ABSTRACT
Cell division cycle and apoptosis regulator 1 (CCAR1) is a deleted in breast cancer domain containing protein and their molecular roles in the animal system are well characterized. So far study on CCAR1 protein has not available in the plant system. The domain-based phylogenetic study clearly indicates that the CCAR1 protein has widely distributed throughout the plant kingdom. Intron-less CCAR1 gene encoded protein comprises five conserved domain and huge intrinsic disorder. Functional annotation of cis-regulatory elements (CREs) reveales that a wide range of potential transcription factor binding sites (TFBSs) are present in CCAR1 gene promoter. Besides that potential miRNA targets those control post-transcriptional regulations are also identified in the present study. Furthermore, gene ontogeny (GO) analysis revealed that CCAR1 have dynamic role in a wide number of cellular and metabolic processes. In this study, we first report the regulation, functional and structural property of CCAR1 transcript and protein which will help to assist crop improvement by manipulating CCAR1.
Data-mining bioinformatics and CCAR1
In animal system, the significant role of CCAR1 in programmed cell death and other cellular processes have been reported.Citation1 In plant system, we have first emphasized the characterization, regulation and function of CCAR1 in plant using various data-mining bioinformatics. Using the NCBI protein database we harvest CCAR1 protein sequences (homologs to Morus notabilis CCAR1 amino acid sequence, NCBI accession number: EXC20006.1) and construct a phylogenetic tree ().Citation2,Citation3 The CCAR1 proteins have grouped into 10 clades, from which none of them are characterized previously. In brief, clade I family members have comprised Fagaceae, Moraceae, and other putative CCAR1 proteins. Clade 2 consisted Vitaceae, Rutaceae, and other CCAR1 putative like proteins. Clade 5 comprises Fabaceae putative CCAR1 proteins. Clade 6 comprises Cleomaceae, Brassicaceae putative CCAR1 proteins. Clade 8 comprises only Amaranthaceae plant family CCAR1 proteins.
Figure 1. (a) iTOL server has been used to construct the phylogenetic tree of CCAR1 protein sequences. (b) Identification of conserved motifs of CCAR1 protein family. (c) The positions of the motifs are denoted by P1, P2, P3, P4, and P5. Conserved domain structure of M. notabilis CCAR1 protein along with predicted fold/unfold property. Despite Nudix/DBC1 homology domain, MAEBL and calmodulin (CaM) domains are also found in C-terminal of CCAR1 protein. Deleted in breast cancer domain and it homologs from diverse eukaryotes are a catalytically inactive version of the Nudix hydrolase (MutT) domain. It is predicted to bind NAD metabolites and regulate the activity of SIRT1 or related deacetylases by sensing the soluble products or substrates of the NAD-dependent deacetylation reaction.
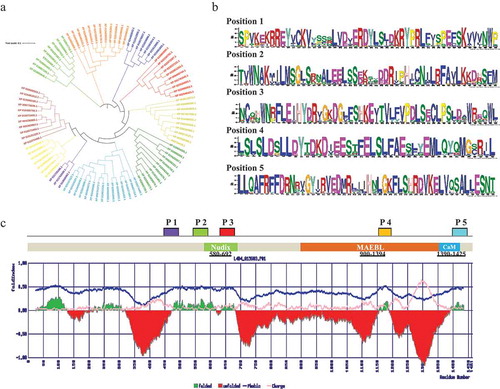
MeMe Suite toolCitation4 is implemented to predict the conserved motif CCAR1 family proteins. Analyzed data indicate five conserved sites are present across the protein sequence length (). Out of five motifs, three are located around the Nudix domain and the remaining two are situated at the C-terminal site of CCAR1 protein. Besides that, to understand the folding nature or disorder state of the CCAR1 protein, here we have predicted the folding nature using the FoldIndex© online tool.Citation5 The findings of this analysis clearly elucidated that M. notabilis CCAR1 protein has most of the regions are intrinsically disorder/unfold. Intrinsically disordered regions have a significant contribution to a wide range of cellular processes via molecular interaction.Citation6 Thus, the CCAR1 protein may have a potential fundamental role in the plant system.
