ABSTRACT
The plant-specific TCP transcription factors play pivotal roles in various processes of plant growth and development. However, little is known regarding the functions of TCPs in plant oil biosynthesis. Our recent work showed that TCP4 mediates oil production via interaction with WRINKLED1 (WRI1), an essential transcription factor governing plant fatty acid biosynthesis. Arabidopsis WRI1 (AtWRI1) physically interacts with multiple TCPs, including TCP4, TCP10, and TCP24. Transient co-expression of AtWRI1 with TCP4, but not TCP10 or TCP24, represses oil accumulation in Nicotiana benthamiana leaves. Increased TCP4 in transgenic plants overexpressing a miR319-resistant TCP4 (rTCP4) decreased the expression of AtWRI1 target genes. The tcp4 knockout mutant, the jaw-D mutant with significant reduction of TCP4 expression, and a tcp2 tcp4 tcp10 triple mutant, display increased seed oil contents compared to the wild-type Arabidopsis. The APETALA2 (AP2) transcription factor WRI1 is characterized by regulating fatty acid biosynthesis through cross–family interactions with multiple transcriptional, post-transcriptional, and post-translational regulators. The interacting regulator modules control the range of AtWRI1 transcriptional activity, allowing spatiotemporal modulation of lipid production. Interaction of TCP4 with AtWRI1, which results in a reduction of AtWRI1 activity, represents a newly discovered mechanism that enables the fine-tuning of plant oil biosynthesis.
Introduction
The plant-specific TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR (TCP) transcription factors play pivotal roles in various physiological processes, such as leaf development, cell cycle, plant hormone signaling, defense responses, and the circadian clock.Citation1–3 The 24 Arabidopsis TCPs are divided into class I and class II sub-families.Citation2–5 The Class II TCP2-4, TCP10, and TCP24 are post-transcriptionally controlled by microRNA319 (miR319), and the jaw-D mutant overproducing miR319 significantly reduces the expression of the five Class II TCPs.Citation5 Yeast-two-hybrid (Y2H) assays show that several miRNA319-regulated TCPs are ASYMMETRIC LEAVES 2 (AS2)-interacting proteins, which play roles in repressing class-I KNOX genes.Citation6 TCP3 interacts with R2R3-MYB transcription factors that regulate flavonoid biosynthesis.Citation7 A subset of TCPs are SUPPRESSOR OF rps4-RLD1 (SRFR1)-interacting transcriptional regulators, and the SRFR1-TCP interacting modules equilibrate plant development and immunity.Citation8 Despite numerous known roles of TCPs in plant physiological and biochemical processes, participation of TCPs in plant oil accumulation was not reported.
WRINKLED1 (WRI1; )), a member of APETALA2 (AP2) transcription factor family,Citation9,Citation10 plays an essential role in the transcriptional regulation of plant oil biosynthetic pathways.Citation11–13 Arabidopsis WRI1 (AtWRI1) loss-of-function mutant (wri1-1) displays an approximately 80% reduction in seed oil content compared to wild-type (WT).Citation14 Transcriptomic analysis of developing seeds revealed that a majority of the down-regulated genes in wri1-1 encode fatty acid biosynthetic and glycolytic enzymes.Citation15 Numerous genes encoding enzymes in the late glycolysis and fatty acid biosynthesis are validated as AtWRI1 targets in subsequent studies.Citation16–18 The AW-box, [CnTnG](n)7[CG], in the gene promoters is characterized as the binding sequence of AtWRI1.Citation17 WRI1 orthologs have been discovered in diverse monocot and dicot species, and have shown to be functional in mediating plant oil accumulation.Citation19–27 Ectopic expression of AtWRI1 or WRI1 orthologs leads to elevated oil accumulation in seeds and vegetative tissues of the transgenic plants.Citation9,Citation13,Citation20,Citation23,Citation24,Citation28,Citation29 Transient overexpression