ABSTRACT
Protein phosphatases 2C (PP2Cs) are important regulators of plant responses to abiotic stress. It is established that clade A PP2Cs inhibit ABA-activated SNF1-related protein kinases 2 (SnRK2s). Our recently published results show that ABI1, a member of clade A of PP2C is also a negative regulator of SnRK2.4, a kinase not activated in response to ABA. Here, we show that another member of this clade - PP2CA, interacts with and inhibits SnRK2.4. The salt-induced SnRK2.4/SnRK2.10 activity is higher in abi1–2 pp2ca-1 mutant than in wild type or single abi1 or pp2ca mutants, indicating that both phosphatases are inhibitors of SnRK2.4 and are at least partially redundant. Moreover, PP2CA together with ABI1 and SnRK2.4 regulates root growth in response to salinity.
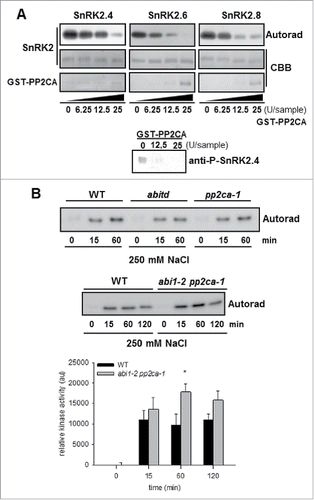
Plants possess multiple defense mechanisms, which enable them to cope with harsh environmental conditions. SNF1-related protein kinases 2 (SnRK2s) are plant specific enzymes involved in ABA-dependent plant development and in responses to environmental stresses, mainly drought and salinity (for review see refs.Citation1-5 and references herein). All SnRK2s, except Arabidopsis SnRK2.9, are activated rapidly and transiently in response to osmotic stress (water deficit, salt stress) and some of them are also activated by ABA treatment. Based on phylogenetic analysis SnRK2s are classified into 3 groups. The classification correlates with their response to exogenously applied ABA; group 1 consists of kinases non-activated by ABA, group 2 - kinases non-activated or weakly activated by ABA (depending on the plant species), and group 3 - strongly activated by this hormone.Citation6,7 The activity of SnRK2s is dependent on phosphorylation of specific Ser/Thr residues within their activation loops.Citation7-11 It is well established that phosphoprotein phosphatases type-2C (PP2C) belonging to clade A are negative regulators of ABA-activated SnRK2s.Citation12,13 In control conditions, when ABA level in plant cells is low, the phosphatases interact with ABA-activated SnRK2s blocking their activity. With the rise of the level of ABA (e.g., during seed maturation or in response to stress) ABA receptors - RCAR/PYR/PYL (RCAR, Regulatory Component of ABA Receptor/PYR1, Pyrabactin Resistance 1/PYL, PYR1-like) – bind the hormone, which results in interaction with clade A PP2Cs and their inhibition, allowing ABA-dependent SnRK2 activation.Citation14-19 This mechanism of PP2Cs inhibition is strongly dependent on ABA. It was recently shown that also Type One Protein Phosphatase 1 (TOPP1) regulates the activity of ABA-activated SnRK2s,Citation20 and it was proposed that regulation of TOPP1 activity and subsequently that of SnRK2s was dependent on PYL11 and ABA. On the other hand, the SnRK2 OST1 interacts with regulatory PP2AA- and PP2AB’-subunits and pp2aa regulatory subunit double mutants show reduced ABA sensitivity in stomatal closure, although pp2a catalytic subunit double mutants show enhanced ABA-dependent SnRK2 activation.Citation21 Much less is known on enzymes controlling the activity of ABA-non-activated SnRK2, from group 1. Our recently published results showed that at least one ABA-non-activated SnRK2, namely SnRK2.4 is inhibited by ABI1, a phosphatase from clade A of the PP2C family.Citation22 Moreover, we showed that ABI1 and SnRK2.4 regulate primary root growth in response to salinity; the phenotype of ABI1 knockout mutant (abi1td) exposed to salt stress was opposite to that of the snrk2.4 mutant. Now, in order to check whether the differences in the phenotypes of the snrk2.4 and abi1 mutants correlate with differences in SnRK2 activity, we monitored the SnRK2.4/SnR2.10 activity in WT and abi1td mutant seedlings exposed to NaCl. The SnRK2.4/SnRK2.10 activity was measured by immunocomplex in gel-kinase activity assay using MBP as substrate. We measured simultaneously the activity of SnRK2.4 and SnRK2.10, due to the specificity of antibodies used. Unfortunately, we did not observe any significant differences in the kinase activity