ABSTRACT
Post-translational modification of proteins by small ubiquitin-like modifier (SUMO) plays essential roles in a large variety of cellular and developmental processes. While SUMO conjugation to target proteins has been reported in numerous studies in animals and human, and partly also in the model plant Arabidopsis, little is known about the specific roles of SUMO in crop plants. Here, we report about the maize SUMO family and show that the highly conserved core isoform SUMO1 predominately locates to the nucleus where it marks euchromatin rather than heterochromatin. Moreover, SUMO1 is especially present in nuclei of small dividing cells. Strong overexpression of SUMO1 caused a severe dwarf phenotype and abnormalities in floral organ structures. Defects in anther development and female gametogenesis occurred similar to null-mutant phenotypes reported in Arabidopsis. Taken together, these studies imply that precise and fine-tuned conjugation of the highly conserved plant SUMO1 isoform to target proteins is required for vegetative and reproductive development. Mis-regulation by overexpression or knock-out is deleterious, strongly affecting fertility in both dicots and monocots, including the crop plant maize.
Text
Being exposed to constant environmental changes and developmental cues, plants have evolved diverse molecular mechanisms to rapidly sense and respond at various transcriptional, translational and post-translational levels. Post-translational modifications (PTMs) are now increasingly considered as core components in cell signaling serving as fine-tuning mechanisms to adjust cellular responses to changes in the environment and during developmental progression.Citation1 Compared with other PTMs like phosphorylation and ubiquitination occurring predominately in the cytoplasm, SUMOylation targets have been shown to be especially localized to the nucleus being involved especially in DNA repair, chromatin modification and gene regulation. Moreover, it has been further shown that SUMOylation appears to play a key role in abiotic and biotic stress responses, while its role in development and reproduction is less investigated.Citation2,Citation3 Most studies have been performed using the model plant Arabidopsis and therefore little is known about SUMO conjugation (SUMOylation) in agricultural crops, especially in the economically important grasses. To contribute to our understanding about SUMOylation in grasses, we report here about SUMOs in maize and extent two recent reports about the enzymatic SUMO systemCitation4 and its targets during embryogenesisCitation5 in maize.
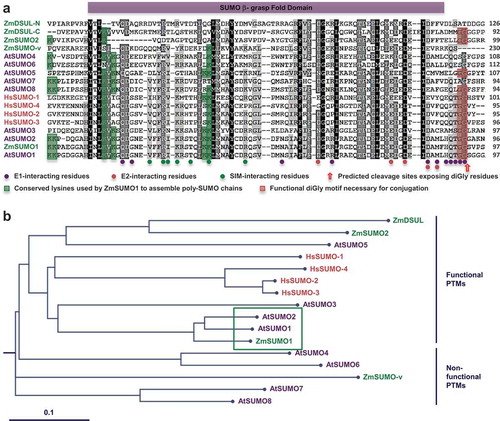
SUMO family modifiers generally contain at least one characteristic β-grasp fold domain that harbors the E1-, E2-, and SIM (SUMO-interacting motif)-interacting residues ()). These amino acid residues perform key molecular functions as they determine the efficiency (SUMO-E1 interactions), pattern (SUMO-E2 interactions) and molecular basis (SUMO-SIM interactions) of SUMO conjugation.Citation6 To visualize the different structural domains, we have aligned all SUMO protein sequences from Arabidopsis (AtSUMO1-8), maize (ZmSUMO1 and ZmSUMO2, the non-functional variant ZmSUMO-v and the diSUMO-like protein ZmDSUL) and human (HsSUMO1-4). Remarkably, AtSUMO1, AtSUMO2 and ZmSUMO1 contain identical amino acid residues at above described positions being involved in non-covalent interactions with E1, E2 and SIM indicating conserved functions and targets. Phylogenetic analysis confirmed this observation showing that the three proteins cluster together ()). Notably, ZmSUMO1 was previously reported to use nearly all accessible Lys residues (Lys-9, Lys-10, Lys-21, Lys-23, Lys-41 and Lys-42) to assemble poly-SUMO chains in the E. coli system.Citation4 Conservation of the non-covalent interaction sites and Lys residues is reduced in other SUMO isoforms and nearly absent in ZmDSUL, suggesting diversification of SUMO-/DSUL-conjugation patterns and confirming the observation that there is almost no overlap between SUMO1 and DSUL targets of maize.Citation5 Phylogenetic and expression analyses further show that AtSUMO4, AtSUMO6-8 (transcriptionally inactive) and ZmSUMO-v (lacking the diGly motif necessary for conjugation) belong to the so-called non-functional PTMs ()).Citation4 Functional PTMs are generated by transcriptionally active members of the SUMO family with obvious C-terminal diGly motifs present in AtSUMO1, AtSUMO2 and ZmSUMO1 that form their own clade, which is related to all human SUMOs. These likely represent the highly conserved and predominantly nuclear-localized isoforms in plants and they share largely overlapping substrates and similar subcellular localization patterns.Citation5,Citation7 Moreover, SUMOylation by AtSUMO1/2, OsSUMO1/2, ZmSUMO1a/b as well as GmSUMO1/2 are rapidly induced by stressCitation4,Citation7–Citation9 suggesting that these likely represent the core SUMOs in plants that are very likely regulated by similar mechanisms and perform analogous (nuclear) functions. AtSUMO5 forms an own clade together with maize ZmSUMO2 and ZmDSUL. It will now be interesting to elucidate whether ZmSUMO2 fulfills ZmDSUL functions outside female gametophyte and early embryo development, which are the only tissues expressing ZmDSUL,Citation10 and to which extent AtSUMO5 and ZmSUMO2 as well as ZmDSUL targets overlap.
