, we have shown that treatment of 28-homobrassinolide leads to higher level of hydrogen peroxide in sensitive cultivar Pusa Basmati-1 both under the control and salt stress conditions indicating a definitive link between H2O2 and BR signaling.Citation5 This experiment suggests that endogenous BR level and ROS are linked and together regulate the stress responsive pathways leading to salt stress adaptation in salt tolerant cultivars of rice.
Figure 2. Real-time PCR analysis of BR biosynthesis and signaling pathway genes in 2 salt sensitive (IR64 and Pusa Basmati-1) and 2 salt tolerant (Luna Sankhi and Luna Suvarna) cultivars of rice. Bars represent mean ± SE (n = 3). (IR: IR64, PB: Pusa Basmati-1, SA: Luna Sankhi, SU: Luna Suvarna). Different letters (a, b, c) within cultivars are significantly different (Fisher LSD, p ≤ 0.05).
Proline metabolism in salt sensitive and tolerant cultivars of rice
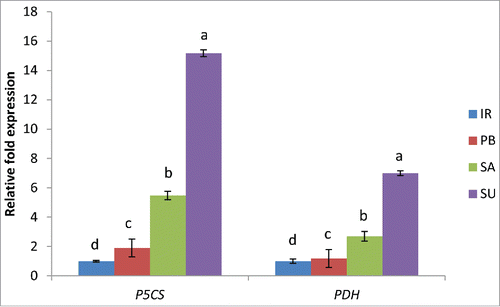
Proline is an important amino acid involved in osmoprotection and ROS scavenging.Citation1 Proline content in the cell is tightly coordinated at the level of its synthesis in the chloroplast or cytoplasm and degradation in the mitochondria.Citation11 Δ-1-pyrroline-5-carboxylate synthase (OsP5CS, Os05g38150.2) and proline dehydrogenase (OsPDH, Os10g40360.1) are the key genes involved in the biosynthesis and degradation of proline, respectively. In our earlier study, we indicated that the level of proline was significantly higher in salt tolerant as compared to salt sensitive cultivars. To further validate this data, we analyzed the expression of these genes in contrasting salt tolerant cultivars. Results indicate a 5.47 and 15.16 fold increase in the expression of OsP5CS in tolerant cultivars Luna Sankhi and Luna Suvarna, respectively with respect to IR64, whereas expression level in Pusa Basmati-1 was around 2 fold higher than IR64 (). Further, the OsPDH gene showed 2.70 and 7.00 fold increase in Luna Sankhi and Luna Suvarna, respectively whereas it was nearly same in Pusa Basmati-1 in comparison to IR64 (). The results indicate that the higher proline content observed in tolerant cultivar is at least in part resulting of a higher expression level of genes involved in proline synthesis such as OsP5CS.
Figure 3. Real-time PCR analysis of proline metabolism genes in 2 salt sensitive (IR64 and Pusa Basmati-1) and 2 salt tolerant (Luna Sankhi and Luna Suvarna) cultivars of rice. Bars represent mean ± SE (n = 3). (IR: IR64, PB: Pusa Basmati-1, SA: Luna Sankhi, SU: Luna Suvarna). Different letters (a, b, c, d) within cultivars are significantly different (Fisher LSD, p ≤ 0.05).
Analysis of chlorophyllase gene expression
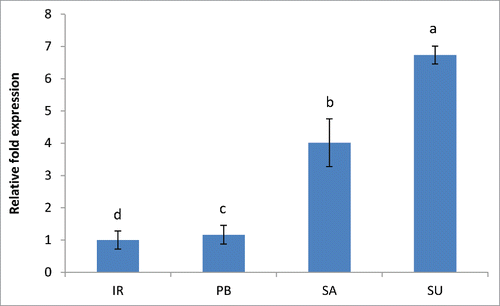
Interestingly, the content of chlorophyll was observed to be more in the sensitive cultivars than the tolerant cultivars.Citation1 Salt stress leads to destruction of thylakoids in the chloroplast causing release of chlorophyll from the thylakoid membranes.Citation6,12 This released chlorophyll needs to be degraded immediately to protect the plants from the cellular damage that may occur because of the photodynamic nature of chlorophylls.Citation12 The major enzyme involved in chlorophyll degradation is chlorophyllase.Citation13 Therefore, we analyzed the gene expression of chlorophyllase gene (Os10g28370.1) in the salt sensitive and tolerant cultivars. The expression of chlorophyllase gene was found to be 4.01 and 6.73 fold higher in Luna Sankhi and Luna Suvarna than the sensitive IR64 and Pusa Basmati-1 which were at par with each other (). This substantiates our earlier finding where we have reported reduced chlorophyll content in tolerant cultivars as compared to the sensitive ones and hence opens up an area for further investigation on the role of pigments in salt stress mechanisms.
