ABSTRACT
GmGASA is the GASA gibberellin regulated cysteine-rich protein family. The expression of GmGASA is up-regulated by gibberellin, which is the longest plant hormone in plants playing vital roles in plant development. However, very few reports explaining the direct regulation of downstream genes by GASA gene are available. In the current study, the GmGASA32, a member of the GASA family affecting soybean height was identified. In the early stage, preliminary verification of the response of GmGASA32 to gibberellin through phenotypic experiment was done. The promoter activity analysis confirmed that GmGASA32 was induced by gibberellin. Subcellular localization showed that GmGASA32-GFP fusion protein enriched in the nucleus after gibberellin treatment. In order to confirm the function of GmGASA32 in the nucleus, we confirmed that the GASA domain in the C terminal of GmGASA32 can interact with GmCDC25 (cell cycle-associated protein) through the bimolecular fluorescence complementation (BiFC) assay.
Introduction
Members of the plant-specific gibberellic acid-stimulated Arabidopsis (GASA) gene family play roles in various developmental programs, including seed germination, root formation, establishment of seed size, stem growth and flowering time, fruit development and ripening, fiber development, and leaf expansion.Citation1,Citation2 So far, 14 GASA genes have been identified in Arabidopsis thaliana and 37 in soybean.Citation3
Most of the GASA genes are induced by a variety of hormones, including gibberellic acid (GA), abscisic acid (ABA), auxin, brassinosteroids (BR), and salicylic acid (SA).Citation4 For example, AtGASA6 links RGL2 and AtEXPA1 functions and plays a role as an integrator of gibberellin, abscisic acid, and Glc signaling, resulting in the regulation of seed germination through promotion of cell elongation.Citation2 In addition, GASA also participates in the regulation of flowering. Expression analysis of apple (Malus domestica) GASA genes revealed at least seven potentially flowering-related genes within the family.Citation5 But interestingly, there is an opposite mechanism in Arabidopsis thaliana. AtET2 acts as a negative regulator of the GA-induced GASA4, which is known to be involved in the control of cell division.Citation6,Citation7 The effect of ET proteins on GASA gene expression is not only specific to Arabidopsis, as the Brassica napus homolog (BnET) also represses the GA-induced GASA4 promoter.Citation8 In addition to participating in the gibberellin signaling pathway, GASA was found to have enzyme activity. For example, GASA4 and GASA5 were shown to have redox activity, potentially acting on specific targets, not as general ROS scavengers.Citation9 Therefore, GASA proteins could act as integrators of internal and environmental cues participating in hormone homeostasis to modulate plant development and stress tolerance through adjusting the balance of cell growth promotion and cell growth inhibition.Citation4
However, these studies remain at the transcriptional level and are limited to the model plant Arabidopsis thaliana. Although soybean is an important cash crop, the signal transduction of GASA in soybean is very scarce,Citation10 and this adds to the importance of a GmGASA study. In this study, we preliminarily confirmed that GmGASA32 was induced by gibberellin through phenotypic experiments and promoter activity analysis. A cell cycle associated protein GmCDC25 was then identified by interaction analysis. This work laid a foundation for further advancing the analysis of soybean gibberellin signal regulation pathway.