Figure 1. Alignment and phylogenetic relationship of SUMO proteins from maize (Zm), Arabidopsis (At) and human (Hs). (a) Sequence alignment showing all SUMO proteins and variants from the three species. Conserved and similar amino acids are highlighted in black and grey, respectively. DiSUMO-LIKE (DSUL) from maize was split into an N-terminal and a C-terminal SUMO domain for alignment. The SUMO β-grasp fold domain is indicated by a purple box. E1-, E2-, and SIM-interacting residues are indicated by purple, red and green dots, respectively. Conserved lysines used by ZmSUMO1 to assemble poly-SUMO chains are highlighted in green. Predicted cleavage sites (red arrow) exposing a C-terminal diGly motif (GG highlighted in red) essential for conjugation are indicated. (b) Phylogenetic analysis of SUMO proteins reveals a functional PTM group and a non-functional PTM group. Protein sequences were aligned by Clustal Omega and the rooted tree was drawn by TreeView. Branch lengths are proportional to phylogenetic distances and the scale bar represents 10% substitutions per site. The highly conserved core clade formed by ZmSUMO1, AtSUMO1 and AtSUMO2 is boxed in green.
To characterize maize core SUMO1 in more detail, we performed immunofluorescence microscopy to investigate its subcellular localization in different cell types. Triple-staining of maize root tips was conducted as described previously:Citation5 a ZmSUMO1-specific antibody (@SUMO1) was used in combination with a microtubule-specific antibody (@β-Tubulin) and the DNA-marker DAPI (4′,6-diamidino-2-phenylindole). Notably, strong ZmSUMO1 signals were detected in all nuclei of fast dividing cells in the root meristem, while a significant decrease in signal intensity was detected in nuclei of outer cell layers and signals were almost lacking in root cap cells ()). Remarkably, ZmSUMO1 was not detectable in the quiescent center. Similarly, cross sections of the root maturation zone showed stronger ZmSUMO1 signals at the stele compared to the cortex, which represents a highly differentiated storage tissue ()). We thus conclude that core SUMOylation is required in small dividing cells, while differentiated cells show less SUMO conjugation. High resolution imaging of nuclei showed that ZmSUMO1 is almost excluded from heterochromatic domains of the nucleus and appears in many small euchromatic foci ()), which might represent active sites of transcription. This confirms our previous work showing that most SUMO1 substrates are involved in chromatin modification and transcriptional regulation.Citation5 It should be noted that studies in mammals and other species revealed that in many cases SUMOylation is also linked to heterochromatin and gene inactivation.Citation11 However, ChIP experiments on a small scale in yeast showed that SUMO is associated to promoters of active, but not of repressed genes.Citation12
Figure 2. ZmSUMO1 localizes to euchromatin and small cytoplasmic granules in small dividing cells. Triple-staining of root tips from wild-type maize plants (inbred line B73), using anti-SUMO1, anti-β-Tubulin antibodies and DAPI. ZmSUMO1 signals are shown in green, the tubulin network in red and nucleus in blue. (a) Longitudinal section of a root tip and (b) cross section of the root maturation zone. The quiescent center is indicated by a dashed circle in (a). (c) Single CLSM section of an interphase nucleus showing ZmSUMO1 localization mainly at euchromatic regions. (d) Overview of an interphase cell showing that ZmSUMO1 accumulates also in small cytoplasmic granules. Heterochromatic domains and ZmSUMO1 granules are indicated by open arrowheads and arrowheads, respectively. VCL, vascular cell lineage; OCL, outer cell layers; RC, root cap; ST, stele; CO, cortex; NU, nucleus; CY, cytoplasm. Scale bars represent 200 μm in (a) and (b) and 10 μm in (c) and (d).
