ABSTRACT
CAPRICE (CPC), an R3-type MYB transcription factor, is known to promote root hair differentiation in the root epidermis of Arabidopsis. CPC moves from non-hair cells to adjacent hair-forming cells. In contrast, we have previously shown that there is no movement of the CPC homologs, ENHANCER OF TRY AND CPC 1, 2, and 3 (ETC1, 2, and 3), and TRYPTICHON (TRY) between root epidermal cells. We also identified a tomato homolog of CPC, named Solanum lycopersicumTRYPTICHON (SlTRY).SlTRY-introduced transgenic Arabidopsis produced many root hairs, like CPC-introduced transgenic Arabidopsis. To clarify the cell-to-cell movement ability of the SlTRY protein, in this study, we observed the distribution of GFP fluorescence in CPCp:SlTRY:GFP transgenic Arabidopsis. Unexpectedly, SlTRY moved from non-hair cells to adjacent root hair cells, like CPC, in Arabidopsis root epidermis. SlTRY does not have the cell-to-cell movement sequence (S1) defined in CPC, and the mechanism of movement is still unknown. Further investigation is necessary to elucidate the mechanism of cell-to-cell movement of SlTRY. Our results will help in the further unraveling of the functions of these MYB transcription factors in determining cell fate.
KEYWORDS:
CAPRICE (CPC) was identified as a gene that induces the root hairs of Arabidopsis thaliana.Citation1 CPC encodes an R3-type MYB transcription factor.Citation1 Homologous genes in Arabidopsis with sequences similar to CPC include TRYPTICHON (TRY), ENHANCER OF TRY AND CPC 1, 2, and 3 (ETC1, ETC2, and ETC3), and TRICHOMELESS 1 and 2 (TCL1 and TCL2).Citation2–Citation10 These six CPC homologs induce root hairs similar to CPC in Arabidopsis. Arabidopsis roothair cells are over two underlying cortical cells, whereas non-hair cells are over a single cortical cell.Citation11,Citation12 This position-determined developmental manner results in lines of roothair cells along the longitudinal root axis and has been found in Brassicaceae and other eudicot families.Citation13–Citation16 This stripy root hair pattern (type 3) is one of the three types of root hair cell distribution patterns.Citation13–Citation15,Citation17–Citation20 Previously, we reported that CPC is expressed in non-hair cells of the root epidermis, and the translated CPC protein then migrates from non-hair cells to hair-forming cells in Arabidopsis.Citation21 This ability of CPC may determine the type 3 root hair pattern. The cell-to-cell movement of transcription factors, through plasmodesmata, is important for plant cell differentiation.Citation22,Citation23 On the contrary, four CPC-like MYB proteins, including TRY, ETC1, ETC2, and ETC3, do not seem to move from non-hair cells to root hair cells.Citation24–Citation26 Although TRY has been shown to move between leaf epidermal cells,Citation27 the cell-to-cell movement ability is likely to be specific to CPC among R3-type MYB transcription factors in Arabidopsis.
Previously, we identified tomato (Solanum lycopersicum) homolog of Arabidopsis CPC and named it Solanum lycopersicumTRYPTICHON (SlTRY).Citation28 SlTRYwas so named because its encoded protein is more closely related to TRY than to CPC.Citation28 Tomato shows a root hair pattern different from Arabidopsis (type 3). Tomato belongs to the most prevalent type 1 root hair pattern group in which all root epidermal cells can generate root hairs.Citation29 SlTRY-introduced transgenic Arabidopsis plants produce many root hairs, a phenotype similar to CPC or CPC-like MYB-introduced transgenic Arabidopsis plants.Citation29 In this study, we explored protein localization and cell-to-cell movement of SlTRY in Arabidopsis.