Results
GmGASA32 can be positively regulated by gibberellin in soybean
In our previous study, we found that the expression of GmGASA32 (Gene ID: 100787539; Locus tag: GLYMA_18G259400) is relatively high in the whole GASA gene family of soybeans. This result is consistent with previous research by retrieved from phytozome.Citation3 The response of GASA to gibberellin has two possibilities, positive and (or not and as only one possibility is there at a time) negative regulation.Citation11,Citation12 We used soybean hairy root transformation (Agrobacterium rhizogenes K599 induced) to preliminarily identify the response pattern of GmGASA32 to gibberellin (). The results demonstrated that in control growth conditions, the plant height of GmGASA32 overexpressed lines (OE lines) were significantly higher than WT lines (Williams 82 transformed with empty vector). After gibberellin treatment, the height of GmGASA32 OE and WT lines increased, but the increase of OE lines was greater than that of WT lines (). We can preliminarily infer that GmGASA32 has a promoting effect on the plant height of soybean through the gibberellin pathway. This is similar to GASA14 regulated leaf expansion.Citation13
Figure 1. GmGASA32 can be up-regulated by gibberellin (GA). (a) The function of GmGASA32 was verified by soybean hairy root transformation. The left was the picture of WT and GmGASA32 overexpressed lines before treated with 10 μM gibberellin (pour 50 ml into the soil for one time). The right was the picture of WT and GmGASA32 overexpressed lines treated with gibberellin for 5 days. (b) Plant height of WT and GmGASA32 overexpressed lines after treatment with gibberellin was measured. Means and standard deviations (SDs) were derived from four independent samples (n = 4). The statistical significance of differences in gene expression level was determined by Tukey’s honestly significant difference (HSD) test (P < .01). (c) GUS activity assay. The pGAMBIA1305 vector was used to construct the new vector. The 35S promoter was replaced with the self-promoter of GmGASA32. The soybean hairy root transformation system was used for transformation. The positive roots were treated with 10 μM gibberellin for 1 h. The image on the left was untreated as control and the image on the right is of the treated roots. The images were viewed using Olympus optical microscope BX53M. Bar = 1 mm. (d) Real-time PCR analysis was carried out to determine the expression of GUS. Each experiment was repeated three times. Values are reported as mean, and error bars represent SDs. Statistical significance was determined using two-tail Student’s t-test. **, P < .01
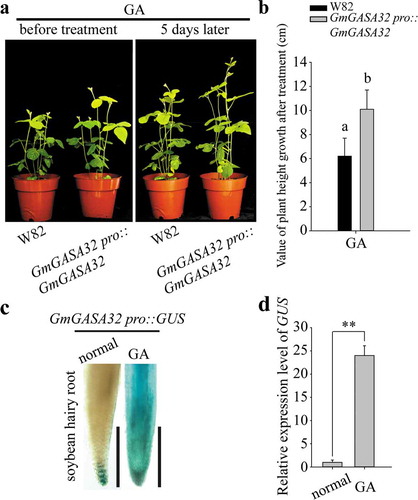
To further confirm that GmGASA32 can be induced by gibberellin, GUS activity analysis was performed (). The promoter of GmGASA32 was connected to the front of the GUS reporter gene, and also used the transformation of soybean hairy roots to obtain positive GUS roots. The figure depicted that under in control conditions, GmGASA32 was expressed only at the root tip. However, after gibberellin treatment, GmGASA32 was expressed in the whole root including the elongation area. Real time-PCR results showed that under the action of gibberellin, the expression of GmGASA32 increased over 23 times than the control condition (). Further confirmation of positive regulation of GmGASA32 by gibberellin in soybean was also done.
The C’ terminal GASA domain of GmGASA32 can enter the nucleus and interact with GmCDC25
In order to understand how GmGASA32 responds to gibberellin, the intracellular location of GmGASA32 was determined by protoplast transient transformation (). In control (normal) conditions, GmGASA32 got distributed in the cytoplasm and nucleus, whereas under gibberellin treatment, GmGASA32 enriched in the nucleus (). Enrichment of GmGASA32 in the nucleus after gibberellin treatment is quite an interesting process. To dig out more about the above-mentioned phenomenon, previous studies were used as a basis for proposing the hypothesis.Citation14 GASA family members encode small polypeptides sharing common structural features: a N-terminal signal peptide, a highly divergent intermediate region, and a conserved C-terminal domain containing 12 cysteine residues conserved within the alignment giving the potential for these proteins to possess 6 disulfide bonds.Citation14 The N-terminal signal peptide is critical for the subcellular localization of GASA proteins. Both AtGASA4 and AtGASA6 were localized to the cell peripheries. But once the N-terminal signal peptide is cleaved, it will affect the position of C-terminal domain. Both C-terminal domains of AtGASA4 and AtGASA6 were localized to the nucleus.Citation14 The domain of GmGASA32 was also analyzed. Primary, SMART (Simple Modular Architecture Research Tool) (http://smart.embl-heidelberg.de/) analysis showed that there is a signal peptide at the N-terminal of GmGASA32, starting at 1 and ending at 24 amino acids (). Second time, Protter (interactive protein feature visualization) (http://wlab.ethz.ch/protter/#) analysis showed a similar result to (). Furthermore, the result also showed that GmGASA32 is not a membrane protein because it does not have a transmembrane domain. This finding has further demonstrated that GmGASA32 has the potential to get into the nucleus.