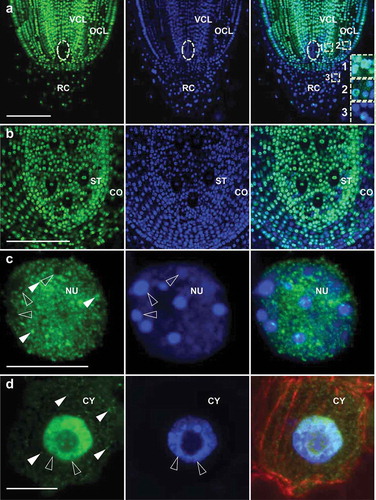
SUMO1 also localizes to small cytoplasmic granules in interphase cells ()). Studies in animals indicates that besides nuclear functions, SUMO also plays extranuclear roles in regulating, for example, channel activity, receptor function, G-protein signaling, enzyme activity, cytoskeletal organization, exocytosis, autophagy and mitochondrial dynamics.Citation13 The observation of SUMO1 signals in maize cytoplasmic granules suggests that SUMOylation of cytoplasmic and membrane associated proteins may be conserved, though SUMO1 signals were relatively low and often difficult to detect in the cytoplasm. It is thus also possible that SUMO2 and DSUL of maize possess predominant cytoplasmic functions compared with SUMO1 localizing mainly in the nucleus.
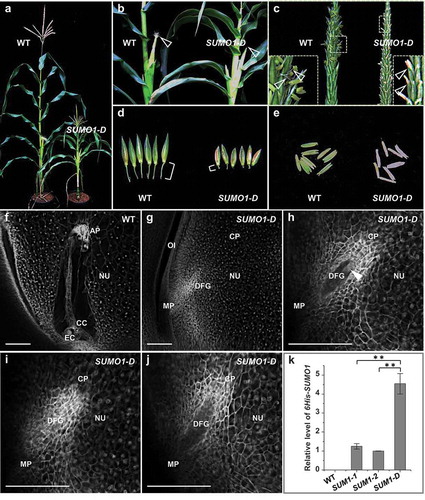
To further deepen our understanding about the importance of SUMOylation in maize, we have generated transgenic plants expressing 6His-tag-fused SUMO1 from the ubiquitin promoter, which had been shown previously to be constitutively active in vegetative as well as reproductive tissues of maize.Citation10 Whereas most 6His-SUMO1 plants were not distinguishable from wild-type (WT) plants grown under standard greenhouse conditions, 8.3% transgenic plants displayed a severe dwarf phenotype ()). This phenotype was strongly correlated with 6His-SUMO1 transcript amount ()). In contrast to WT and phenotypically normal 6His-SUMO1 plants, silks of dwarf plants (SUMO1-D) never emerged from ear hull leaves ()). In addition, male florets were significantly shorter compared to the WT and anthers did not fully protrude from spikelets (a pair of two male florets) due to insufficient filament length ()). The pedicel (stem of the spikelet) of SUMO1-D male flowers was significantly shorter ()). Although anthers of SUMO1-D plants exhibited a similar size compared to WT anthers, they failed to develop to maturity and instead became brown and shriveled, and failed to shed pollens for fertilization ()). Confocal microscopy of SUMO1-D mature ovules showed severely affected ovule development including smaller nucellus cells and a degenerated female gametophyte (–)). Quantitative RT-PCR showed that the abnormal phenotypes observed in SUMO1-D plants were strongly associated with the degree of up-regulated SUMO1 transcript level ()). A slight increase in SUMO1 level as observed, for example in SUM1-1 and SUM1-2 plants, did not affect vegetative and reproductive maize development. However, a stronger increase (>4 times compared with SUM1 plants) in SUMO1 transcript probably severely interferes with fine-tuned SUMO1 conjugation and thus significantly affects development and reproduction. Similar effects have been reported for SUMO null-mutants in Arabidopsis.Citation5 Future studies should now aim to elucidate the individual effect of SUMOylation on the various target proteins localized in the cytoplasm and/or nucleus to understand the deleterious effects of SUMO1 overexpression. Although this is challenging in a crop system like maize, and the effects are likely modified under changing environmental conditions and during stress, interesting target proteins have been identified already in maize and will serve as a valuable start to further our understanding about SUMOylation in grasses and in plants in general.
