ABSTRACT
Autophagy, which is one of the self-degradation systems, promotes intracellular zinc (Zn) recycling under Zn deficiency (−Zn) in plants. Therefore, autophagy defective plants show severe chlorosis under −Zn. Root is the plant organ which directly exposed to Zn deficient environment, however, in our recent study, −Zn symptom was prominently observed in leaves as chlorosis. Here, we conducted micrografting to determine which organ’s autophagic activities are important to suppress the −Zn induced chlorosis. Grafted plants that have autophagic activities only in roots or leaves were grown under −Zn and then compared chlorotic phenotypes among them. As a result, regardless of the autophagic activities in rootstocks, −Zn induced-chlorosis in leaves was occurred only when autophagy in scion was defective. This data indicates that Zn resupplied by autophagic degradation in root cells could not contribute to suppress the chlorosis in leaves. Thus, autophagy in the aerial part is critical for controlling −Zn induced-chlorosis in leaves. Taken together, along with our recently reported data, we conclude that the mechanism of Zn resupply by autophagic degradation is not systemic throughout the plant but rather a local system.
Plants cannot move from sprouted ground, therefore, nutrient deficiencies in the soil environment are major factors for inhibition of growth. Plants have various countermeasures to adapt to nutrient deficiencies, one of which is the recycling of nutrient elements via autophagy. Autophagy is one of the major intracellular degradation mechanisms in eukaryote. During plant autophagy, intracellular components such as proteins and organelles are encapsulated in an autophagosome. The outer membrane of the autophagosome fuses with the tonoplast, and the inner membrane and its contents are released into the vacuolar lumen for degradation by vacuolar hydrolases, and then recycled as nutrients.Citation1 Autophagic processes are driven by autophagy-related (ATG) proteins.Citation2,Citation3 In Arabidopsis thaliana (Arabidopsis), knockout mutants of ATG genes were identified and the mutants have been proved to be defective in autophagy.Citation4 These plants are called “atg mutants”.
Recently, we reported that autophagy plays an important role in zinc (Zn) recycling under Zn deficiency (−Zn).Citation5 Under −Zn, autophagy is induced and degrades various intracellular structures to recover mobile Zn2+ ions and thus resupply to required places. Since atg mutants cannot resupply Zn, they show severe Zn deficiency symptoms represented by chlorosis in leaves. Interestingly, −Zn induced-chlorosis in the atg mutants was suppressed by limiting the iron (Fe) concentration in −Zn media.Citation5 These results indicated that the increased Zn deficiency levels due to defective of autophagy changes the balance of the Fe-mediated reactive oxygen species (ROS) production in chloroplasts, leading to the induction of chlorosis.Citation5 Zn2+ ions are absorbed as an essential nutrition from soils. Therefore, roots are the organ which directly exposed to −Zn environments. However, the −Zn symptoms are remarkably observed in leaves as chlorosis. Although autophagy was found to play an important role in suppressing −Zn induced-chlorosis, it is still unclear which plant organ’s autophagic activities are critical for suppression of the chlorosis.
One of the functions of plant autophagy is suppression of salicylic acid (SA) signaling. We previously reported that atg mutants to accumulate higher amounts of SA than wild-type, resulting accelerated natural senescence.Citation6 Overexpression of NahG gene from Pseudomonas putida, which encodes SA hydroxylase, reduces endogenous SA levels in Arabidopsis atg5 mutant and then suppresses the early senescence phenotype.Citation6,Citation7 Under −Zn, NahG atg5 plants showed more severe chlorosis than NahG plants. This result indicates that excessive SA signaling of the atg mutant is not a major factor of the −Zn induced-chlorosis.Citation5 In this experiment, we used NahG as a control plant which has normal autophagic activities and NahG atg5 as an autophagy defective plant to avoid secondary effects of excessive SA in senescence phenotypes. The use of NahG plants could clearly distinguish chlorosis by natural senescence from −Zn symptoms.