Figure 4. Real-time PCR analysis of chlorophyllase gene in 2 salt sensitive (IR64 and Pusa Basmati-1) and 2 salt tolerant (Luna Sankhi and Luna Suvarna) cultivars of rice. (IR: IR64, PB: Pusa Basmati-1, SA: Luna Sankhi, SU: Luna Suvarna).Bars represent mean ± SE (n = 3). Different letters (a, b, c, d) within cultivars are significantly different (Fisher LSD, p ≤ 0.05).
Conclusions
It is evident that ROS plays a critical role in salt stress adaptive mechanisms of plant. The homeostasis of ROS is dependent on its generating and scavenging system which is tightly regulated in plants. In the present experiment, direct and indirect evidence obtained from the factors controlling the dynamics of ROS machinery provides valuable clues regarding acquisition of salt stress tolerance in plants.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
Navdeep Kaur is thankful to the Department of Biotechnology (Govt. of India) for Senior Research Fellowship. We thank the Director, CRRI, Odisha, India for providing the seeds of Luna Sankhi and Luna Suvarna and Department of Genetics IARI, New Delhi, India for providing the seeds of IR64 and Pusa Basmati-1. Authors also acknowledge the Department of Biotechnology, Government of India for financial support (Project No. BT/PR13965/BRB/10/883/2010).
References
- Kaur N, Dhawan M, Sharma I, Pati PK. Interdependency of reactive oxygen species generating and scavenging system in salt sensitive and salt tolerant cultivars of rice. BMC Plant Biol 2016; 16:1; PMID:26728271; http://dx.doi.org/10.1186/s12870-016-0824-2
- Agarwal P, Parida SK, Raghuvanshi S, Kapoor S, Khurana P, Khurana JP, Tyagi AK. Rice improvement through genome-based functional analysis and molecular breeding in India. Rice 2016; 9:1; PMID:26743769; http://dx.doi.org/10.1186/s12284-015-0073-2
- Gupta B, Huang B. Mechanism of salinity tolerance in plants: physiological, biochemical, and molecular characterization. Int J Genomics 2014; 18; PMID:701596; http://dx.doi.org/10.1155/2014/701596; PMID:701596
- Kaur G, Sharma A, Guruprasad K, Pati PK. Versatile roles of plant NADPH oxidases and emerging concepts. Biotechnol Adv 2014; 32:551-563; PMID:24561450; http://dx.doi.org/10.1016/j.biotechadv.2014.02.002
- Sharma I, Bhardwaj R, Pati PK. Exogenous application of 28-homobrassinolide modulates the dynamics of salt and pesticides induced stress responses in an elite rice variety pusa basmati-1. J Plant Growth Regul 2015; 34:509–518; http://dx.doi.org/10.1007/s00344-015
- Shu S, Guo SR, Sun J, Yuan LY. Effects of salt stress on the structure and function of the photosynthetic apparatus in Cucumis sativus and its protection by exogenous putrescine. Physiol Plant 2012; 146 (3):285-96; PMID:22452600; http://dx.doi.org/10.1111/j.1399-3054.2012.01623.x
- Wang GF, Li WQ, Li WY, Wu GL, Zhou CY, Chen KM. Characterization of rice NADPH oxidase genes and their expression under various environmental conditions. Int J Mol Sci 2013; 14:9440-9458; PMID:23629674; http://dx.doi.org/10.3390/ijms14059440
- Sharma I, Ching E, Saini S, Bhardwaj R, Pati PK. Exogenous application of brassinosteroid offers tolerance to salinity by altering stress responses in rice variety Pusa Basmati-1. Plant Physiol Biochem 2013; 69:17-26; PMID:23707881; http://dx.doi.org/10.1016/j.plaphy.2013.04.013
- Chung Y, Choe S. The regulation of brassinosteroid biosynthesis in Arabidopsis. Crit Rev Plant Sci 2013; 32 (6):396-410; http://dx.doi.org/10.1080/07352689.2013.797856
- Li QF, He JX. Mechanisms of signaling crosstalk between brassinosteroids and gibberellins. Plant Signal Behav 2013; 8 (7):e24686; PMID:23603943; http://dx.doi.org/10.4161/psb.24686
- Verslues PE, Sharma S. Proline metabolism and its implications for plant-environment interaction. The Arabidopsis book 2010: p.e0140.
- Kariola T, Brader G, Li J, Palva ET. Chlorophyllase 1, a damage control enzyme, affects the balance between defense pathways in plants. Plant Cell 2005; 17 (1):282-294; PMID:15598807; http://dx.doi.org/10.1105/tpc.104.025817
- Li J, Hu L, Zhang L, Pan X, Hu X. Exogenous spermidine is enhancing tomato tolerance to salinity–alkalinity stress by regulating chloroplast antioxidant system and chlorophyll metabolism. BMC Plant Biol 2015; 15:1; PMID:25592487; http://dx.doi.org/10.1186/s12870-014-0410-4
