ABSTRACT
Feeding by Spodoptera exigua (beet armyworm) larvae on Zea mays (maize) induces expression of 9-lipoxygenases to a greater extent than 13-lipoxygenases. Whereas 13-lipoxygenases have an established role in the synthesis of jasmonates that serve as defense signaling molecules in many plant species, relatively little is known about the role of 9-lipoxygenases in herbivore defense. Phylogenetic analysis of lipoxygenases from maize inbred lines B73 and W22 shows that, although most Lox genes are present in both lines, Lox12, a 9-lipoxygenase that has been implicated in fungal defense, is truncated and unlikely to encode a functional protein in W22. Two independent Mutator transposon insertions in another 9-lipoxygenase, Lox4, caused improved S. exigua growth on the mutant lines relative to wildtype W22. This observation suggests a function in herbivore defense for metabolic products downstream of maize Lox4, either through direct toxicity or a perhaps an as yet unknown signaling function.
KEYWORDS:
Introduction
Plants respond to insect herbivory with a large variety of metabolic changes. Some induced metabolites have toxic or deterrent effects against the herbivores, whereas others act as signaling molecules to induce defense responses in parts of the plant that are not yet under attack. Two well-studied plant metabolic pathways, those of indole glucosinolates in Arabidopsis thalianaCitation1 and benzoxazinoids in Zea mays (maize),Citation2,Citation3 produce metabolites with both direct defensive properties and defense signaling molecules.
Jasmonic acid and its derivatives methyl jasmonate and jasmonate-isoleucine are well-studied components of the defense signaling pathway in many plant species.Citation4 The first reaction in the synthesis of jasmonic acid from α-linolenic acid is catalyzed by 13-lipoxygenases ().Citation5 In the sequenced B73 maize genome,Citation6 six genes are predicted to encode 13-lipoxygenases (Lox7, Lox8, Lox9, Lox10, Lox11, and Lox13; ). Lox7 and Lox8, are involved in jasmonate biosynthesis, thereby contributing to inflorescence development and plant defense, respectively.Citation7,Citation8 Lox10 activity leads to the release of herbivore-induced plant volatiles.Citation8
Figure 1. 9-Lipoxygenase and 13-lipoxygenase oxygenate α-linolenic acid at different positions to produce (10E,12Z)-9-Hydroperoxy-10,12,15-octadecatrienoic acid (9-HPOT and (10E,12Z)-13-hydroperoxy-10,12,15-octadecatrienoic acid (13-HPOT), respectively. 9-HPOT and 13-HPOT serve as precursors for differing sets of bioactive plant metabolites. 10-OPDA = 10-oxophytodienoic acid;12-OPDA = 12-oxophytodienoic acid; DES = divinyl ether synthase; AOS = allene oxide synthase; HPL = hydroperoxide lysase. Modified from Schiller et al. (2015).
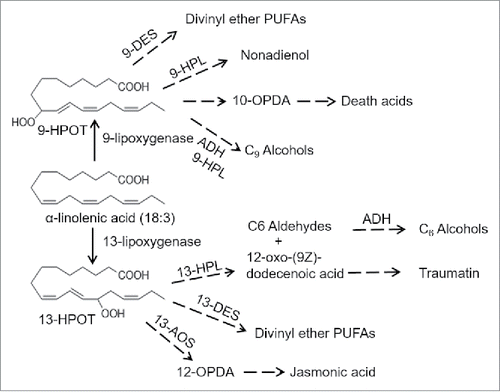
In addition to 13-lipoxygenases, the B73 genome contains seven genes encoding predicted 9-lipoxygenases (Lox1, Lox2, Lox3, Lox4, Lox5, Lox6, and Lox12; ), which oxidize 18:3 α-linolenic acid and 18:2 α-linoleic acid to produce 10-oxo-11-phytodienoic acid (10-OPDA; ) and 10-oxo-11-phytoenoic acid (10-OPEA), respectively. Fungal infection, mechanical wounding, and feeding by Helicoverpa zea (corn earworm) induce 10-OPEA accumulation in maize.Citation9,Citation10 Furthermore, in vitro experiments show that 10-OPEA inhibits the growth of fungal pathogens (Aspergillus flavus and Fusarium verticillioides) and H. zea at physiologically relevant concentrations. Similarly, assays with maize 9-lipoxygenase mutants have demonstrated functions in plant defense. For instance, a lox12 mutation reduced F. verticilloides resistanceCitation11 and Meloidogyne incognita (root-knot nematode) grew better on a lox3 mutant line.Citation12
