ABSTRACT
Aminoacyl-tRNA synthetases play a critical role in protein synthesis by catalyzing the covalent attachment of amino acids to their cognate tRNAs. However, the role of aminoacyl-tRNA synthetases in the transition from vegetative to reproductive growth in plants remains poorly understood. In this study, a rice (Oryza sativa) glycyl-tRNA synthetase 3, OsGlyRS3, was found to impact heading date in rice. Flowering in osglyrs3, a mutant line containing a T-DNA insertion in OsGlyRS3, was advanced by approximately 2 weeks compared to wild type. Expression analysis of flowering regulator genes showed that transcript levels of Heading date 1 (Hd1), Heading date 3a (Hd3a), and OsMADS51 were elevated in osglyrs3. These data indicate that the loss of OsGlyRS3 activity induces the expression of flowering-activating genes, resulting in early flowering.
Result and discussion
The timing of floral transition is a key determinant of rice adaptation and production. Because flowering time also affects other agronomic traits, the molecular mechanisms underlying the timing of the transition from vegetative to reproductive growth have been extensively studied in rice.Citation1,Citation2,Citation3 As a result, several flowering-related genes, including OsGIGANTEA (OsGI, a rice homolog of Arabidopsis GIGANTEA gene), Hd1 (a rice ortholog of Arabidopsis CO gene), Hd3a (a rice ortholog of Arabidopsis FT gene), OsMADS50 (a rice ortholog of Arabidopsis SUPPRESOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1)), OsMADS51 (a type I MADS-box gene), GRAIN NUMBER PLANT HEIGHT AND HEADING DATE 7 (Ghd7, a CCD domain transcription factor), and EARLY HEADING DATE 1 (Ehd1, a B-type response regulator), have been identified in rice.Citation1,Citation4,Citation5,Citation6,Citation7,Citation8,Citation9,Citation10,Citation11 These flowering-related genes regulate heading date under short-day (SD) and/or long-day (LD) conditions.
GlyRS catalyzes the covalent attachment of glycine to its cognate tRNA. Arabidopsis EMBRYO DEFECTIVE DEVELOPMENT1 (EDD1) is a nuclear gene that encodes a glycyl-tRNA synthetase that localizes to plastids and mitochondria, and loss-of-function mutations in EDD1 are embryonic lethal.Citation12,Citation13,Citation14 In addition, edd1 mutants also display abnormal leaf development and patterning.Citation15 In rice, the Rice Albino 1 (RA1) gene (LOC_Os06g01400) encodes a GlyRS that localizes to chloroplasts. The ra1 mutant has decreased chlorophyll and carotenoid levels as well as plastidic structural defects, including abnormal thylakoid membrane structures and increased numbers of osmiophilic particles, which produce albino phenotypes in seedlings.Citation16
Previously, we examined proteins that accumulated specifically in the seeds of a low eating-quality rice cultivar, Dobong, and a high eating-quality cultivar, Gopum, and found that OsGlyRS3 accumulated in immature seeds of Dobong and accumulated at high levels in mature seeds of both Dobong and Gopum.Citation17 However, OsGlyRS3 levels in mature seeds were two-fold higher in Gopum than in Dobong seeds, suggesting that OsGlyRS3 might be a positive regulator of high eating quality. Thus, we further examined the effect of OsGlyRS3 on vegetative and reproductive growth during rice development.
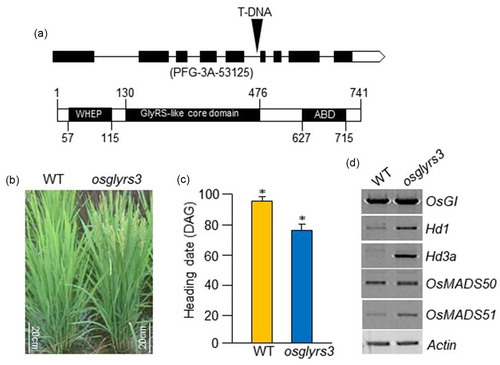
The effects of OsGlyRS3 were investigated using mutant analysis of the encoding gene, OsGlyRS3 (LOC_ Os08g42560). The OsGlyRS3 gene contains nine exons and eight introns (), upper), and the OsGlyRS3 protein contains three conserved domains: a plant-specific helix-turn-helix WHEP domain (57–115 aa), a glycyl-tRNA synthetase-like core domain (130–476 aa), and an anticon binding domain (627–715 aa) (), lower). An OsGlyRS3 mutant was identified from T-DNA insertion mutant lines (provided by the GynHeung An laboratory, Kyung Hee University, Korea). The mutant (PFG-3A-53125) harbored a T-DNA insertion in the fifth intron of the OsGlyRS3 gene, and was named osglyrs3 (), upper). Growth was assessed in Dongjin (wild type, WT) and osglyrs3 (T-DNA insertion mutant) plants. No significant differences were observed between WT and osglyrs3 during the germination and vegetative phases, although osglyrs3 appeared to grow slightly faster than WT. However, substantial differences were observed during the heading stage. Flowering time was estimated using days-to-heading, as determined by the appearance of the first panicle. Flowering in osglyrs3 mutants occurred approximately 2 weeks earlier than in WT plants ()), indicating that loss of OsGlyRS3 induced the transition from the vegetative to the reproductive phase. Early flowering in osglyrs3 also suggested that the expression of flowering-related genes was upregulated or downregulated in the mutant. The expression of five flowering-related genes, OsGI, Hd1, Hd3a, OsMADS50, and OsMADS51 genes was investigated using RT-PCR with gene-specific primers. Transcript levels of OsGI and OsMADS50 were similar between WT and osglyrs3, but expression of Hd1, Hd3a, and OsMDS51 was upregulated in osglyrs3.