of WRI1s in tobacco leaves also stimulates oil accumulation.Citation27,Citation30-33 Recent progress advanced our understanding of the structure/function of WRI1, particularly in the functional domains/motifs, protein structural features, and the interacting regulators (summarized in ). AtWRI1 interacts with CULLIN3-based E3 ligase adaptor BTB/POZMATH (BPM) proteins which mediate 26S proteasomal degradation of AtWRI1.Citation34 In silico analysis discovered that AtWRI1 protein contains three intrinsically disordered regions (IDRs).Citation30 Particularly, the IDR3 comprises a PEST motif that mediates AtWRI1 stability.Citation30 When the IDR3-PEST is removed or the putative phosphorylation residues in IDR3-PEST are mutated, the variants are more stable and capable of inducing a higher oil accumulation in plant cells compared to the native AtWRI1, suggesting a possible regulation of AtWRI1 at the IDR3-PEST motif by phosphorylation.Citation30 AtWRI1 also interacts with 14-3-3 proteins in yeast and plant cells.Citation35 Co-expression of 14-3-3 with AtWRI1 enhances AtWRI1-mediated oil production, likely by increasing the stability and transcriptional activity of AtWRI1.Citation35,Citation36 AtWRI1 is a substrate of the SNF1-related protein kinase KIN10, which catalyzes AtWRI1 phosphorylation that mediates AtWRI1 degradation.Citation33 Phosphorylation deficient mutations at T70 and S166 positions abolish KIN10-triggered phosphorylation and enhance the stability of AtWRI1.Citation33 Further study found that trehalose 6-phosphate (T6P) plays a role in stabilizing AtWRI1 protein and enhancing fatty acid production through repression of KIN10.Citation37 Recruitment of the mediator subunits by transcriptional regulators to initiate transcription is a conserved mechanism in eukaryotic cells.Citation38 Arabidopsis mediator subunit MED15 physically interacts with AtWRI1, and the transgenic Arabidopsis plants overexpressing MED15 shows elevated expression of AtWRI1 targets.Citation39
Figure 1. Structural features of WRI1 and TCP4 and molecular mechanisms of WRI1-regulated oil biosynthesis. (a) Schematic diagram of AtWRI1, including two AP2 domains, three intrinsically disordered regions (IDRs), a nuclear localization signal (NLS), a functional motif of “VYL”, the 14-3-3 and E3 ligase adaptor (BPM) binding motifs, TCP4-interacting domain, the transactivation domain (TAD), the ubiquitination sites, and the KIN10 phosphorylation sites. The schematic diagram of TCP4 shows the basic region and the helix-loop-helix (HLH) region of the TCP domain. (b) The fine-tuning of lipid biosynthesis through modulating WRI1 activity leads to an increase (large oil drop) or a decrease (small oil drop) of oil accumulation. In response to environmental or developmental signals, various WRI1 modules are formed, which either positively or negatively regulate the transcriptional activity of WRI1. Interaction of WRI1 with 14-3-3s or MED15 leads to enhanced protein stability, transcriptional activity, or assembly of the transcriptional machinery, resulting in higher oil accumulation. T6P stabilizes WRI1 by inhibition of KIN10, which positively mediates oil biosynthesis. Modules formed between WRI1 and TCP4, BPMs, or KIN10 reduce transcriptional activity or protein stability, resulting in lower oil accumulation. 14-3-3s, 14-3-3 proteins; MED15, Mediator subunit 15; T6P, trehalose 6-phosphate; BPMs, CULLIN3-based E3 ligase adaptor BTB/POZMATH (BPM) proteins
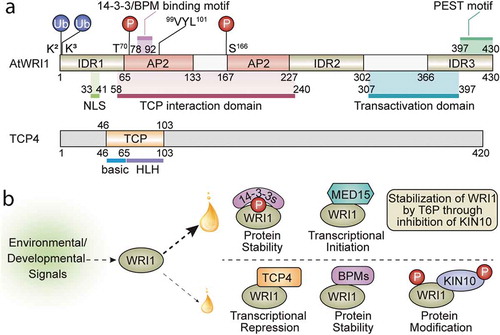
The involvement of TCP4 in WRI1-mediated plant oil accumulation
