ABSTRACT
Arabidopsis MAP KINASE17 (MPK17) was recently identified as a novel regulator of peroxisome division in response to salt stress. Further, the known peroxisome division factor PEROXISOME AND MITOCHONDRIAL DIVISION FACTOR1 (PMD1) genetically acts downstream of MPK17. We previously showed that mutants defective in either MPK17 or PMD1 fail to proliferate peroxisomes in response to NaCl stress. Here, we show that, unlike their abnormal NaCl responses, mpk17 and pmd1 mutants display wild type responses to other stresses known to alter peroxisome proliferation, suggesting that plants distinguish among peroxisome division-inducing stresses and alter the peroxisome division pathway based on the stress applied.
Introduction
Peroxisomes, small organelles conserved throughout all eukaryotes, perform a wide variety of essential functions including fatty acid β-oxidation and hydrogen peroxide detoxification [reviewed inCitation1]. Plant peroxisomes house additional specialized functions, including conversion of hormone precursors into their active forms, some steps of vitamin synthesis, and branched chain amino acid synthesis [reviewed inCitation1]. These highly dynamic organelles divide through a constriction-and-fission process [reviewed inCitation2] and track rapidly throughout the cell via the actin cytoskeleton.Citation3 Peroxisomes in Arabidopsis proliferate in response to a variety of both biotic and abiotic stresses, including salt,Citation4,Citation5 pathogens,Citation6 high light,Citation7 cadmium,Citation8,Citation9 and general ROS stress.Citation10 Multiple lines of evidence suggest that stress induction of peroxisome proliferation is differentially triggered by each stress. First, plants differentially upregulate peroxisome biogenesis gene expression in response to distinct stresses. For example, the PEX1 transcript increases in response to light, pathogen, and salt stresses, but remains unchanged in response to osmotic stress.Citation11,Citation12 In contrast, the PEX10 transcript increases in response to both salt stress and osmotic stress.Citation11 Second, whereas the number of peroxisomes is reported to increase in response to all the above stresses, peroxisome populations do not behave identically after division. Pathogen attack not only increases the number of peroxisomes but also reorients peroxisomes to the site of pathogen attack.Citation6,Citation13 Under high light stress, plants proliferate peroxisomes and extend peroxules from these peroxisomes, which associate with mitochondria.Citation14 Peroxules also form under cadmium stress,Citation8 but haven't been reported under high salt conditions or pathogen attacks. Together, these data suggest plants distinguish among these stresses and trigger distinct peroxisome responses for each of them.
Previously, we reported a novel peroxisome division regulator in Arabidopsis, MAP KINASE17 (MPK17), which represses peroxisome division with the peroxisome division factor PEROXISOME AND MITOCHONDRIAL DIVISION FACTOR1 genetically acting downstream of MPK17.Citation15 Neither mpk17 nor pmd1 increase peroxisome division in response to salt stress.Citation15 Here, we show that mpk17-1 and pmd1-1 display normal peroxisome responses to other division-inducing stresses, supporting previous findings that plants distinguish among peroxisome division-inducing stresses and may utilize different division pathways depending on the stress condition.
Results
Because mpk17-1 and pmd1-1 do not respond normally to NaCl stress by increasing peroxisome division,Citation15 we examined mpk17-1 and pmd1-1 peroxisome numbers in response to a variety of other stresses. In all examined conditions, both mpk17-1 and pmd1-1 display wild-type responsiveness in peroxisome proliferation, suggesting that MPK17 and PMD1 are important for salt-induced peroxisome proliferation but not in peroxisome proliferation in response to general stresses.
mpk17 and pmd1 respond normally to light exposure
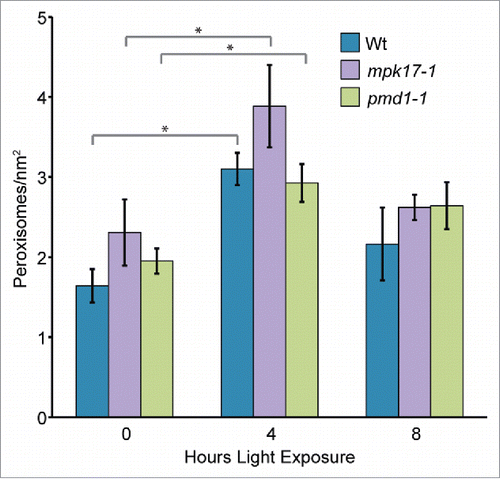
Sudden light exposure is reported to induce peroxisome division,Citation16 likely by increasing transcription of PEX11B.Citation7 To determine whether MPK17 and/or PMD1 might also act in this pathway, we examined peroxisome proliferation in mpk17-1 and pmd1-1 in response to sudden light exposure. Similar to previous reports,Citation7-16 wild type rapidly and transiently displayed increased peroxisome numbers upon the sudden light exposure of dark-grown seedlings (). mpk17-1 and pmd1-1 also rapidly and transiently displayed increased peroxisome numbers upon sudden light exposure of dark-grown seedlings (), suggesting that MPK17 and PMD1 are not necessary for this response.
mpk17 and pmd1 respond normally to cadmium stress
Figure 1. Peroxisomes in mpk17-1 and pmd1-1 respond normally to sudden light exposure. Mean number of peroxisomes in dark grown hypocotyls from wild type (Wt; Col-0), mpk17-1, and pmd1-1 after the indicated length of light exposure. * indicates p value < 0.05.