To understand the transcriptional regulation and promoter structure we have harvested 1 kb upstream promoter sequences from the NCBI database. PlantRegMap databaseCitation7 has been used to characterize the CCAR1 gene promoter. Cis-regulatory elements (CREs) annotation reveales several potential transcription factors such as AP2, C2H2, CAMTA, Dof, GRAS, ERF, EIL, HD-ZIP, HSF, MIKC_MADS and MYB binding sites (see the supplementary file table 1). Those transcription factors are controlled by various environmental factors (light, salt, heat, drought, wounding), developmental factors (flowering, gravitropism), and cellular metabolites (H2O2, auxin, ethylene, gibberellin, cytokinin, salicylic acid, jasmonic acid, and cadmium ion, etc.).
Furthermore, all putative microRNAs that target to CCAR1 transcript of M. notabilis is identified using psRNATarget serverCitation8 (expectation value >3.5). Identified putative miRNAs are gra-miR8737, gma-miR9752, aly-miR156 h-3p, ath-miR836, bdi-miR7735-5p, cca-miR6108e-3p, gma-miR5041-5p, gma-miR9752, gra-miR3267, rgl-miR5577, rgl-miR7800, stu-miR5303e, stu-miR5303 f, and stu-miR8022 that regulates post-transcription of the CCAR1 by translation and cleavage processes (see the supplementary file table 2). Thus, both CREs and miRNA have potential impacts on regulation at the transcription level.
Gene ontogeny (GO) data have been collected from MorusDB databaseCitation9 to understand the biological and molecular function of the CCAR1 protein. Analyzed data revealed that CCAR1 involves in several developments and stress-related fundamental processes such as programmed cell death, single-organism process, single-organism cellular process, gene expression, heterocycle metabolic process, nitrogen compound metabolic process, cellular macromolecule metabolic process, organic cyclic compound metabolic process, metabolic process, RNA metabolic process, cellular nitrogen compound metabolic process, primary metabolic process, cellular aromatic compound metabolic process, nucleic acid metabolic process, RNA processing, regulation of biological process. Hence, CCAR1 plays a crucial role in cellular, molecular, and biological processes according to in silico GO analysis.
In the present study, we have emphasized on the phylogenetic relationship, transcriptional regulation, structural integrity, and functional aspects of M. notabilis CCAR1 protein by harvesting good-quality data from publically available databases. Moreover, present in silico investigation has been briefly represented the novelty of CCAR1 protein, which will provide a basic platform for future-advance biotechnological research for crop improvement.
Conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental Material
Download MS Word (25.9 KB)Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Brunquell J, Yuan J, Erwin A, Westerheide SD, Xue B. DBC1/CCAR2 and CCAR1 are largely disordered proteins that have evolved from one common ancestor. Biomed Res Int. 2014;(18)1. doi:10.1155/2014/418458.
- Federhen S. The NCBI taxonomy database. Nucleic Acids Res. 2012;(40)136–3. doi:10.1093/nar/gkr1178.
- Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;(44)242–W245. doi:10.1093/nar/gkw290.
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;(37)202–208. doi:10.1093/nar/gkp335.
- Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex©: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21(16):3435–3438. doi:10.1093/bioinformatics/bti537.
- Hsu WL, Oldfield C, Meng J, Huang F, Xue B, Uversky VN, Romero P, Dunker AK. Intrinsic protein disorder and protein-protein interactions. Biocomputing. 2012:116–127. doi:10.1142/9789814366496_0012.
- Tian F, Yang DC, Meng YQ, Jin J, Gao G. PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 2020;48(D1):1104–1113. doi:10.1093/nar/gkz1020.
- Dai X, Zhuang Z, Zhao PX. psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018;46(W1):49–54. doi:10.1093/nar/gkr319.
- Li T, Qi X, Zeng Q, Xiang Z, He N. MorusDB: a resource for mulberry genomics and genome biology. Database. 2014:bau054. doi:10.1093/database/bau054.