Our recent study uncovered a previously unknown relationship between WRI1 and TCP4 that affects the plant oil biosynthesis.Citation40 We screened an Arabidopsis transcription factor library and identified several TCP transcription factors (including TCP4, TCP10, and TCP24) as new AtWRI1-interacting partners. We validated the interaction between AtWRI1 and TCPs in yeast and plant cells. In a plant transient expression system, TCP4 represses AtWRI1-mediated oil production, as well as the transcriptional activity of AtWRI1.Citation40 Overexpressing miR319-resistant TCP4 (rTCP4) reduced expression of AtWRI1 targets in transgenic plants. TCP4 represses the transcriptional activity of AtWRI1, and TCP4 mutants, including tcp4 T-DNA knockout, the jaw-D mutant with reduced TCP4 expression, and a tcp2 tcp4 tcp10 triple mutant, display increased seed oil content compared to WT.Citation40
The WRI1-TCP4 module: combinatorial transcriptional control of plant oil biosynthesis
Intra- and inter-family transcription factor interactions greatly expand the complexity of combinatorial transcriptional regulation. In plant cells, protein complex formation within a transcription factor family has been well documented; however, cross-family transcription factor interactions are increasingly recognized.Citation41 A transcription factor family comprises a group of sequence-specific DNA-binding factors with distinct or overlapping functions. Different families vary in their transactivation activity (e.g. as activators or repressors), DNA-binding specificity, and response to various biotic and abiotic stimuli. Cross-family transcription factor interactions provide multi-layer regulatory mechanisms that are required for fine-tuning gene expression in response to various developmental and environmental signals. Increasing evidence demonstrates that TCP transcription factors serve as transcriptional hubs via interaction with diverse transcriptional regulators in numerous biological processes.Citation41 However, only recently we revealed the involvement of TCP in plant lipid biosynthesis through interaction with the AP2 transcription factor WRI1.Citation40
The involvement of TCP4, acting as a transcriptional repressor, in lipid biosynthesis is intriguing. Fatty acid biosynthesis is tightly controlled, especially during the seed development to maintain the proportional balance of lipids, proteins, and carbohydrates. As such, plants have evolved mechanisms that allow dialing the amplitudes of transactivation by critical fatty acid regulator, such as WRI1. While several activators of WRI1 activity have been characterized, negative regulation of AtWRI1 activity, for example, through phosphorylation and proteasomal degradation, have also been reported.Citation13,Citation25,Citation30,Citation33,Citation34 However, little is known regarding transcriptional repressors that negatively regulate the AtWRI1 activity. Our recent discovery highlights the possible role of TCP4 as a transcriptional repressor of AtWRI1 to modulate seed oil biosynthesis.Citation40 Nevertheless, our understanding of the WRI1-TCP4 regulatory module is by no means complete. Using in silico phosphorylation analysisCitation40 and mass spectrometry,Citation42 multiple phosphorylation residues in TCP4 have been identified. Given the importance of phosphorylation in mediating protein functions, such as protein–protein interaction, subcellular localization, and transcriptional activity, we speculate that phosphorylation might play a role in regulating the AtWRI1-TCP4 module. Questions remain as to how the upstream developmental or environmental signals trigger the phosphorylation and what kinases are involved in these processes.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The WRI1 work in the Ma Lab was supported by a Nanyang Technological University Startup grant and a Ministry of Education (MOE) of Singapore Tier 1 to W.M. (2018-T1-002-019).
Additional information
Funding
References
- Danisman S. TCP transcription factors at the interface between environmental challenges and the plant’s growth responses. Front Plant Sci. 2016;7:1. doi:https://doi.org/10.3389/fpls.2016.01930.