In this study, we performed grafting. Grafting is an experimental technique in which plants from different individuals are artificially adhered to each other by cut surfaces to generate one individual plant. In Arabidopsis, micrografting technology using seedlings has been established.Citation8 To generate NahG and NahG atg5 grafted plants, we used a micrografting device.Citation9 Surfaces sterilized and vernalized NahG and NahG atg5 seeds were sown on the device, grown for 2 days in the dark, then moved to a light place, and further grown for 2 days. The hypocotyl was cut with a precision knife, then the aerial part of the plant was removed, and the aerial part collected from another plant (scion) was fitted into the device and brought into close contact to rootstock (, lower parts). After grafting, plants were incubated for 6 days at 27°C under low light intensity to promote bonding. Then, plants were incubated on a plate another 4 days.Citation9 Successful-grafted plants were set to hydroponic system, and grown for 7 days under normal Zn (+Zn, 1 μM ZnSO4) media, and then transplanted to −Zn (0 μM ZnSO4) media.Citation5
Figure 1. Zn starvation induced-chlorosis of NahG and NahG atg5 grafted plants. An overview of the micrograftingis shown in the lower part. Hypocotyl of NahG (showed in black shape) and NahG atg5 (showed in gray shape) seedlings were cut to separate scions and rootstocks. The scions and rootstocks were grafted in all 4 combinations. Upper part showed phenotypes of leaves of each plant at 7 days after transplant to −Zn media. NahG/NahG (scion/rootstock) and NahG/NahG atg5 plants did not show chlorosis, on the other hand, NahG atg5/NahG atg5 and NahG atg5/NahG plants showed remarkable Zn starvation induced-chlorosis. Scale bar indicates 2 cm.
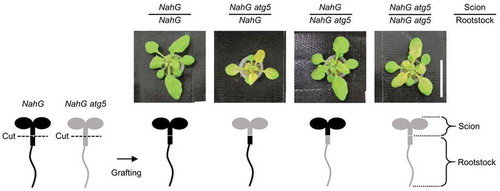
Phenotypes of each grafted plant at 7 days after transplant to −Zn were shown in upper parts of . Expectedly, when both scion and rootstock were NahG (NahG/NahG), −Zn induced-chlorosis was not observed. Contrary, when both scion and rootstock were NahG atg5 (NahG atg5/NahG atg5), significant chlorosis was appeared, again as expected. Those data are suggesting that the manipulation of seedlings by the grafting does not affect −Zn induced-chlorosis. Most important point is that, in NahG scion and NahG atg5 rootstock plant (NahG/NahG atg5), −Zn induced-chlorosis was not observed similar to the NahG/NahG control plant. On the other hand, NahG atg5 scion and NahG rootstock plant (NahG atg5/NahG) showed remarkable chlorosis similar to the NahG atg5/NahG atg5 plant, the negative control plant (). These results indicate that −Zn induced-chlorosis correlated with defect of autophagy is prescribed by autophagic activity in scion regardless of rootstock. The fact that the autophagy in scion determines the leaf phenotypes is same as the case of natural senescence (Supplementary Figure S1).
Our current data clearly indicated that autophagy in the rootstock did not recover the −Zn symptoms in leaves. This suggests that tolerance to −Zn via autophagy is not the systemic but relatively local system. We recently reported that autophagy is induced by −Zn in both roots and leaves.Citation5 Given that −Zn response via autophagy is local, a model of −Zn tolerance via autophagic local Zn recycling is considered as shown in . In leaves, autophagy is induced according to the levels of Zn starvation to recover Zn2+ ions from intracellular constituents. Suppression of the −Zn state in leaves by this feedback mechanism leads to reduce −Zn induced-chlorosis (). Since Zn deficiency induced-chlorosis is caused by Fe-mediated ROS production in chloroplasts.Citation5 Resupplied Zn by autophagy in leaves may contributes to suppress this ROS production.
Figure 2. Mechanism of Zn deficiency tolerance via autophagic local Zn recycling. In leaves, Zn starvation induces autophagy which degrades intracellular structures and resupplies mobile Zn2+ ions. Zn deficiency promotes Fe uptake/translocation and induces chlorosis by light– and Fe-mediated ROS production. Autophagy in leaves recovers Zn2+ ions from intracellular constituents and thus alleviates the Zn starvation levels, suppressing this ROS production process. Similarly, in roots, there is the Zn homeostasis controlling cycle via autophagy, however, Zn2+ ions resupplied by root autophagy is not transported to leaves and therefore Fe uptake/translocation is not suppressed. Zn2+ ions resupplied in roots are probably used only in the roots, which suppresses root growth reduction under Zn starvation.