As previously reported, the SlTRY-encoded protein is more closely related to TRY, than CPC or ETC1 ().Citation28 However, the total number of amino acids in SlTRY protein is 94, which is the same as that in CPC (). CPCp:SlTRY:GFP in Col-0 transgenic plants produce about 1.6-times more root hairs than are present in the wild-type Col-0.Citation28 CPCp:SlTRY:GFP in cpc-2 transgenic plants also produce a greater number of root hairs and completely complement the cpc-2 mutant phenotype with decreased number of root hairs.Citation28 We observed the GFP distribution of CPCp:SlTRY:GFP in Col-0 and CPCp:SlTRY:GFP in cpc-2 transgenic Arabidopsis roots ().GFP fluorescence was observed in the nuclei of nearly all cells of the root epidermis of CPCp:SlTRY:GFP in Col-0 transgenic plants (), suggesting that tomato SlTRY is a transcription factor localized in the nucleus. Furthermore, it was revealed that SlTRY moved from non-hair cells, where SlTRY was expressed under the CPC promoter, to root hair cells (). GFP fluorescence was also observed in the nuclei of almost all the root epidermis cells of CPCp:SlTRY:GFP in cpc-2 transgenic plants (). Even in the cpc-2 mutant background, SlTRY showed nuclear localization and cell-to-cell movement.
Figure 1. The amino acid sequence of R3 MYB proteins. Sequence alignment of SlTRY (Solyc01g095640.1.1), CPC (FJ268773), TRY (AC007288), and ETC1 (NM100020). Black shaded letters indicate identical residues. Gray shaded letters pointed by arrows indicate identical residues where only SlTRY and CPC are identical. The R3 MYB domain is indicated by a line above the sequences. The amino acids comprising the S1 and S2 domains are indicated with boxes outlined in gray. CPC, CAPRICE; SlTRY, Solanum lycopersicum TRYPTICHON; TRY, TRYPTICHON.
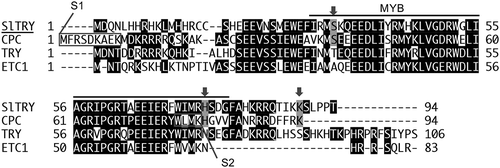
Figure 2. Distribution of GFP fluorescence in CPCp:SlTRY:GFP in Col-0 and CPCp:SlTRY:GFP in cpc-2 transgenic plants. Confocal laser scanning microscopy images showing GFP (green) and PI (red) fluorescence in the root epidermis of 10-d-old seedlings. The CPCp:SlTRY:GFP in Col-0 (a) and CPCp:SlTRY:GFP in cpc-2 (b) transgenic plants were observed. CPC, CAPRICE; SlTRY, Solanum lycopersicum TRYPTICHON.
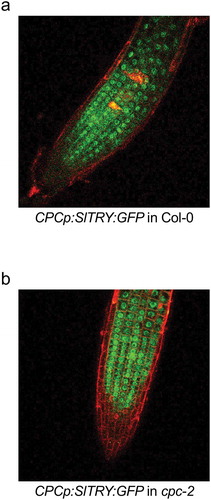
When CPC-like MYBof Arabidopsis (TRY, ETC1, ETC2, and ETC3) was expressed under the CPC promoter, the translated proteins were mainly localized in non-hair cells.Citation25,Citation30-Citation32 In contrast, CPC localized in both non-hair cells and migrated root hair cells, evenly, that is, in all the root epidermal cells.Citation30 Both CPC and CPC-like MYBs induce root hairs, but only CPC shows an obvious movement between root epidermal cells. We confirmed SlTRY:GFP fusion protein nuclear localization in all root epidermal cells for five CPCp:SlTRY:GFP in Col-0 independent homozygous transgenic lines and six CPCp:SlTRY:GFP in cpc-2 independent homozygous transgenic lines. Our results suggest that tomato SlTRY retains cell-to-cell movement ability from non-hair cells to root hair cells.
Kurata et al. identified the S1 (Sequence 1) and S2 regions in CPC, which are active sites for cell-to-cell movement of CPCin Arabidopsis epidermal cells ().Citation33 The conservation of S2 (WxM) in all R3 MYBs () suggests that S2 is necessary but not sufficient for cell-to-cell movement ability in root epidermal cells. Surprisingly, SlTRY did not have S1, as in the case of non-mobile proteins TRY and ETC1 (). Previously, we reported that the addition of S1 sequence to ETC1 does not allow cell-to-cell movement of ETC1.Citation34 Besides S1 and S2, there may be amino acids crucial for cell-to-cell movement. Comparing the migrating group (SlTRY and CPC) with the non-migrating group (TRY and ETC1), S, H, and K (indicated by gray arrows in ) were amino acids specific to the migrating group. These amino acids might be involved in the cell-to-cell movement of SlTRY and CPC.SlTRY and CPC, which belong to the migrating group, both have 94amino acids, so their size may be important for cell-cell movement (). The mechanism of CPCp:SlTRY:GFPin cpc-2 mutant might be more complicated than that of Col-0.The CPC promoter might work in all epidermal cells in the cpc-2 mutant. Eventually, however, SlTRY protein moved from neighboring cells and was localized in all epidermal cells ().