between WT and abi1td mutant. Since most of clade A phosphatases are redundant in respect to inhibition of ABA-activated SnRK2s, we considered that it might also be the case for ABA-non-activated SnRK2s. Therefore, we decided to analyze the salt stress-induced SnRK2.4/SnRK2.10 activity in seedlings of double abi1–2 pp2ca-1 mutant plants, with insertions in both ABI1 and PP2CA genes, and additionally in the pp2ca-1 single mutant. We have chosen pp2ca-1 mutants for our studies since our results showed that in vitro recombinant PP2CA phosphatase dephosphorylates and inhibits the activity of SnRK2.4 similarly to ABI1 (). Measurement of the kinase activity in seedlings exposed to salt stress showed that the activity of SnRK2.4/SnRK2.10 was higher by about 50% after 60 min of salt treatment in the abi1–2 pp2ca-1 mutant in comparison to wild type Arabidopsis (), whereas we did not observe higher SnRK2.4/SnRK2.10 activity in the single pp2ca-1 mutant, as it was in the case of abi1td. The enhanced activity of the ABA-non-activated SnRK2s in the double abi1–2 pp2ca-1 mutant indicates that not only ABI1, but also PP2CA, might be a physiological regulator of SnRK2.4. Moreover, their function is at least to some extent redundant.
Figure 1. GST-PP2CA dephosphorylates and inhibits SnRK2.4 in vitro (A). Recombinant SnRK2.4 (1 µg) and in parallel SnRK2.6 (1 µg) and SnRK2.8 (1 µg) as controls were incubated with increasing amounts of recombinant GST-PP2CA for 30 min at 30°C and kinase activity was analyzed by in-gel kinase activity assay using MBP as substrate. Dephosphorylation of SnRK2.4 after treatment with GST-PP2CA (0, 12.5, and 25 U/sample) was monitored by immunoblotting using specific anti-P-SnRK2 antibodies recognizing a phosphorylated residue in the kinase activation loop (Ser-158). U, amount of enzyme, which releases 1 picomole of phosphate per minute; Autorad, autoradiograph; CBB, Coomassie brilliant blue. Data represent one of 3 independent experiments showing similar results. Salt-induced SnRK2.4/SnRK2.10 activity is increased in the abi1–2 pp2ca-1 double mutant but not in the abi1td or pp2ca-1 single mutants (B). Activity of SnRK2.4 and SnRK2.10 in extracts was monitored simultaneously by immunocomplex kinase in gel activity assay, using antibodies recognizing both kinases and MBP as substrate. Autoradiographs (Autorad) represent one of 5 independent experiments.
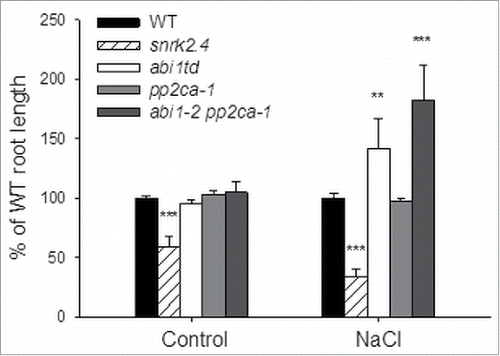
To check whether PP2CA phosphatase, similarly to ABI1, is involved in the regulation of root growth in response to salinity we analyzed the phenotypes of abi1td, double abi1–2 pp2ca-1, and pp2ca-1 mutants and in parallel the snrk2.4 mutant and wild type (as controls). We compared the primary root length of seedlings exposed to 115 mM NaCl (and grown on control medium), as in the experiments described in refs.Citation22,23 The roots of snrk2.4 seedlings grown on medium supplemented with NaCl were significantly shorter than those of all other lines, confirming previous results showing the positive role of SnRK2.4 in the control of root growth in these conditions.Citation23 In contrast, the roots of the abi1–2 pp2ca-1 mutant were significantly longer, by 80% comparing with WT, and by about 40% comparing with to abi1td (). However, we did not observe any differences in root growth between WT and the pp2ca-1 mutant either in control conditions or under salinity; PP2CA function in root growth regulation was only apparent in the double loss-of-function mutant abi1–2 pp2ca-1. The results indicate that PP2CA together with ABI1 is involved in the regulation of root growth under salinity stress and their effect antagonizes that of SnRK2.4. The phenotype of the abi1–2 pp2ca-1 mutant (longer primary roots in comparison to WT seedlings grown on medium supplemented with NaCl) is most probably caused by the release of SnRK2.4 from negative inhibition by the phosphatases and its higher activity. However, having in mind that PP2CA regulates several different signaling pathways, we cannot exclude that some of them have also an impact on root growth.