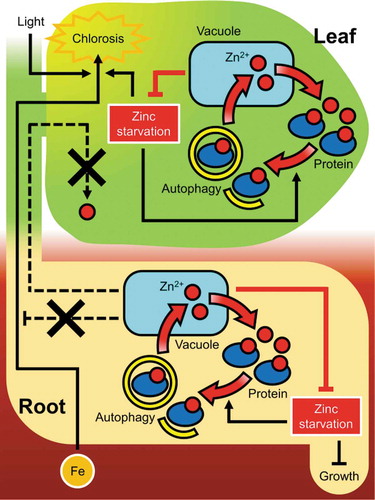
In a split-root assay using +Zn and −Zn separate plate we performed recently, autophagic activities were not increased on the +Zn side, but were increased in the roots of −Zn side.Citation5 This data indicate that autophagy in root under −Zn is not systemic and Zn is not resupplied to the Zn deficient site from the Zn sufficient site. The phenotype of NahG atg5/NahG plant in also suggests that resupplied Zn via autophagy in root is unlikely to be remobilized to leaves and cannot suppress Fe uptake/translocation under −Zn (). Given that root elongation of atg mutants was suppressed compared to wild-type on −Zn media.Citation5 The resupplied Zn via root autophagy seems to be locally used in the root to alleviate the inhibition of root growth by Zn starvation (). There may be a feedback loop mechanism in roots that senses the levels of Zn deficiency and regulates the activity of autophagy independent from leaves (). In summary, the adaptation system against Zn starvation via autophagy probably functions independently in roots and leaves to maintain Zn homeostasis in each organ to avoid each Zn deficiency symptom, respectively ().
Materials and methods
Details of NahG and NahG atg5 plants were described in our report.Citation5 Micrografting was performed according to the method described in our preprint in bioRxiv.Citation9 After grafting, the roots of 14-day-old-grafted plants were sandwiched by rockwool and set it in a hydroponic system with +Zn molecular genetics research laboratory (MGRL) media.Citation5,Citation10 After growing for 7 days in +Zn MGRL media, plants were transplanted to −Zn MGRL media and grown for another 7 days, and then observed.Citation5
Abbreviations
| ATG | = | autophagy-related |
| MGRL | = | molecular genetics research laboratory |
| NahG | = | bacterial salicylate hydroxylase |
| ROS | = | reactive oxygen species |
| SA | = | salicylic acid |
| −Zn | = | zinc deficiency |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental Material
Download MS Word (379.7 KB)Supplemental material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Chen Q, Shinozaki D, Luo J, Pottier M, Havé M, Marmagne A, Reisdorf-Cren M, Chardon F, Thomine S, Yoshimoto K, et al. Autophagy and nutrients management in plants. Cells. 2019;8(11):1. PMID:31726766. doi:10.3390/cells8111426.
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27(1):107–4. PMID:21801009. doi:10.1146/annurev-cellbio-092910-154005.
- Yoshimoto K, Ohsumi Y. Unveiling the molecular mechanisms of plant autophagy – from autophagosomes to vacuoles in plants. Plant Cell Physiol. 2018;59(7):1337–1344. PMID:29893925. doi:10.1093/pcp/pcy112.
- Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002;129(3):1181–1193. PMID:12114572. doi:10.1104/pp.011024.
- Shinozaki D, Merkulova EA, Naya L, Horie T, Kanno Y, Seo M, Ohsumi Y, Masclaux Daubresse C, Yoshimoto K. Autophagy increases zinc bioavailability to avoid light-mediated reactive oxygen species production under zinc deficiency. Plant Physiol. 2020;182(3):1284–1296. PMID:31941669. doi:10.1104/pp.19.01522.
- Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell. 2009;21(9):2914–2927. PMID:19773385. doi:10.1105/tpc.109.068635.
- Delaney TP, Uknes S, Vemooij B, Friedrich L, Weymann K, Nerrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, et al. A central role of salicylic acid in plant disease resistance. Science. 1994;266(5188):1247–1250. PMID:17810266. doi:10.1126/science.266.5188.1247.
- Turnbull CGN, Booker JP, Leyser HMO. Micrografting techniques for testing long-distance signaling in Arabidopsis. Plant J. 2002;32:255–262. PMID:12383090. doi:10.1046/j.1365-313x.2002.01419.x.
- Tsutsui H, Yanagisawa N, Kawakatsu Y, Ikematsu S, Sawai Y, Tabata R, Arata H, Higashiyama T, Notaguchi M. Micrografting device for testing environmental conditions for grafting and systemic signaling in Arabidopsis. bioRxiv. 2019. doi:10.1101/2019.12.20.885525.
- Fujiwara T, Hirai MY, Chino M, Komeda Y, Naito S. Effects of sulfur nutrition on expression of the soybean seed storage protein genes in transgenic petunia. Plant Physiol. 1992;99(1):263–268. PMID:16668860. doi:10.1104/pp.99.1.263.