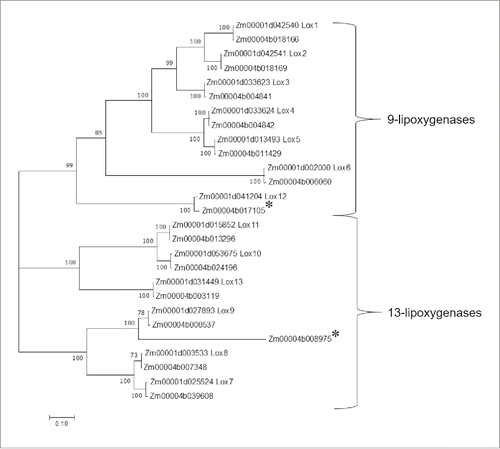
Figure 2. Dendrogram of predicted protein sequences of 27 lipoxygenases from the maize inbred line B73 v4 assembly (Zm00001d numbers) and the inbred line W22 v2 assembly (Zm00004b numbers). The evolutionary history was inferred by using the Maximum Likelihood method based on the JTT matrix-based model. Evolutionary analyses were conducted in MEGA7. The tree with the highest log likelihood is shown. Bootstrap values are based on 1000 replications. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. All positions with less than 80% site coverage were eliminated. There were a total of 835 positions in the final dataset. Proteins that are truncated and likely non-functional in W22 are marked with asterisks.
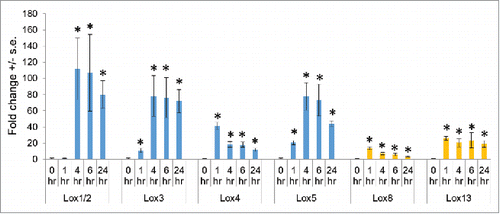
In recent research, we investigated the transcriptional and metabolic responses of inbred line B73 to feeding by Spodoptera exigua (beet armyworm) caterpillars.Citation13 As expected, insect herbivory induced the expression of 13-lipoxygenases () and increased the accumulation of jasmonic acid and its derivatives. However, S. exigua growth was not affected by a lox8 mutation, suggesting possible redundancy in maize 13-lipoxygenase activity. Compared to 13-lipoxygenases, 9-lipoxygenases were induced to an even greater extent by S. exigua feeding (). Expression of Lox1/2, Lox3, Lox4, and Lox5 was increased up to 100-fold within an hour of caterpillar feeding and remained at a high level for the duration of the 24-hour experiment. In other research, Lox1 and Lox5 expression was induced by Ostrinia furnicalis (Asian corn borer) feeding.Citation14 Previous experiments using RNA blots to measure induction of gene expression in response to Spodoptera frugiperda (fall armyworm) feeding showed increased expression of Lox5, but not Lox4 after 1, 4, and 8 days of caterpillar feeding.Citation15 Differences in the observed induction of Lox4 expression ( vs.Citation15) could result from the use of different maize lines, different herbivore species, or perhaps a transient expression of Lox4 early during herbivory. Together, these gene expression results suggested that the activity of 9-lipoxygenases might also make significant contributions to maize defense against caterpillar herbivory.
Results and discussion
Whereas our gene expression analysisCitation13 was conducted with maize inbred line B73, better genetic resources, in particular, collections of Mutator (Mu), Activator (Ac) and Dissociation (Ds) transposon insertions are available for the W22 inbred line.Citation16-20 To facilitate the identification of a 9-lipoxygenase knockout mutation in W22, we first compared the complement of Lox proteins encoded in the B73 genomeCitation6 and the recently assembled W22 genome.Citation21
The protein sequences of 14 predicted W22 lipoxygenases and 13 predicted B73 lipoxygenases were aligned (). This showed that about the W22 homolog of B73 Lox12 (Zm00004b017105) is truncated by about 50% and is likely to be non-functional. As a Lox12 mutation in the B73 genetic background increases susceptibility to Fusarium verticilloides,Citation11 W22 might also show susceptibility to this fungal pathogen. Zm00004b008975, a Lox9-like protein that is present in W22 but not in B73 is similarly truncated and likely to be non-functional. A phylogenetic analysis of the B73 and W22 lipoxygenase proteins () showed that, with the exception of Lox12, there is a full-length W22 homolog for each of the 13 known B73 lipoxygenases (). The two pairs of tandem-duplicated lipoxygenases in B73, Lox1/2 and Lox3/4 are similarly localized as gene pairs in the W22 genome.
