ABSTRACT
Pre-mRNA splicing is a vital step in the posttranscriptional regulation of gene expression. Splicing is catalyzed by the spliceosome, a multidalton RNA–protein complex, through two successive transesterifications to yield mature mRNAs. In Arabidopsis, more than 61% of all transcripts from intron-containing genes are alternatively spliced, thereby resulting in transcriptome and subsequent proteome diversities for cellular processes. Moreover, it is estimated that more alternative splicing (AS) events induced by adverse stimuli occur to confer stress tolerance. Recently, increasing AS variants encoding normal or novel proteins, or degraded by nonsense-mediated decay (NMD) and their corresponding splicing factors or regulators acting at the posttranscriptional level have been functionally characterized. This review comprehensively summarizes and highlights the advances in our understanding of the biological functions and underlying mechanisms of AS events and their regulators in Arabidopsis and provides prospects for further research on AS in crops.
1 Introduction
In eukaryotes, the process that the introns are precisely recognized and removed from the nascent mRNAs (pre-mRNAs) and the exons are ligated in the right order to yield translatable mRNAs is called pre-mRNA splicing.Citation1 Splicing is catalyzed by the spliceosome, which is mainly composed of five small RNAs with their specific accessory proteins (snRNPs) and multiple splicing factors.Citation2
In many cases, the splicing process is flexible enough to generate multiple mature mRNAs by alternative splice site selections from one single gene, known as AS.Citation3 AS regulates gene expression at the posttranscriptional level, which exaggerates the transcriptome and proteome diversities with limited gene numbers.Citation4 In 1977, adenovirus late mRNA synthesis involving multiple splicing during maturation was observed, representing the first characterized example of alternative modes of RNA splicing.Citation5 In 1989, an alternative splicing event of rubisco activase in plants was first identified.Citation6 With sequencing technology development, genome-wide transcriptome analysis revealed that more than 61% of all transcripts are subjected to AS under normal growth conditions in Arabidopsis thaliana.Citation7 To date, substantial research has demonstrated that AS provides versatility for gene expression, in order to fine-tune the function of regulatory factors and modulate the responses to the developmental and physiological signals in Arabidopsis.Citation8
2 Alternative splicing and the spliceosome
2.1 Basic types of alternative splicing events
High-throughput sequencing revealed that up to 95% of intron-containing genes in humans are subject to AS.Citation9 In Arabidopsis, at least 61% of genes undergo AS, followed by 46% in rice.Citation10 In addition, extensive studies indicate that the estimated figures will increase when more RNA-seq data are available, particularly from specific tissues/cells at different developmental stages and under stressful conditions.Citation4 Based on splice site selection, AS can be classified into four main types: (1) intron retention (IR); (2) exon skipping (ES); (3) alternative 5’splice sites (Alt 5’ss); and (4) alternative 3’splice sites (Alt 3’ss). In humans, ES accounts for 38.4%, followed by Alt 3’ss (18.4%) and Alt 5’ss (7.9%), and IR is the least (2.8%). In contrast, IR is the most frequent AS event in plants. In Arabidopsis, IR accounts for 39.6%, followed by Alt 3’ss (15.5%) and Alt 5’ss (7.5%), and ES is the rarest (2.7%).Citation4
2.2 Characterized spliceosome components in Arabidopsis
Alternative splicing takes place in the spliceosome. A vast number of genetic analysis and biochemical studies, especially in yeast and humans, has led to a consensus view that pre-mRNA splicing is largely conserved in all organisms.Citation11 The spliceosome is assembled dynamically in a stepwise manner, depending on multiple interactions among RNAs and/or proteins, affording the flexibility and fidelity required for splicing. Several distinct sub-complexes (i.e. E, A, B, Bact, B*, C, P and ILS complex), which represent spliceosome assembly, activation, splicing and disassembly stages, sequentially form along with specific proteins dissociated and recruited during the splicing process.
The spliceosome in plants is also composed of five snRNAs, consisting of U1, U2, U4, U5, and U6, which are packaged as small ribonucleoprotein particles with respective sets of proteins (snRNPs), plus a variety of non-snRNPs associated with pre-mRNAs.Citation12 To date, multiple sequence conserved splicing factors and regulators have been functionally characterized in Arabidopsis, which regulate many aspects of development and stress responses.Citation13 However, the dynamic interactions among them that are implicated in recruitment and dissociation are much less known than those in yeast and human.
3 Involvement of alternative splicing in plant development and stress response
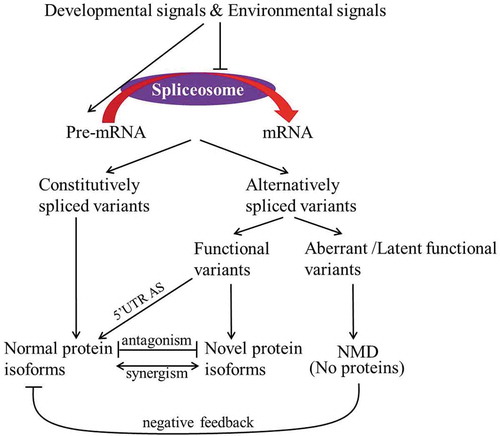
Various mRNAs generated from one single gene via AS may be proceeded by several pathways in plants, mainly as follows: (1) the mRNAs are translated into functional or truncated proteins which display loss or gain of function, providing a means of synergistically or antagonistically fine-tuning biological processes; (2) the AS located in the untranslated region (UTR) affects the stability or translational efficiency of mRNA, showing a rapid means of controlling the protein level; and (3) the non-translated mRNAs containing premature termination codons (PTCs) are targeted for degradation by the nonsense-mediated decay (NMD) pathway, functioning as a negative feedback loop to control the amount of functional proteins (). AS is a crucial regulatory mechanism for almost every aspect of development processes and stress responses in plant lifespan via the above mentioned pathways.