- Lopez JA, Sun Y, Blair PB, Mukhtar MS. TCP three-way handshake: linking developmental processes with plant immunity. Trends Plant Sci. 2015;20(4):238–4. doi:https://doi.org/10.1016/j.tplants.2015.01.005.
- Li LS. The Arabidopsis thaliana TCP transcription factors: A broadening horizon beyond development. Plant Signal Behav. 2015;10(7):e1044192. doi:https://doi.org/10.1080/15592324.2015.1044192.
- Cubas P, Lauter N, Doebley J, Coen E. The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 1999;18(2):215–222. doi:https://doi.org/10.1046/j.1365-313X.1999.00444.x.
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. Control of leaf morphogenesis by microRNAs. Nature. 2003;425(6955):257–263. doi:https://doi.org/10.1038/nature01958.
- Li Z, Li B, Shen W-H, Huang H, Dong A. TCP transcription factors interact with AS2 in the repression of class-I KNOX genes in Arabidopsis thaliana. Plant J. 2012;71(1):99–107. doi:https://doi.org/10.1111/j.1365-313X.2012.04973.x.
- Li S, Zachgo S. TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J. 2013;76(6):901–913. doi:https://doi.org/10.1111/tpj.12348.
- Kim SH, Son GH, Bhattacharjee S, Kim HJ, Nam JC, Nguyen PD, Hong JC, Gassmann W. The Arabidopsis immune adaptor SRFR1 interacts with TCP transcription factors that redundantly contribute to effector-triggered immunity. Plant J. 2014;78(6):978–989. doi:https://doi.org/10.1111/tpj.12527.
- Cernac A, Benning C. WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 2004;40(4):575–585. doi:https://doi.org/10.1111/j.1365-313X.2004.02235.x.
- Masaki T, Mitsui N, Tsukagoshi H, Nishii T, Morikami A, Nakamura K. ACTIVATOR of Spomin::LUC1/WRINKLED1 of Arabidopsis thaliana transactivates sugar-inducible promoters. Plant Cell Physiol. 2005;46(4):547–556. doi:https://doi.org/10.1093/pcp/pci072.
- Chapman KD, Ohlrogge JB. Compartmentation of triacylglycerol accumulation in plants. J Biol Chem. 2012;287(4):2288–2294. doi:https://doi.org/10.1074/jbc.R111.290072.
- Kong Q, Yuan L, Ma W. WRINKLED1, a “Master Regulator” in transcriptional control of plant oil biosynthesis. Plants (Basel). 2019;8(7):238. doi:https://doi.org/10.3390/plants8070238.
- Kong Q, Yang Y, Guo L, Yuan L, Ma W. Molecular basis of plant oil biosynthesis: insights gained from studying the WRINKLED1 transcription factor. Front Plant Sci. 2020;11:24. doi:https://doi.org/10.3389/fpls.2020.00024.
- Focks N, Benning C. WRINKLED1: a novel, low-seed-oil mutant of arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 1998;118(1):91–101. doi:https://doi.org/10.1104/pp.118.1.91.
- Ruuska SA, Girke T, Benning C, Ohlrogge JB. Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell. 2002;14(6):1191–1206. doi:https://doi.org/10.1105/tpc.000877.
- Baud S, Mendoza MS, To A, Harscoet E, Lepiniec L, Dubreucq B. WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 2007;50(5):825–838. doi:https://doi.org/10.1111/j.1365-313X.2007.03092.x.
- Maeo K, Tokuda T, Ayame A, Mitsui N, Kawai T, Tsukagoshi H, Ishiguro S, Nakamura K. An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J. 2009;60(3):476–487. doi:https://doi.org/10.1111/j.1365-313X.2009.03967.x.
- Marchive CNK, Nikovics K, To A, Lepiniec L, Baud S. Transcriptional regulation of fatty acid production in higher plants: molecular bases and biotechnological outcomes. Eur J Lipid Sci Technol. 2014;116(10):1332–1343. doi:https://doi.org/10.1002/ejlt.201400027.