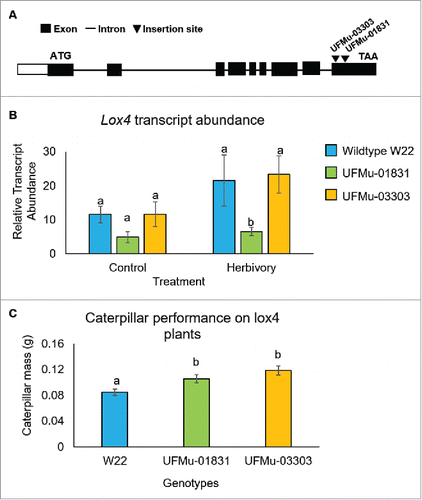
Analysis of publicly available Mu transposon insertions identified lines UFMu-01831 and UFMu-03303 as having an insertion in the W22 Lox4 gene (). Homozygous Mu insertions in Lox4 were verified by PCR amplification and DNA sequencing. Both transposon insertions are in coding regions and lead to predicted C-terminal truncations of the conserved lipoxygenase domain of the protein. Analysis of Lox4 gene expression by quantitative PCR using primers that bind upstream of the transposon insertion sites showed reduced transcript levels in UFMu-01831 but not UFMu-03303, with and without caterpillar feeding. It is possible that the two Mu-induced truncations of the gene have differing effects on Lox4 transcript stability. Relative to W22, S. exigua caterpillars gained more mass over a five-day period on both UFMu-01831 and UFMu-03303, indicating that maize defense responses are compromised by the Lox4::Mu mutations that truncate the C-terminus of this protein.
Figure 4. Analysis of a W22 Lox4 Mu insertion lines (A) Location of the UFMu-01831and UFMu-03303 insertions in the Lox4 gene and positions of primers that were used for quantitative RT-PCR gene expression analysis. (B) Expression level of the Lox4 gene, with and without six hours of S. exigua feeding. Mean +/- s.e. of 5 independent samples. (C) Mass of S. exigua caterpillars after five days of feeding on W22, UFMu-01831, and UFMu-03303seedlings. Mean +/- s.e. of 57 samples. *P value < 0.05, ANOVA followed by Tukey's HSD test.
Compared to the extensive research on the role of 13-lipoxygenases for jasmonate-mediated plant defense signaling, there is relatively little published information on the role that 9-lipoxygenases play in defense against insect herbivory. Altered caterpillar performance due to the lox4 mutation could be due to either changes in the production of toxic or deterrent metabolites, e.g. 10-OPEA.Citation9 or perhaps changes in plant defense signaling. For instance, just as 12-OPDA is a precursor for the production of jasmonic acid, 10-OPDA or 10-OPEA could lead to the formation of mobile signaling molecules that lead to the induction of defense responses in maize.
Given the large caterpillar-induced changes in gene expression () and the improved growth of S. exigua on lox4 mutants (), further research on the role of 9-lipoxygenase in maize defense against insects is warranted. In addition to Lox4::Mu insertion lines that we have investigated, Mu insertions in the W22 genetic background are available for two other 9-lipoxygenases, Lox2 and Lox5, in public collections. Investigation of herbivore resistance, gene expression changes, and metabolite accumulation in these transposon insertion lines likely will identify new pathways of maize defense against insect herbivory.
Methods
Molecular Phylogeny. Protein sequences of the 13 known lipoxygenase proteins (Lox1 – Lox13) in maize inbred line B73 RefGen v4 were downloaded from the Maize Genetics and Genomics Database (www.maizegdb.org). Sequence comparisons of B73 genes from the v2 assembly of the W22 genomeCitation21 identified 14 predicted lipoxygenase genes. Protein sequences of these 27 identified lipoxygenases were aligned with MuscleCitation22 using the following program parameters: gap penalties – gap open -2.9, gap extend 0, hydrophobicity multiplier 1.2; max iterations 8; clustering method UPGMB; and min diag length 24. A dendrogram of protein sequences was prepared using MEGA7.Citation23 The evolutionary history was inferred using the Maximum Likelihood method based on the JTT matrix-based model.Citation24 Initial trees for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. All amino acid positions with less than 80% site coverage among the 27 analyzed proteins were eliminated. There were a total of 835 positions were included in the final dataset.