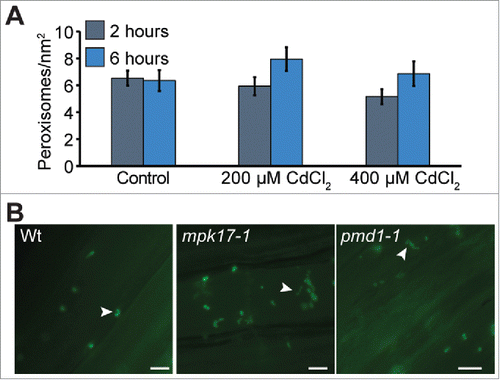
Peroxisomes are reported to divide rapidly and form peroxules when grown on elevated levels of cadmium.Citation8 We examined peroxisome numbers in wild type, mpk17-1, and pmd1-1 in response to heavy metals. Seedlings were exposed to a short term CdCl2 stress, then imaged. Although we did not observe increased peroxisome numbers under these conditions, wild type, mpk17-1, and pmd1-1 formed peroxules in response to this cadmium stress (B).
mpk17 displays normal responses to clofibrate
Figure 2. Peroxisomes under cadmium stress A) Mean number of perosixomes per unit area do not increase significantly in wild type under short term cadmium stress. B) mpk17-1 and pmd1-1 respond normally to cadmium by forming peroxules after 6h treatment with 100 µM CdCl2. Peroxules are indicated with white arrowheads. Scale bars = 5 µm.
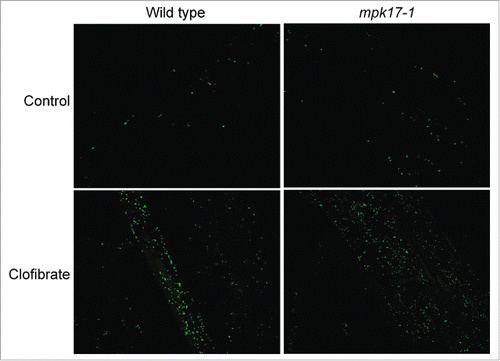
One possible explanation of stress-induced peroxisome division is the observation that all the division-inducing stresses also increase intracellular ROS, so the division could be a result of increased ROS, not a direct response to each individual stress. Peroxisomes break down many species of ROS, so a quick increase in peroxisome number could help remove ROS after the stress signal has been perceived, but before ROS can damage cellular components. If this hypothesis is true, mutants impaired in stress-induced peroxisome division should display increased ROS during the stress and should further display increased peroxisomes in response to all ROS-generating stresses, including chemicals known to increase intracellular ROS. To test whether altered ROS responses might contribute to the lack of response to salt, peroxisome division in mpk17-1 was evaluated on ROS-producing chemical clofibrate, which increases peroxisome division in mammalian cells.Citation17 Similar to wild type, mpk17-1 responds to clofibrate with increased peroxisome numbers (), suggesting that ROS-induced peroxisome proliferation does not require MPK17.
Discussion
The data above, together with our prior work,Citation15 demonstrate that plants distinguish between different types of stress and use varied pathways depending on the stress to induce peroxisome division. But beyond “how”, questions about “why” remain.
An adaptive benefit from peroxisome proliferation remains elusive for most stresses, except pathogen attack. Under biotic stress, peroxisomes directly produce anti-fungal compounds,Citation13 and the rice PEX5 peroxisome receptor is an active anti-fungal protein.Citation18 No direct benefit to the plant from increasing peroxisome division during salt stress has been observed. Artificially increasing peroxisome number by overexpressing peroxisome division factors fails to appreciably increase abiotic stress tolerance in Arabidopsis.Citation5,Citation6 Notably, it has not been shown that overexpressing peroxisome division factors in otherwise salt-hypersensitive backgrounds can rescue the salt hypersensitivity.Citation4 Thus, while we now know that plants utilize different signaling and peroxisome division pathways based on the type of stress they are facing, whether this increase in division aids in mitigating the effects of stress in ways we have been unable to accurately measure, or whether this division is an unintended side effect of stress signaling pathways remains an open question.
Abbreviations and acronyms
| MPK17 | = | Arabidopsis Map Kinase17 |
| PMD1 | = | PEROXISOME AND MITOCHONDRIAL DIVISION FACTOR1 |
| ROS | = | Reactive oxygen species |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Hongwei Jing, Samantha Powers, and Ryan Emenecker for critical comments on the manuscript. This work was supported by the National Science Foundation (IOS-1453750 to L.C.S.), the United States Department of Agriculture-National Institute of Food and Agriculture Fellowship Program (2016-67011-25096 to E.M.F.) and the National Institutes of Health (R01 GM112898 to L.C.S.).