Figure 2. ABI1 and PP2CA double mutant (abi1–2 pp2ca-1) plants display opposite phenotypes to snrk2.4 in terms of root growth under salt stress. Five-day-old seedlings grown vertically on ½ MS media were transferred into square Petri plates with control media or 115 mM NaCl. After seven days of growth, the increase in the length of roots was measured and calculated as percentage of primary root length in wild type plants. The graph presents mean values (± SE) from 3 independent experiments with 4 biological repeats per condition and mutant line each. Statistical analysis was done by t-test.
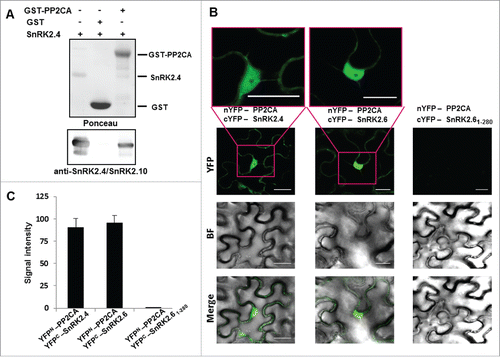
All these results indicate that SnRK2.4 is a cellular substrate of both ABI1 and PP2CA. However, our previous yeast 2-hybrid assay data did not show any interaction between SnRK2.4 and PP2CA. Taking into consideration that the yeast 2-hybrid assay quite often gives false positive or negative results we checked the binding between SnRK2.4 and the PP2CA using a pull down assay. The results show clear in vitro interaction between proteins studied (). To verify if SnRK2.4 and PP2CA interact in plant cells we analyzed complex formation between them in Nicotiana benthamiana leaves transiently transformed by agroinfiltration with constructs encoding PP2CA fused to the N-terminal fragment of YFP (nYFP-PP2CA) and SnRK2.4 in fusion with the C-terminal fragment (cYFP-SnRK2.4), or cYFP-SnRK2.6 as a positive control, or cYFP-SnRK2.61–280, cYFP fused to a truncated form of SnRK2.6 lacking the C-terminal part (amino acids 281–362), as a negative control of the assay. The results of bimolecular fluorescence complementation (BiFC) assays confirmed the in vitro binding data () and showed that SnRK2.4 interacts with PP2CA in plant cells.
Figure 3. SnRK2.4 interacts with ABI1 and PP2CA in pull down assay (A) GST-fused PP2CA or GST alone (as negative control) bound to glutathione-sepharose beads, were incubated with recombinant SnRK2.4 (tag free). The presence of bound SnRK2.4 was monitored by immunoblotting using anti-SnRK2.4/SnRK2.10 antibodies. Data represent one of 2 independent experiments showing similar results. Interaction of the proteins was analyzed by BiFC assay (B,C) Nicotiana benthamiana leaves were transiently co-transformed with pairs of plasmids encoding: nYFP-PP2CA and cYFP-SnRK2.4, nYFP-PP2CA and cYFP-SnRK2.6, or nYFP-PP2CA and cYFP-SnRK2.61–280 (as controls). The leaf epidermal cells were co-infiltrated with Agrobacterium suspensions containing the indicated constructs and the silencing suppressor p19. A positive control of the BiFC assay is provided by the nYFP-PP2CA/cYFP-SnRK2.6 interaction, which is abolished when the C-truncated form of SnRK2.6, cYFP-SnRK2.61–280 was used. The BiFC assay was performed as described in refs 24 and 25. BiFC images were analyzed using ImageJ software and signal intensity (arbitrary fluorescence units) was calculated after subtracting the mean background. Data are averages ± SD (n = 20).