Mu transposon insertions in Lox4. Line UFMu-01831 and UFMu-03303 were obtained from Maize Genetics COOP (http://maizecoop.cropsci.uiuc.edu/). Mu transposon insertions in the Lox4 gene were confirmed by PCR amplification using a Biorad C1000 thermal cycler the gene-specific primer (GGCGACGATAATCCTGGACCATACAAGG) and the transposon-specific primer TIR6 primer (AGAGAAGCCAACGCCAWCGCCTCYATTTCGTC), followed by DNA sequencing. Lox4 expression levels were measured by quantitative RT-PCR using an Applied Biosystems 7900HT instrument and the primers GCTGAACCTGAACATCTACG and GTAGAGCCCAAGGATGTCTT.
Insect bioassays. Eggs of S. exigua were obtained from Benzon Research (www.benzonresearch.com). Eggs were hatched at 28ºC and moved to S. exigua artificial diet (Southland Products, http://www.tecinfo.com/∼southland/). After five days, larvae were moved onto 17-day-old maize plants that were grown in a Conviron walk-in growth room at 23ºC with16:8 h light:dark cycle and 180 mmol photons m−2s−1 light intensity. After 5 days on the plants, caterpillars were removed and their mass was measured using a Sartorius precision balance.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
supp_data_1422462.pptx
Download MS Power Point (40.5 KB)Acknowledgments
This research was funded by US National Science Foundation awards 1139329 and 1339237 to GJ, and Vaadia-BARD Postdoctoral Fellowship Award FI-471-2012 to VT. We thank Kokulapalan Wimalanathan for assistance in identifying W22 Lox sequences.
Additional information
Funding
References
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science. 2009;323:95–101. doi:10.1126/science.1164627.
- Meihls LN, Handrick V, Glauser G, Barbier H, Kaur H, Haribal MM, Lipka AE, Gershenzon J, Buckler ES, Erb M, et al. Natural variation in maize aphid resistance is associated with 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one glucoside methyltransferase activity. Plant Cell. 2013;25:2341–2355. doi:10.1105/tpc.113.112409.
- Ahmad S, Veyrat N, Gordon-Weeks R, Zhang Y, Martin J, Smart L, Glauser G, Erb M, Flors V, Frey M, et al. Benzoxazinoid metabolites regulate innate immunity against aphids and fungi in maize. Plant Physiol. 2011;157:317–27. doi:10.1104/pp.111.180224.
- Howe GA, Jander G. Plant immunity to insect herbivores. Ann Rev Plant Biol. 2008;59:41–66. doi:10.1146/annurev.arplant.59.032607.092825.
- Schiller D, Contreras C, Vogt J, Dunemann F, Defilippi BG, Beaudry R, Schwab W. A dual positional specific lipoxygenase functions in the generation of flavor compounds during climacteric ripening of apple. Hortic Res. 2015;2:15003. doi:10.1038/hortres.2015.3.
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei FS, Pasternak S, Liang CZ, Zhang JW, Fulton L, Graves TA, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi:10.1126/science.1178534.
- Acosta IF, Laparra H, Romero SP, Schmelz E, Hamberg M, Mottinger JP, Moreno MA, Dellaporta SL. Tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science. 2009;323:262–265. doi:10.1126/science.1164645.
- Christensen SA, Nemchenko A, Borrego E, Murray I, Sobhy IS, Bosak L, DeBlasio S, Erb M, Robert CA, Vaughn KA, et al. The maize lipoxygenase, ZmLOX10, mediates green leaf volatile, jasmonate and herbivore-induced plant volatile production for defense against insect attack. Plant J. 2013;74:59–73. doi:10.1111/tpj.12101.