Figure 3. Phenotypes of SUMO1 overexpression plants. (a) A subset of SUMO1 over-expression plants showed a severe dwarf phenotype. (b) Silks of dwarf plants (SUMO1-D) never emerged from ear hull leaves. (c) SUMO1-D male florets and filaments are significantly shorter compared to WT. (d) Pedicels of SUMO1-D male flowers were significantly shorter. (e) Anthers of SUMO1-D plants became brown and shriveled during development. (f and g) Longitudinal optical sections of ovules dissected from WT (f) and SUMO1-D plants (g). Single CLSM sections are shown. (h–j) Single focus planes of SUMO1-D ovules showing degenerated female gametophytes. (k) Quantitative RT-qPCR of 6His-SUMO1 expression in WT and leaves of 6His-SUMO1 plants. Values are mean values ± SD. Two asterisks (**) represent P < 0.01. EC, egg cell; CC, central cell; AP, antipodal cells; NU, nucellus; OI, outer integument; MP, micropylar pole; DFG, degenerated female gametophyte; CP, chalazal pole. Scale bars represent 50 μm.
Acknowledgments
We are grateful to Ursula Wittmann and Armin Hildebrand for plant care and thank Ulrich Z. Hammes for advice on SUMO1 immunolocalization. Funding through the Collaborate Research Center SFB960 and the DFG Priority Program SPP1365/2 (grant DR334/10-1) to T.D. is gratefully acknowledged.
Additional information
Funding
References
- Vu LD, Gevaert K, De Smet I. Protein language: post-translational modifications talking to each other. Trends Plant Sci. 2018;23:1068–1080. doi:10.1016/j.tplants.2018.09.004.
- Augustine RC, Vierstra RD. SUMOylation: re-wiring the plant nucleus during stress and development. Curr Opin Plant Biol. 2018;45:143–154. doi:10.1016/j.pbi.2018.06.006.
- Elrouby N. Regulation of plant cellular and organismal development by SUMO. Adv Exp Med Biol. 2017;963:227–247. doi:10.1007/978-3-319-50044-7_14.
- Augustine RC, York SL, Rytz TC, Vierstra RD. Defining the SUMO system in maize: SUMOylation is up-regulated during endosperm development and rapidly induced by stress. Plant Physiol. 2016;171:2191–2210. doi:10.1104/pp.16.00353.
- Chen J, Kalinowska K, Muller B, Mergner J, Deutzmann R, Schwechheimer C, Hammes UZ, Dresselhaus T. DiSUMO-LIKE interacts with RNA-binding proteins and affects cell-cycle progression during maize embryogenesis. Curr Biol. 2018;28:1548–60 e5. doi:10.1016/j.cub.2018.03.066.
- Castano-Miquel L, Segui J, Lois LM. Distinctive properties of Arabidopsis SUMO paralogues support the in vivo predominant role of AtSUMO1/2 isoforms. Biochem J. 2011;436:581–590. doi:10.1042/BJ20101446.
- Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc Natl Acad Sci U S A. 2010;107:16512–16517. doi:10.1073/pnas.1004181107.
- Park HC, Kim H, Koo SC, Park HJ, Cheong MS, Hong H, Baek D, Chung WS, Kim DH, Bressan RA. Functional characterization of the SIZ/PIAS-type SUMO E3 ligases, OsSIZ1 and OsSIZ2 in rice. Plant Cell Environ. 2010;33:1923–1934. doi:10.1111/j.1365-3040.2010.02195.x.
- Li Y, Wang G, Xu Z, Li J, Sun M, Guo J, Ji W. Organization and regulation of soybean SUMOylation system under abiotic stress conditions. Front Plant Sci. 2017;8:1458. doi:10.3389/fpls.2017.01458.
- Srilunchang KO, Krohn NG, Dresselhaus T. DiSUMO-like DSUL is required for nuclei positioning, cell specification and viability during female gametophyte maturation in maize. Development. 2010;137:333–345. doi:10.1242/dev.035964.
- Cubenas-Potts C, Matunis MJ. SUMO: a multifaceted modifier of chromatin structure and function. Dev Cell. 2013;24:1–12. doi:10.1016/j.devcel.2012.11.020.
- Rosonina E, Duncan SM, Manley JL. SUMO functions in constitutive transcription and during activation of inducible genes in yeast. Genes Dev. 2010;24:1242–1252. doi:10.1101/gad.1917910.
- Wasik U, Filipek A. Non-nuclear function of sumoylated proteins. Biochim Biophys Acta. 2014;1843:2878–2885. doi:10.1016/j.bbamcr.2014.07.018.