Figure 2. The C-terminal of GmGASA32 enters into the nucleus, and interacts with GmCDC25. (a) Subcellular localization of GmGASA32. We connected the full-length cDNA (non-terminating codon) of GmGASA32 to p16318 vector, and GFP gene was located at the 3’ end of GmGASA32. 35S promoter was replaced with the self-promoter of GmGASA32. The young leaves of Arabidopsis thaliana (14 days old) were enzymatically decomposed by cellulase and pectinase to form protoplasts for subsequent transformation. Three groups of experiments were conducted, namely, the empty control group (35S::GFP), the experimental group (GmGASA32 pro::GmGASA32-GFP) and the treatment group which was treated with 10 μM gibberellin after dark culture for 16 h. The four panels in the upper two rows are green GFP, white bright field, red chlorophyll fluorescence, and merged signals. On the bottom row the nucleus indicating DAPI (4’,6-Diamidino-2-Phenylindole, Dihydrochloride) fluorescence was added. The images were observed using the laser confocal microscope ZEISS LSM 980 with Airyscan 2. The white arrow points to the nucleus. Bar = 10 μm. (b) SMART (Simple modular architecture research tool)(http://smart.embl-heidelberg.de/) analysis of GmGASA32. Δ1 (Red represents the signal peptide.)starts at 1 and ends at 24 amino acids. Δ2 starts at 48 and ends at 107 amino acids. (c) Protter (interactive protein feature visualization) (http://wlab.ethz.ch/protter/#) analysis of GmGASA32. Red represents the signal peptide. (d) According to the different peptide segments of GmGASA32, BiFC experiments were conducted to verify their interaction with GmCDC25. pSPYNE-35S and pSPYCE-35S vectors were used to construct the transformed vectors. Top to bottom are the results of the interactions between full length GmGASA32, N-terminal of GmGSAS32 (Δ1), and C-terminal of GmGASA32 (Δ2) with GmCDC25. There are five panels, yellow YFP fluorescence, white bright field, red chlorophyll fluorescence, blue DAPI fluorescence and merged signals. The white arrow points to the nucleus. These red arrows indicate a larger view of the nucleus. Bar = 10. (e) Diagram depicting inferred signal path pattern. Experiments emphasized that after treating with gibberellin, the C-terminal of GmGASA32 (Δ2) was promoted to enter the nucleus and interact with GmCDC25, thus affecting the process of cell division
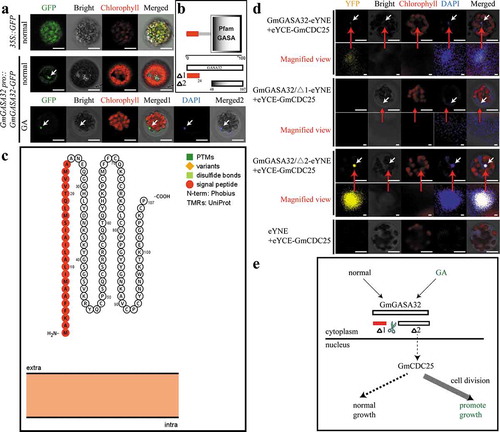
In order to investigate the functions of GmGASA32 after entering the nucleus, an interesting interactional protein GmCDC25 (the protein associated with cell cycle) was selected for further experimentation by BiFC (Bimolecular Fluorescence Complementation) (). Since the signal peptide of GmGASA32 has an effect on its position in the cell, three separate experiments were conducted. In the initial experiments, the vector with the full length of GmGASA32 was constructed. The results demonstrated that GmGASA32 and GmCDC25 had very weak interaction signals in the nucleus. Subsequently, we demonstrated that the signal peptide of GmGASA32 (Δ1:1–24amino acids) () could not interact with GmCDC25. But interestingly, the only one domain GASA on the C-terminal (Δ2: 48–107 amino acids) of GmGASA32 could interact strongly with GmCDC25 in the nucleus.
To recapitulate, preliminarily results confirmed that the C-terminal of GmGASA32 can enter the nucleus and interact with GmCDC25 in the nucleus.