- Christensen SA, Huffaker A, Kaplan F, Sims J, Ziemann S, Doehlemann G, Ji L, Schmitz RJ, Kolomiets MV, Alborn HT, et al. Maize death acids, 9-lipoxygenase-derived cyclopente(a)nones, display activity as cytotoxic phytoalexins and transcriptional mediators. Proc Natl Acad Sci U S A. 2015;112:11407–11412. doi:10.1073/pnas.1511131112.
- Christensen SA, Huffaker A, Hunter CT, Alborn HT, Schmelz EA. A maize death acid, 10-oxo-11-phytoenoic acid, is the predominant cyclopentenone signal present during multiple stress and developmental conditions. Plant Signal Behav. 2016;11:e1120395. doi:10.1080/15592324.2015.1120395.
- Christensen SA, Nemchenko A, Park YS, Borrego E, Huang PC, Schmelz EA, Kunze S, Feussner I, Yalpani N, Meeley R, et al. The novel monocot-specific 9-lipoxygenase ZmLOX12 is required to mount an effective jasmonate-mediated defense against Fusarium verticillioides in maize. Mol Plant Microbe Interact. 2014;27:1263–1276. doi:10.1094/MPMI-06-13-0184-R.
- Gao X, Starr J, Gobel C, Engelberth J, Feussner I, Tumlinson J, Kolomiets M. Maize 9-lipoxygenase ZmLOX3 controls development, root-specific expression of defense genes, and resistance to root-knot nematodes. Mol Plant Microbe Interact. 2008;21:98–109. doi:10.1094/MPMI-21-1-0098.
- Tzin V, Hojo Y, Strickler SR, Bartsch LJ, Archer CM, Ahern KR, Zhou S, Christensen SA, Galis I, Mueller LA, et al. Rapid defense responses in maize leaves induced by Spodoptera exigua caterpillar feeding. J Exp Bot. 2017;68:4709–4723. doi:10.1093/jxb/erx274.
- Yang F, Zhang Y, Huang Q, Yin G, Pennerman KK, Yu J, Liu Z, Li D, Guo A. Analysis of key genes of jasmonic acid mediated signal pathway for defense against insect damages by comparative transcriptome sequencing. Sci Rep. 2015;5:16500. doi:10.1038/srep16500.
- Park YS, Kunze S, Ni XZ, Feussner I, Kolomiets MV. Comparative molecular and biochemical characterization of segmentally duplicated 9-lipoxygenase genes ZmLOX4 and ZmLOX5 of maize. Planta. 2010;231:1425–1437. doi:10.1007/s00425-010-1143-8.
- McCarty DR, Latshaw S, Wu S, Suzuki M, Hunter CT, Avigne WT, Koch KE. Mu-seq: sequence-based mapping and identification of transposon induced mutations. PLoS One. 2013;8:e77172. doi:10.1371/journal.pone.0077172.
- Kolkman JM, Conrad LJ, Farmer PR, Hardeman K, Ahern KR, Lewis PE, Sawers RJ, Lebejko S, Chomet P, Brutnell TP. Distribution of Activator (Ac) throughout the maize genome for use in regional mutagenesis. Genetics. 2005;169:981–95. doi:10.1534/genetics.104.033738.
- Vollbrecht E, Duvick J, Schares JP, Ahern KR, Deewatthanawong P, Xu L, Conrad LJ, Kikuchi K, Kubinec TA, Hall BD, et al. Genome-wide distribution of transposed dissociation elements in maize. Plant Cell. 2010;22:1667–1685. doi:10.1105/tpc.109.073452.
- Brutnell T, Conrad LJ. Transposon tagging using Activator (Ac) in maize. Methods in Molecular Biology. 2003;236:157–176.
- Hunter CT, Suzuki M, Saunders J, Wu S, Tasi A, McCarty DR, Koch KE. Phenotype to genotype using forward-genetic Mu-seq for identification and functional classification of maize mutants. Front Plant Sci. 2014;4:545. doi:10.3389/fpls.2013.00545.
- Springer NM, Anderson SN, Andorf CM, Ahern K, Bai F, Barad O, Brad Barbazuk WB, Bass HW, Baruch K, Ben-Zvi G, et al. The W22 genome: a foundation for maize functional genomics and transposon biology. Nat Genet. 2017; in review.
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi:10.1093/nar/gkh340.
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi:10.1093/molbev/msw054.
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282.