Figure 1. Early flowering in the osglyrs3 rice mutant. (a) Schematic representation of OsGlyRS3. The OsGlyRS3 (Os08g42560) gene consists of nine exons (boxes) and eight introns (lines between boxes). The osglyrs3 mutant (PFG-3A-53125) contains a T-DNA insertion in the fifth intron (upper). The OsGlyRS3 protein (lower) contains a helix-turn-helix WHEP domain, a glycyl-tRNA synthetase-like core domain, and an anticon binding domain. (b) Phenotypes of wild-type (WT) and osglyrs3 mutant plants at the heading stage under long day (LD; 13.5 light/10.5 dark) conditions in the field. (c) Heading date in osglyrs3 and WT plants under LD conditions. Error bars indicate standard deviations. DAG, days after germination. Asterisks denote P-value <0.01. (d) Expression of floral regulators in osglyrs3 mutant plants. Total RNAs were prepared from second leaf blades of WT and osglyrs3 plants grown under LD conditions. Transcript levels of the OsGI, Hd1, Hd3a, OsMADS50, and OsMADS51 genes were assessed using RT-PCR with gene-specific primers. OsGI, a rice ortholog of the Arabidopsis GIGANTEA gene, Hd1, a rice ortholog of the Arabidopsis CO gene; Hd3a, a rice ortholog of the Arabidopsis FT gene; OsMADS50, a rice ortholog of the Arabidopsis SUPPRESOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) gene; and OsMADS51, a type I MADS-box gene involved in the SD promotion pathway in rice
Previous research showed that OsGI upregulated the expression of Hd1, which activated expression of Hd3a, promoting rice heading under both short day (SD) and long day (LD) conditions.Citation18,Citation19 OsMADS50 acted as an upstream regulator of Hd3a and stimulated flowering,Citation6 and OsMADS51 was also shown to act as a flowering promoter, particularly under SD conditions.Citation7 Mutation of the OsGI gene was found to significantly delay flowering under SD conditions and slightly delay flowering under LD conditions.Citation11 However, other studies showed that hd1-1 mutants experienced late flowering under SD conditions and early flowering under LD conditions,Citation9,Citation20 prompting the hypothesis that Hd1 acted as a flowering activator under SD conditions and as a flowering repressor under LD conditions. Notably, Hd1 overexpression caused constitutive late flowering under both SD and LD conditions,Citation21 suggesting that Hd1 bound to different partners under different conditions.
In this study, OsGI and OsMADS50 expression was not affected by the loss of OsGlyRS3 activity, whereas Hd1, Hd3a, and OsMADS51 transcript levels were substantially elevated in osglyrs3, suggesting an additional mechanism or pathway for the regulation of OsGlyRS3-mediated gene expression. Taken together, our findings and results from earlier research suggest the existence of other regulatory pathways involved in modulating flowering time in rice.
Two genes encoding GlyRS were identified in the rice genome (http://ricexpro.dna.affrc.go.jp): OsGlyRS1 (LOC_Os04g32650) and OsGlyRS3. OsGlyRS1 was highly expressed in leaf and inflorescence tissues, and OsGlyRS3 was mainly expressed in ovaries and anthers, but no information regarding subcellular localization was available. Recent research indicated that RA1 was also a GlyRS and was involved in chloroplast development,Citation16 but RA1 was not homologous to OsGlyRS1 and OsGlyRS3 at the amino acid level. Further studies, such as expression analysis of OsGlyRS1 and OsGlyRS3 and assessment of the effects of OsGlyRS1 and OsGlyRS3 on tissue and organ development, will help elucidate their roles in vegetative and reproductive growth.
Disclosure of potential conflicts of interest
The authors report no conflict of interest.
Additional information
Funding
References
- Xue WY, Xing YZ, Weng XY, Zhao Y, Tang WJ, Wang L, Zhou HJ, Yu SB, Xu CG, Li XH, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40:1–3. doi:10.1038/ng.143.