Since GmCDC25 plays an important roleCitation15–17 in the cell cycle pathway, we can speculate that after the signal peptide is removed, the C-terminal of GmGASA32 enters into the nucleus, and interacts with GmCDC25 when treated with gibberellin. This phenomenon will affect the cell division, further affecting the plant growth ().
Materials and methods
Plant materials and growth conditions
Seeds of Arabidopsis thaliana ecotype Columbia-0 (Col-0) were obtained from the European Arabidopsis Stock Center and were germinated on 1/2 Murashige Skoog (MS)agar medium and seedlings were transferred to soil and grown in a chamber with controlled conditions at (16-h light/8-h dark cycle, 22°C and 70% relative humidity).Citation18
Soybean (Glycine max (L.) Merr.) cultivar Williams 82 was used in current study. Plants were grown in growth cabinets under short-day conditions (8 h light/16 h dark), at a light intensity of 120–180 µmol m−2 s−1 and a temperature of 25°C.Citation19
Soybean hairy roots transformation
The transformation method of soybean hair root was based on our previous research.Citation20 No less than ten seedlings were guaranteed for each line.
BiFC assay
This BiFC assay method was based on our previous research.Citation20 The images were observed using the laser confocal microscope ZEISS LSM 980 with Airyscan 2.
Statistical analysis of the data
The statistical methods of phenotypic data are as follows. Means and standard deviations (SDs) were derived from four independent samples (n = 4). The statistical significance of differences in gene expression level was determined by Tukey’s honestly significant difference (HSD) test (P < .01). GUS gene expression data were analyzed as follows. Each experiment was repeated three times. Values are reported as mean, and error bars represent SDs. Statistical significance was determined using two-tail Student’s t-test. **, P < .01.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
K.C. and H.Y.L. conceived the research. K.C. performed the experiments. K.C. and H.Y.L. wrote the manuscript.
Supplemental Material
Download MS Word (15.3 KB)Acknowledgments
We are grateful to Dr Lijuan Qiu of the Institute of Crop Science, Chinese Academy of Agricultural Sciences for kindly providing soybean seeds. The authors would also like to thank all the reviewers who participated in the review and MJEditor (www.mjeditor.com) for its linguistic assistance during the preparation of this manuscript.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Roxrud I, Lid SE, Fletcher JC, Schmidt ED, Opsahl-Sorteberg HG. GASA4, one of the 14-member Arabidopsis GASA family of small polypeptides, regulates flowering and seed development. Plant Cell Physiol. 2007;48:1–5. doi:10.1093/pcp/pcm016.
- Zhong C, Xu H, Ye S, Wang S, Li L, Zhang S, Wang X. Gibberellic acid-stimulated arabidopsis6 serves as an integrator of gibberellin, abscisic acid, and glucose signaling during seed germination in arabidopsis. Plant Physiol. 2015;169:2288–2303. doi:10.1104/pp.15.00858.
- Ahmad MZ, Sana A, Jamil A, Nasir JA, Ahmed S, Hameed MU. Abdullah. A genome-wide approach to the comprehensive analysis of GASA gene family in glycine max. Plant Mol Biol. 2019;100:607–620. doi:10.1007/s11103-019-00883-1.
- Nahirñak V, Almasia NI, Hopp HE, Vazquez-Rovere C. Snakin/GASA proteins: involvement in hormone crosstalk and redox homeostasis. Plant Signal Behav. 2012;7:1004–1008. doi:10.4161/psb.20813.
- Fan S, Zhang D, Zhang L, Gao C, Xin M, Tahir MM, Li Y, Ma J, Han M. Comprehensive analysis of GASA family members in the Malus domestica genome: identification, characterization, and their expressions in response to apple flower induction. BMC Genomics. 2017;18:827. doi:10.1186/s12864-017-4213-5.
- Ivanov R, Tiedemann J, Czihal A, Schallau A, Diep le H, Mock HP, Claus B, Tewes A, Bäumlein H. EFFECTOR OF TRANSCRIPTION2 is involved in xylem differentiation and includes a functional DNA single strand cutting domain. Dev Biol. 2008;313:93–106. doi:10.1016/j.ydbio.2007.09.061.