- Chen B, Zhang G, Li P, Yang J, Guo L, Benning C, Wang X, Zhao J. Multiple GmWRI1s are redundantly involved in seed filling and nodulation by regulating plastidic glycolysis, lipid biosynthesis and hormone signalling in soybean (Glycine max). Plant Biotechnol J. 2020;18(1):155–171. doi:https://doi.org/10.1111/pbi.13183.
- Liu J, Hua W, Zhan G, Wei F, Wang X, Liu G, Wang H. Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant Physiol Biochem. 2010;48(1):9–15. doi:https://doi.org/10.1016/j.plaphy.2009.09.007.
- Ma W, Kong Q, Arondel V, Kilaru A, Bates PD, Thrower NA, Benning C, Ohlrogge JB. Wrinkled1, a ubiquitous regulator in oil accumulating tissues from Arabidopsis embryos to oil palm mesocarp. PLoS One. 2013;8(7):e68887. doi:https://doi.org/10.1371/journal.pone.0068887.
- Pouvreau B, Baud S, Vernoud V, Morin V, Py C, Gendrot G, Pichon J-P, Rouster J, Paul W, Rogowsky PM, et al. Duplicate maize wrinkled1 transcription factors activate target genes involved in seed oil biosynthesis. Plant Physiol. 2011;156(2):674–686. doi:https://doi.org/10.1104/pp.111.173641.
- Shen B, Allen WB, Zheng P, Li C, Glassman K, Ranch J, Nubel D, Tarczynski MC. Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol. 2010;153(3):980–987. doi:https://doi.org/10.1104/pp.110.157537.
- Yang Y, Munz J, Cass C, Zienkiewicz A, Kong Q, Ma W, Sanjaya, Sedbrook J, Benning C. Ectopic expression of WRINKLED1 affects fatty acid homeostasis in brachypodium distachyon vegetative tissues. Plant Physiol. 2015;169(3):1836–1847. doi:https://doi.org/10.1104/pp.15.01236.
- Kong Q, Ma W. WRINKLED1 transcription factor: how much do we know about its regulatory mechanism? Plant Sci. 2018;272:153–156. doi:https://doi.org/10.1016/j.plantsci.2018.04.013.
- Sun R, Ye R, Gao L, Zhang L, Wang R, Mao T, Zheng Y, Li D, Lin Y. Characterization and ectopic expression of CoWRI1, an AP2/EREBP domain-containing transcription factor from coconut (Cocos nucifera L.) endosperm, changes the seeds oil content in transgenic arabidopsis thaliana and rice (Oryza sativa L.). Front Plant Sci. 2017;8:63. doi:https://doi.org/10.3389/fpls.2017.00063.
- An D, Kim H, Ju S, Go YS, Kim HU, Suh MC. Expression of camelina WRINKLED1 isoforms rescue the seed phenotype of the arabidopsis wri1 mutant and increase the triacylglycerol content in tobacco leaves. Front Plant Sci. 2017;8:34. doi:https://doi.org/10.3389/fpls.2017.00034.
- Sanjaya, Durrett TP, Weise SE, Benning C. Increasing the energy density of vegetative tissues by diverting carbon from starch to oil biosynthesis in transgenic Arabidopsis. Plant Biotechnol J. 2011;9(8):874–883. doi:https://doi.org/10.1111/j.1467-7652.2011.00599.x.
- An D, Suh MC. Overexpression of Arabidopsis WRI1 enhanced seed mass and storage oil content in Camelina sativa. Plant Biotechnol Rep. 2015;9(3):137–148. doi:https://doi.org/10.1007/s11816-015-0351-x.