Tomato and Arabidopsis have different root hair patterns, type1 and type 3, respectively. The function of SlTRY in tomato is still unclear. This study revealed that the unique property of cell-to-cell movement, such as in CPC, is maintained in SlTRY. Further investigation is necessary to elucidate the mechanism of cell-to-cell movement of CPC and SlTRY.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Wada T, Tachibana T, Shimura Y, Okada K. Epidermal cell differentiation in arabidopsis determined by a Myb homolog, CPC. Science. 1997;277:1–4. doi:10.1126/science.277.5329.1113.
- Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, Thumfahrt J, Jürgens G, Hülskamp M. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in arabidopsis. Embo J. 2002;21:5036–5046. doi:10.1093/emboj/cdf524.
- Kirik V, Simon M, Huelskamp M, Schiefelbein J. The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in arabidopsis. Dev Biol. 2004;268:506–513. doi:10.1016/j.ydbio.2003.12.037.
- Kirik V, Simon M, Wester K, Schiefelbein J, Hulskamp M. ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of arabidopsis. Plant Mol Biol. 2004;55:389–398. doi:10.1007/s11103-004-0893-8.
- Esch JJ, Chen MA, Hillestad M, Marks MD. Comparison of TRY and the closely related At1g01380 gene in controlling arabidopsis trichome patterning. Plant J. 2004;40:860–869. doi:10.1111/j.1365-313X.2004.02259.x.
- Simon M, Lee MM, Lin Y, Gish L, Schiefelbein J. Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev Biol. 2007;311:566–578. doi:10.1016/j.ydbio.2007.09.001.
- Tominaga R, Iwata M, Sano R, Inoue K, Okada K, Wada T. Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development. 2008;135:1335–1345. doi:10.1242/dev.017947.
- Wang S, Kwak SH, Zeng Q, Ellis BE, Chen XY, Schiefelbein J, Chen JG. TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in arabidopsis. Development. 2007;134:3873–3882. doi:10.1242/dev.009597.
- Gan L, Xia K, Chen JG, Wang S. Functional characterization of TRICHOMELESS2, a new single-repeat R3 MYB transcription factor in the regulation of trichome patterning in arabidopsis. BMC Plant Biol. 2011;11:176. doi:10.1186/1471-2229-11-176.
- Tominaga-Wada R, Nukumizu Y. Expression analysis of an R3-type MYB transcription factor CPC-LIKE MYB4 (TRICHOMELESS2) and CPL4-related transcripts in arabidopsis. Int J Mol Sci. 2012;13:3478–3491. doi:10.3390/ijms13033478.
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. Cellular organisation of the arabidopsis thaliana root. Development. 1993;119:71–84.
- Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein JW. The TTG gene is required to specify epidermal cell fate and cell patterning in the arabidopsis root. Dev Biol. 1994;166:740–754. doi:10.1006/dbio.1994.1352.
- Cormack RGH. A comparative model of developing epidermal cells in white mustard and tomato roots. Am J Bot. 1947;34:310–314. doi:10.1002/j.1537-2197.1947.tb12994.x.
- Clowes FAL. Pattern in root meristem development in angiosperms. New Phytologist. 2000;146:83–94. doi:10.1046/j.1469-8137.2000.00614.x.
- Dolan L, Costa S. Evolution and genetics of root hair stripes in the root epidermis. J Exp Bot. 2001;52:413–417. doi:10.1093/jxb/52.suppl_1.413.
- Kim DW, Lee SH, Choi SB, Won SK, Heo YK, Cho M, Park YI, Cho HT. Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell. 2006;18:2958–2970. doi:10.1105/tpc.106.045229.
- Leavitt RG. Trichomes of the root in vascular cryptograms and angiosperms. Proc Boston Soc Nat Hist. 1904;31:273–313.