- Huang XH, Zhao Y, Wei XH, Li CY, Wang A, Zhao Q, Li WJ, Guo YL, Deng LW, Zhu CR, et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat Genet. 2012;44:32–39. doi:10.1038/ng.1018.
- Yano K, Yamamoto E, Aya K, Takeuchi H, Lo PC, Hu L, Yamasaki M, Yoshida S, Kitano H, Hirano K, et al. Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat Genet. 2016;48:927–934. doi:10.1038/ng.3596.
- Lin HX, Yamamoto T, Sasaki T, Yano M. Characterization and detection of epistatic interactions of three QTLs, Hd1, Hd2 and Hd3, controlling heading date in rice using nearly isogenic lines. Theor Appl Genet. 2000;101:1021–1028. doi:10.1007/s001220051576.
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the arabidopsis flowering time gene CONSTANS. Plant Cell. 2000;12:2473–2483. doi:10.1105/tpc.12.12.2473.
- Lee S, Kim J, Han JJ, Han MJ, An G. Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. Plant J. 2004;38(5):754–764. doi:10.1111/j.1365-313X.2004.02082.x.
- Kim SL, Lee S, Kim HJ, Nam HG, An G. OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol. 2007;145(4):1484–1494. doi:10.1104/pp.107.103291.
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi:10.1126/science.1141753.
- Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K. Hd3a and RFT1 are essential for flowering in rice. Development. 2008;135:767–774. doi:10.1242/dev.008631.
- Ryu CH, Lee S, Cho LH, Kim SL, Lee YS, Choi SC, Jeong HJ, Yi J, Park SJ, Han CD, et al. OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant Cell Environ. 2009;32(10):1412–1427. doi:10.1111/j.1365-3040.2009.02008.x.
- Lee YS, An G. OsGI controls flowering time by modulating rhythmic flowering time regulators preferentially under short day in rice. J Plant Biol. 2015;58:137–145. doi:10.1007/s12374-015-0007-y.
- Uwer U, Willmitzer L, Altmann T. Inactivation of a glycyl-tRNA synthetase leads to an arrest in plant embryo development. Plant Cell. 1998;10:1277–1294. doi:10.1105/tpc.10.8.1277.
- Duchene AM, Peeters N, Dietrich A, Cosset A, Small ID, Wintz H. Overlapping destinations for two dual targeted glycyl-tRNA synthetases in Arabidopsis thaliana and Phaseolus vulgaris. J Biol Chem. 2001;276:15275–15283. doi:10.1074/jbc.M011525200.
- Berg M, Rogers R, Muralla R, Meinke D. Requirement of aminoacyl-tRNA synthetases for gametogenesis and embryo development in Arabidopsis. Plant J. 2005;44:866–878. doi:10.1111/j.1365-313X.2005.02580.x.
- Moschopoulos A, Derbyshire P, Byrne ME. The Arabidopsis organelle-localized glycyl-tRNA synthetase encoded by EMBRYO DEFECTIVE DEVELOPMENT1 is required for organ patterning. J Exp Bot. 2012;63(14):5233–5243. doi:10.1093/jxb/ers184.
- Zheng H, Wang Z, Tian Y, Liu L, Lv F, Kong W, Bai W, Wang P, Wang C, Yu X, et al. Rice albino 1, encoding a glycyl-tRNA synthetase, is involved in chloroplast development and establishment of the plastidic ribosome system in rice. Plant Physiol Biochem. 2019;139:495–503. doi:10.1016/j.plaphy.2019.04.008.
- Kim YJ, Choi SH, Park BS, Song JT, Kim MC, Koh HJ, Seo HS. Proteomic analysis of the rice seed for quality improvement. Plant Breed. 2009;128:541–550. doi:10.1111/j.1439-0523.2009.01693.x.
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature. 2003;422:719–722. doi:10.1038/nature01549.
- Zhang Z, Hu W, Shen G, Liu H, Hu W, Zhou X, Liu T, Xing Y. Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions. Sci Rep. 2017;7:5388. doi:10.1038/s41598-017-05873-1.
- Ishikawa R, Tamaki S, Yokoi S, Inagaki N, Shinomura T, Takano M, Shimamoto K. Suppression of the floral activator gene Hd3a is the principal cause of the night break effect in rice. Plant Cell. 2005;17:3326–3336. doi:10.1105/tpc.105.037028.
- Ishikawa R, Aoki M, Kurotani K, Yokoi S, Shinomura T, Takano M, Shimamoto K. Phytochrome B regulates heading date 1 (Hd1)-mediated expression of rice florigen Hd3a and critical day length in rice. Mol Genet Genom. 2011;285:461–470. doi:10.1007/s00438-011-0621-4.