- Aubert D, Chevillard M, Dorne AM, Arlaud G, Herzog M. Expression patterns of GASA genes in Arabidopsis thaliana: the GASA4 gene is up-regulated by gibberellins in meristematic regions. Plant Mol Biol. 1998;36:871–883. doi:10.1023/a:1005938624418.
- Ellerström M, Reidt W, Ivanov R, Tiedemann J, Melzer M, Tewes A, Moritz T, Mock HP, Sitbon F, Rask L, et al. Ectopic expression of EFFECTOR OF TRANSCRIPTION perturbs gibberellin-mediated plant developmental processes. Plant Mol Biol. 2005;59:663–681. doi:10.1007/s11103-005-0669-9.
- Rubinovich L, Ruthstein S, Weiss D. The arabidopsis cysteine-rich GASA5 is a redox-active metalloprotein that suppresses gibberellin responses. Mol Plant. 2014;7:244–247. doi:10.1093/mp/sst141.
- Li KL, Bai X, Li Y, Cai H, Ji W, Tang LL, Wen YD, Zhu YM. GsGASA1 mediated root growth inhibition in response to chronic cold stress is marked by the accumulation of DELLAs. J Plant Physiol. 2011;168:2153–2160. doi:10.1016/j.jplph.2011.07.006.
- Ko CB, Woo YM, Lee DJ, Lee MC, Kim CS. Enhanced tolerance to heat stress in transgenic plants expressing the GASA4 gene. Plant Physiol Biochem. 2007;45:722–728. doi:10.1016/j.plaphy.2007.07.010.
- Rubinovich L, Weiss D. The arabidopsis cysteine-rich protein GASA4 promotes GA responses and exhibits redox activity in bacteria and in planta. Plant J. 2010;64:1018–1027. doi:10.1111/j.1365-313X.2010.04390.x.
- Sun S, Wang H, Yu H, Zhong C, Zhang X, Peng J, Wang X. GASA14 regulates leaf expansion and abiotic stress resistance by modulating reactive oxygen species accumulation. J Exp Bot. 2013;64:1637–1647. doi:10.1093/jxb/ert021.
- Qu J, Kang SG, Hah C, Jang JC. Molecular and cellular characterization of GA-Stimulated Transcripts GASA4 and GASA6 in arabidopsis thaliana. Plant Sci. 2016;246:1–10. doi:10.1016/j.plantsci.2016.01.009.
- Boudolf V, Inzé D, De Veylder L. What if higher plants lack a CDC25 phosphatase? Trends Plant Sci. 2006;11:474–479. doi:10.1016/j.tplants.2006.08.009.
- Zhang K, Diederich L, John PC. The cytokinin requirement for cell division in cultured Nicotiana plumbaginifolia cells can be satisfied by yeast Cdc25 protein tyrosine phosphatase: implications for mechanisms of cytokinin response and plant development. Plant Physiol. 2005;137:308–316. doi:10.1104/pp.104.051938.
- Landrieu I, da Costa M, De Veylder L, Dewitte F, Vandepoele K, Hassan S, Wieruszeski JM, Corellou F, Faure JD, Van Montagu M, et al. A small CDC25 dual-specificity tyrosine-phosphatase isoform in arabidopsis thaliana. Proc Natl Acad Sci U S A. 2004;101:13380–13385. doi:10.1073/pnas.0405248101.
- Liu NJ, Bao JJ, Wang LJ, Chen XY. Arabidopsis leaf extracellular vesicles in wound-induced jasmonate accumulation. Plant Signal Behav. 2020;12:1833142. doi:10.1080/15592324.2020.1833142.
- Xiong L, Li C, Li H, Lyu X, Zhao T, Liu J, Zuo Z, Liu B. A transient expression system in soybean mesophyll protoplasts reveals the formation of cytoplasmic GmCRY1 photobody-like structures. Sci China Life Sci. 2019;62:1070–1077. doi:10.1007/s11427-018-9496-5.
- Chen K, Tang WS, Zhou YB, Xu ZS, Chen J, Ma YZ, Chen M, Li HY. Overexpression of GmUBC9 gene enhances plant drought resistance and affects flowering time via histone H2B monoubiquitination. Front Plant Sci. 2020;11:555794. doi:10.3389/fpls.2020.555794.