- Cormack RGH. The development of root hair by Elodea canadensis. New Phytol. 1937;36:19–25. doi:10.1111/j.1469-8137.1937.tb06900.x.
- Cutter EG, Hung CY. Symmetric and asymmetric mitosis and cytokinesis in the root tip of hydrocharis morsus-ranae L. J Cell Sci. 1972;11:723–737.
- Dolan L. Pattern in the root epidermis: an interplay of diffusible signals and cellular geometry. Ann Bot-London. 1996;77:547–553. doi:10.1093/aob/77.6.547.
- Wada T, Kurata T, Tominaga R, Koshino-Kimura Y, Tachibana T, Goto K, Marks MD, Shimura Y, Okada K. Role of a positive regulator of root hair development, CAPRICE, in arabidopsis root epidermal cell differentiation. Development. 2002;129:5409–5419. doi:10.1242/dev.00111.
- Ruiz-Medrano R, Xoconostle-Cazares B, Kragler F. The plasmodesmatal transport pathway for homoeotic proteins, silencing signals and viruses. Curr Opin Plant Biol. 2004;7:641–650. doi:10.1016/j.pbi.2004.09.012.
- Zambryski P. Cell-to-cell transport of proteins and fluorescent tracers via plasmodesmata during plant development. J Cell Biol. 2004;164:165–168. doi:10.1083/jcb.200310048.
- Tominaga R, Iwata M, Okada K, Wada T. Functional analysis of the epidermal-specific MYB genes CAPRICE and WEREWOLF in Arabidopsis. Plant Cell. 2007;19:2264–2277. doi:10.1105/tpc.106.045732.
- Tominaga-Wada R, Wada T. The ectopic localization of CAPRICE LIKE MYB3 protein in Arabidopsis root epidermis. J Plant Physiol. 2016;199:111–115. doi:10.1016/j.jplph.2016.05.014.
- Tominaga-Wada R, Kurata T, Wada T. Localization of ENHANCER OF TRY AND CPC1 protein in Arabidopsis root epidermis. J Plant Physiol. 2017;214:48–52. doi:10.1016/j.jplph.2017.04.001.
- Digiuni S, Schellmann S, Geier F, Greese B, Pesch M, Wester K, Dartan B, Mach V, Srinivas BP, Timmer J, et al. A competitive complex formation mechanism underlies trichome patterning on Arabidopsis leaves. Mol Syst Biol. 2008;4:217.
- Tominaga-Wada R, Nukumizu Y, Sato S, Wada T. Control of plant trichome and root-hair development by a tomato (Solanum lycopersicum) R3 MYB transcription factor. PLoS One. 2013;8:e54019. doi:10.1371/journal.pone.0054019.
- Pemberton LM, Tsai SL, Lovell PH, Harris PJ. Epidermal patterning in seedlings roots of eudicotyledons. Ann Bot-London. 2001;87:649–654. doi:10.1006/anbo.2001.1385.
- Tominaga-Wada R, Wada T. Extended C termini of CPC-LIKE MYB proteins confer functional diversity in Arabidopsis epidermal cell differentiation. Development. 2017;144:2375–2380. doi:10.1242/dev.149542.
- Tominaga-Wada R, Kurata T, Wada T. Localization of the CAPRICE-ENHANCER OF TRY AND CPC1 chimera protein in arabidopsis root epidermis. Biosci Biotechnol Biochem. 2017;;81:1762–1767.
- Yamada K, Sasabe M, Fujikawa Y, Wada T, Tominaga-Wada R. Amino acid substitutions in CPC-LIKE MYB reveal residues important for protein stability in arabidopsis roots. PLoS One. 2018;13.
- Kurata T, Ishida T, Kawabata-Awai C, Noguchi M, Hattori S, Sano R, Nagasaka R, Tominaga R, Koshino-Kimura Y, Kato T, et al. Cell-to-cell movement of the CAPRICE protein in arabidopsis root epidermal cell differentiation. Development. 2005;132:5387–5398. doi:10.1242/dev.02139.
- Tominaga-Wada R, Wada T. Effect of amino acid substitution of CAPRICE on cell-to-cell movement ability in arabidopsis root epidermis. Dev Biol. 2018;435:1–5. doi:10.1016/j.ydbio.2018.01.002.