Additional information
Funding
References
- Islinger M, Grille S, Fahimi HD, Schrader M. The peroxisome: an update on mysteries. Histochem Cell Biol. 2012;137:547–574. doi:10.1007/s00418-012-0941-4.
- Hu J, Baker A, Bartel B, Linka N, Mullen RT, Reumann S, Zolman BK. Plant peroxisomes: biogenesis and function. Plant Cell. 2012;24:2279–2303. doi:10.1105/tpc.112.096586.
- Mathur J, Mathur N, Hülskamp M. Simultaneous visualization of peroxisomes and cytoskeletal elements reveals actin and not microtubule-based peroxisome motility in plants. Plant Physiol. 2002;128:1031–1045. doi:10.1104/pp.011018.
- Fahy D, Sanad MN, Duscha K, Lyons M, Liu F, Bozhkov P, Kunz HH, Hu J, Neuhaus HE, Steel PG, et al. Impact of salt stress, cell death, and autophagy on peroxisomes: quantitative and morphological analyses using small fluorescent probe N-BODIPY. Sci Rep. 2017;7:39069. doi:10.1038/srep39069.
- Mitsuya S, El-Shami M, Sparkes IA, Charlton WL, Lousa Cde M, Johnson B, Baker A. Salt stress causes peroxisome proliferation, but inducing peroxisome proliferation does not improve NaCl tolerance in Arabidopsis thaliana. PLoS One. 2010;5:e9408. doi:10.1371/journal.pone.0009408.
- Koh S, André A, Edwards H, Ehrhardt D, Somerville S. Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. Plant J. 2005;44:516–529. doi:10.1111/j.1365-313X.2005.02545.x.
- Desai M, Hu J. Light induces peroxisome proliferation in Arabidopsis seedlings through the photoreceptor phytochrome A, the transcription factor HY5 HOMOLOG, and the peroxisomal protein PEROXIN11b. Plant Physiol. 2008;146(3):1117–1127. doi:10.1104/pp.107.113555.
- Rodríguez-Serrano M, Romero-Puertas MC, Sanz-Fernández M, Hu J, Sandalio LM. Peroxisomes Extend Peroxules in a Fast Response to Stress via a Reactive Oxygen Species-Mediated Induction of the Peroxin PEX11a. Plant Physiol. 2016;171:1665–1674. doi:10.1104/pp.16.00648.
- Rodríguez-Serrano M, Romero-Puertas MC, Sparkes I, Hawes C, del Río LA, Sandalio LM. Peroxisome dynamics in Arabidopsis plants under oxidative stress induced by cadmium. Free Radic Biol Med. 2009;47:1632–1639. doi:10.1016/j.freeradbiomed.2009.09.012.
- Lopez-Huertas E, Charlton WL, Johnson B, Graham IA, Baker A. Stress induces peroxisome biogenesis genes. EMBO J. 2000;19:6770–6777. doi:10.1093/emboj/19.24.6770.
- Charlton WL, Matsui K, Johnson B, Graham IA, Ohme-Takagi M, Baker A. Salt-induced expression of peroxisome-associated genes requires components of the ethylene, jasmonate and abscisic acid signaling pathways. Plant, Cell and Environment. 2005;28:513–524. doi:10.1111/j.1365-3040.2004.01293.x.
- Charlton WL, Johnson B, Graham IA, Baker A. Non-coordinate expression of peroxisome biogenesis, beta-oxidation and glyoxylate cycle genes in mature Arabidopsis plants. Plant Cell Rep. 2005;23(9):647–53. doi:10.1007/s00299-004-0879-7.
- Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, Landtag J, Brandt W, Rosahl S, Scheel D, et al. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science. 2005;310:1180–1183. doi:10.1126/science.1119409.
- Jaipargas EA, Mathur N, Bou Daher F, Wasteneys GO, Mathur J. High Light Intensity Leads to Increased Peroxule-Mitochondria Interactions in Plants. Front Cell Dev Biol. 2016;4:6. doi:10.3389/fcell.2016.00006.
- Frick EM, Strader LC. Kinase MPK17 and the peroxisome division factor PMD1 influence salt-induced peroxisome proliferation. Plant Physiol. 2018;176(1):340–351. doi:10.1104/pp.17.01019..
- Orth T, Reumann S, Zhang X, Fan J, Wenzel D, Quan S, Hu J. The PEROXIN11 protein family controls peroxisome proliferation in Arabidopsis. Plant Cell. 2007;19:333–350. doi:10.1105/tpc.106.045831.
- Hess R, Staubli W, Riess W. Nature of the hepatomegalic effect produced by ethyl-chlorophenoxy-isobutyrate in the rat. Nature. 1965;208(5013):856–8. doi:10.1038/208856a0.
- Lee JR, Park SC, Kim MH, Jung JH, Shin MR, Lee DH, Cheon MG, Park Y, Hahm KS, Lee SY. Antifungal activity of rice Pex5p, a receptor for peroxisomal matrix proteins. Biochem Biophys Res Commun. 2007;359:941–946. doi:10.1016/j.bbrc.2007.05.210.