- Ma W, Kong Q, Grix M, Mantyla JJ, Yang Y, Benning C, Ohlrogge JB. Deletion of a C-terminal intrinsically disordered region of WRINKLED1 affects its stability and enhances oil accumulation in Arabidopsis. Plant J. 2015;83(5):864–874. doi:https://doi.org/10.1111/tpj.12933.
- Grimberg Å, Carlsson AS, Marttila S, Bhalerao R, Hofvander P. Transcriptional transitions in Nicotiana benthamiana leaves upon induction of oil synthesis by WRINKLED1 homologs from diverse species and tissues. BMC Plant Biol. 2015;15(1):192. doi:https://doi.org/10.1186/s12870-015-0579-1.
- Vanhercke T, El Tahchy A, Shrestha P, Zhou X-R, Singh SP, Petrie JR. Synergistic effect of WRI1 and DGAT1 coexpression on triacylglycerol biosynthesis in plants. FEBS Lett. 2013;587(4):364–369. doi:https://doi.org/10.1016/j.febslet.2012.12.018.
- Zhai Z, Liu H, Shanklin J. Phosphorylation of WRINKLED1 by KIN10 results in its proteasomal degradation, providing a link between energy homeostasis and lipid biosynthesis. Plant Cell. 2017;29(4):871–889. doi:https://doi.org/10.1105/tpc.17.00019.
- Chen L, Lee JH, Weber H, Tohge T, Witt S, Roje S, Fernie AR, Hellmann H. Arabidopsis BPM proteins function as substrate adaptors to a CULLIN3-based E3 ligase to affect fatty acid metabolism in plants. Plant Cell. 2013;25(6):2253–2264. doi:https://doi.org/10.1105/tpc.112.107292.
- Ma W, Kong Q, Mantyla JJ, Yang Y, Ohlrogge JB, Benning C. 14-3-3 protein mediates plant seed oil biosynthesis through interaction with AtWRI1. Plant J. 2016;88(2):228–235. doi:https://doi.org/10.1111/tpj.13244.
- Kong Q, Ma W. WRINKLED1 as a novel 14-3-3 client: function of 14-3-3 proteins in plant lipid metabolism. Plant Signal Behav. 2018;13(8):e1482176. doi:https://doi.org/10.1080/15592324.2018.1482176.
- Zhai Z, Keereetaweep J, Liu H, Feil R, Lunn JE, Shanklin J. Trehalose 6-phosphate positively regulates fatty acid synthesis by stabilizing WRINKLED1. Plant Cell. 2018;30(10):2616–2627. doi:https://doi.org/10.1105/tpc.18.00521.
- Taatjes DJ. The human mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem Sci. 2010;35(6):315–322. doi:https://doi.org/10.1016/j.tibs.2010.02.004.
- Kim MJ, Jang I-C, Chua N-H. The mediator complex MED15 subunit mediates activation of downstream lipid-related genes by the WRINKLED1 transcription factor. Plant Physiol. 2016;171(3):1951–1964. doi:https://doi.org/10.1104/pp.16.00664.
- Kong Q, Singh SK, Mantyla JJ, Pattanaik S, Guo L, Yuan L, Benning C, Ma W. TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR 4 interacts with WRINKLED1 to mediate seed oil biosynthesis. Plant Physiol. 2020;. (In press). doi:https://doi.org/10.1104/pp.20.00547.
- Bemer M, van Dijk ADJ, Immink RGH, Angenent GC. Cross-family transcription factor interactions: an additional layer of gene regulation. Trends Plant Sci. 2017;22(1):66–80. doi:https://doi.org/10.1016/j.tplants.2016.10.007.
- Kubota A, Ito S, Shim JS, Johnson RS, Song YH, Breton G, Goralogia GS, Kwon MS, Laboy Cintrón D, Koyama T, et al. TCP4-dependent induction of CONSTANS transcription requires GIGANTEA in photoperiodic flowering in Arabidopsis. PLoS Genet. 2017;13(6):e1006856. doi:https://doi.org/10.1371/journal.pgen.1006856.