Concluding, our results show that the activity of SnRK2.4, a member of the ABA-non-activated SnRK2s, in plant response to salt stress is regulated by at least 2 clade A PP2Cs (PP2CA in addition to ABI1 shown previously). All three enzymes are involved in the regulation of root growth in response to salinity. Since we do not observe reduction of the kinase activity in single abi1 or pp2ca mutants we can conclude that both phosphatases are at least partially redundant in respect to their inhibition of ABA-non-activated SnRK2s, similarly as it is in the case of ABA-activated SnRK2s. At this stage the mechanism of regulation the PP2C activity in response to salinity or osmotic stress, which would allow SnRK2s activation in an ABA-independent manner, is not known. Studies on this issue are vital for gaining a better understanding of ABA-independent osmotic stress signal transduction in plants.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank dr. Agnieszka Ludwików for sharing with us abi1td seeds and all members of the laboratory for discussions.
Funding
This work was supported by National Science Center (grant 2011/03/B/NZ3/00297 to GD). Funding in the laboratory of Pedro L. Rodriguez was provided by grant BIO2014–52537-R.
References
- Kulik A, Wawer I, Krzywinska E, Bucholc M, Dobrowolska G. SnRK2 protein kinases–key regulators of plant response to abiotic stresses. OMICS 2011; 15:859-72; PMID:22136638; http://dx.doi.org/10.1089/omi.2011.0091
- Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K. Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol 2010; 51:1821-39; PMID:20980270; http://dx.doi.org/10.1093/pcp/pcq156
- Nakashima K, Yamaguchi-Shinozaki K. ABA signaling in stress-response and seed development. Plant Cell Rep 2013; 32:959-70; PMID:23535869; http://dx.doi.org/10.1007/s00299-013-1418-1
- Fujii H, Zhu JK. Osmotic stress signaling via protein kinases. Cell Mol Life Sci 2012; 69:3165-73; PMID:22828864; http://dx.doi.org/10.1007/s00018-012-1087-1
- Yoshida T, Mogami J, Yamaguchi-Shinozaki K. Omics Approaches Toward Defining the Comprehensive Abscisic Acid Signaling Network in Plants. Plant Cell Physiol 2015; 56:1043-52; PMID:25917608; http://dx.doi.org/10.1093/pcp/pcv060
- Boudsocq M, Barbier-Brygoo H, Lauriere C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J Biol Chem 2004; 279:41758-66; PMID:15292193; http://dx.doi.org/10.1074/jbc.M405259200
- Kobayashi Y, Yamamoto S, Minami H, Kagaya Y, Hattori T. Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. The Plant cell 2004; 16:1163-77; PMID:15084714; http://dx.doi.org/10.1105/tpc.019943
- Belin C, de Franco PO, Bourbousse C, Chaignepain S, Schmitter JM, Vavasseur A, Giraudat J, Barbier-Brygoo H, Thomine S. Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant physiology 2006; 141:1316-27; PMID:16766677; http://dx.doi.org/10.1104/pp.106.079327
- Burza AM, Pekala I, Sikora J, Siedlecki P, Malagocki P, Bucholc M, Koper L, Zielenkiewicz P, Dadlez M, Dobrowolska G. Nicotiana tabacum osmotic stress-activated kinase is regulated by phosphorylation on Ser-154 and Ser-158 in the kinase activation loop. J Biol Chem 2006; 281:34299-311; PMID:16980311; http://dx.doi.org/10.1074/jbc.M601977200
- Boudsocq M, Droillard MJ, Barbier-Brygoo H, Lauriere C. Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol Biol 2007; 63:491-503; PMID:17103012; http://dx.doi.org/10.1007/s11103-006-9103-1
- Vlad F, Droillard MJ, Valot B, Khafif M, Rodrigues A, Brault M, Zivy M, Rodriguez PL, Merlot S, Laurière C. Phospho-site mapping, genetic and in planta activation studies reveal key aspects of the different phosphorylation mechanisms involved in activation of SnRK2s. Plant J 2010; 63:778-90; PMID:20561261; http://dx.doi.org/10.1111/j.1365-313X.2010.04281.x
- Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Laurière C, Merlot S. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 2009; 21:3170-84; PMID:19855047; http://dx.doi.org/10.1105/tpc.109.069179
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci U S A 2009; 106:17588-93; PMID:19805022; http://dx.doi.org/10.1073/pnas.0907095106
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009; 324(5930):1064-8; PMID:19407143; http://dx,doi,org/10.1126/science.1172408
- Melcher K, Ng LM, Zhou XE, Soon FF, Xu Y, Suino-Powell KM, Park SY, Weiner JJ, Fujii H, Chinnusamy V, et al. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 2009; 462:602-8; PMID:19898420; http://dx.doi.org/10.1038/nature08613
- Miyazono K, Miyakawa T, Sawano Y, Kubota K, Kang HJ, Asano A, Miyauchi Y, Takahashi M, Zhi Y, Fujita Y, et al. Structural basis of abscisic acid signalling. Nature 2009; 462:609-14; PMID:19855379; http://dx.doi.org/10.1038/nature08583
- Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science 2009; 326:1373-9; PMID:19933100; http://dx.doi.org/10.1126/science.1181829
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009; 324:1068-71; PMID:19407142; http://dx.doi.org/10.1126/science.1173041
- Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park SY, Márquez JA, Cutler SR, Rodriguez PL. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J 2009; 60:575-88; PMID:19624469; http://dx.doi.org/10.1111/j.1365-313X.2009.03981.x
- Hou YJ, Zhu Y, Wang P, Zhao Y, Xie S, Batelli G, Wang B, Duan CG, Wang X, Xing L, et al. Type One Protein Phosphatase 1 and Its Regulatory Protein Inhibitor 2 Negatively Regulate ABA Signaling. PLoS Genet 2016; 12:e1005835; PMID:26943172; http://dx.doi.org/10.1371/journal.pgen.1005835
- Waadt R, Manalansan B, Rauniyar N, Munemasa S, Booker MA, Brandt B, Waadt C, Nusinow DA, Kay SA, Kunz HH, et al. Identification of open stomata1-interacting proteins reveals interactions with sucrose non-fermenting1-related protein kinases2 and with Type 2A protein phosphatases that function in abscisic acid responses. Plant Physiol 2015; 169:760-79; PMID:26175513; http://dx.doi.org/10.1104/pp.15.00575
- Krzywinska E, Bucholc M, Kulik A, Ciesielski A, Lichocka M, Debski J, Ludwików A, Dadlez M, Rodriguez PL, Dobrowolska G, et al. Phosphatase ABI1 and okadaic acid-sensitive phosphoprotein phosphatases inhibit salt stress-activated SnRK2.4 kinase. BMC Plant Biol 2016; 16:136; PMID:27297076; http://dx.doi.org/10.1186/s12870-016-0817-1
- McLoughlin F, Galvan-Ampudia CS, Julkowska MM, Caarls L, van der Does D, Lauriere C, Munnik T, Haring MA, Testerink C. The Snf1-related protein kinases SnRK2.4 and SnRK2.10 are involved in maintenance of root system architecture during salt stress. Plant J 2012; 72:436-49; PMID:22738204; http://dx.doi.org/10.1111/j.1365-313X.2012.05089.x
- Saez A, Rodrigues A, Santiago J, Rubio S, Rodriguez PL. HAB1-SWI3B interaction reveals a link between abscisic acid signaling and putative SWI/SNF chromatin-remodeling complexes in Arabidopsis. Plant Cell 2008; 20:2972-88; PMID:19033529; http://dx.doi.org/10.1105/tpc.107.056705
- Bueso E, Rodriguez L, Lorenzo-Orts L, Gonzalez-Guzman M, Sayas E, Munoz-Bertomeu J, Ibañez C, Serrano R, Rodriguez PL. The single-subunit RING-type E3 ubiquitin ligase RSL1 targets PYL4 and PYR1 ABA receptors in plasma membrane to modulate abscisic acid signaling. Plant J 2014; 80:1057-71; PMID:25330042; http://dx.doi.org/10.1111/tpj.12708