Figure 1. Pathways that alternative splicing variants are processed in Arabidopsis. Alternative splicing is catalyzed by the spliceosome. The developmental and environmental signals enhance alternatively spliced variants by dramatically increasing development- and stress-responsive pre-mRNAs which compete for the limited splicing machinery or repressing splicing machinery activity. Various mRNAs generated from one single gene by alternative splicing may be translated into normal or novel proteins or be degraded by NMD pathway. Moreover, the normal and novel proteins act in an antagonistic or synergistic manner. The 5ʹUTR-located alternative splicing usually has an effect on translation efficiency. The blunt end represents “repression”. UTR, untranslated region; NMD, nonsense-mediated decay. Alternative splicing is catalyzed by the spliceosome. The developmental and environmental signals enhance alternatively spliced variants by dramatically increasing development- and stress-responsive pre-mRNAs which compete for the limited splicing machinery or repressing splicing machinery activity. Various mRNAs generated from one single gene by alternative splicing may be translated into normal or novel proteins or be degraded by NMD pathway. Moreover, the normal and novel proteins act in an antagonistic or synergistic manner. The 5ʹUTR-located alternative splicing usually has an effect on translation efficiency. The blunt end represents “repression”. UTR, untranslated region; NMD, nonsense-mediated decay
3.1 Alternative splicing in seed dormancy and germination regulation
Seed dormancy is an important adaptive trait. DELAY OF GERMINATION 1 (DOG1), which is the first major QTL associated with seed dormancy, was cloned and identified to be seed-specifically expressed in Arabidopsis.Citation14 The accumulation of DOG1 protein was positively associated with the depth of seed dormancy.Citation15 The homologs of DOG1 in other crops were also subsequently identified to be required for seed dormancy. In Arabidopsis, DOG1 is alternatively spliced and generates five transcripts, namely DOG1-α, DOG1-β, DOG1-γ, DOG1-δ, and DOG1-ε, in which DOG1-β, DOG1-γ, and DOG1-ε encode the same protein.Citation14 Genetic analysis indicates that the expression of single DOG1 variants driven by the 35S promoter leads to protein accumulation to fully complement the seed dormancy defects,Citation16 suggesting that each protein could function properly in vivo. In contrast, the overexpression of a single DOG1 variant driven by the endogenous promoter resulted in a lack of DOG1 protein and failed to restore the dormancy phenotype, while the simultaneous expression of at least two variants from the endogenous promoter accumulated DOG1 protein and largely rescued the dormancy phenotype. These genetic analyses collectively suggest that a single variant is functional, but requires two or more isoforms to protect the DOG1 protein from degradation,Citation16 suggesting the synergistic roles of DOG1 AS variants. Interestingly, the expression of DOG1 is also regulated by alternative polyadenylation and antisense at the posttranscriptional level.Citation17,Citation18
Seed germination is the starting point in the plant life cycle, which is tightly linked with seed dormancy in terms of physiological state. PHYTOCHROME INTERACTING FACTOR 6 (PIF6), highly expressed in late stage of seed maturation, yields two transcripts by AS, namely PIF6-α and PIF6-β, respectively. PIF6-α encodes the full-length protein, including a phytochrome-binding motif and a bHLH motif. In contrast, PIF6-β produces a truncated protein lacking the bHLH motif because of the skipping of the third exon. Overexpression of PIF6-α phenocopied with wild type regarding the germination ratio, while PIF6-β overexpressors enhanced the germination potential,Citation19 indicating an antagonistic effect for PIF6 AS variants on seed germination with an unknown mechanism. AS is regulated by spliceosome components. PTB1 and PTB2, which are two closely related polypyrimidine tract-binding proteins in Arabidopsis, are key splicing regulators that influence widespread AS. Interestingly, PTB misexpression results in an increase of PIF6-β, coinciding with the altered rates of ABA-dependent seed germination. In addition, PIF6-β was elevated in sc35-scl quintuple mutant (scl28 scl30 scl30a scl33 sc35), resulting in a higher seed germination rate.Citation20
3.2 Alternative splicing in circadian clock control
Biological rhythms with a period of about 24 hours are regulated by circadian clock, which not only confers a daily rhythm in growth and metabolism but also interacts with signaling pathways involved in plant responses to the environment to provide plants with an adaptive advantage.Citation21 Circadian clock regulation can be divided into input signals, core oscillators, and output pathways. The core oscillator is mainly composed of feedback regulation loops, including morning factors CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), later-expressed PSEUDO RESPONSE REGULATOR 9 (PRR9) and PRR7, and evening factor TIMING OF CAB EXPRESSION 1 (TOC1).Citation21 Recently, extensive researches have indicated that AS makes an important contribution to the circadian regulation of gene expression. Clock genes such as CCA1, LHY, TOC1, PRRs, and EARLY FLOWERING 3 (ELF3) are subjected to ASCitation22–24 (). Temperature and light are important environmental factors modulating clock function partly by AS (, 2c). Low temperature induces AS events of LHY, PRR3 and PRR5, but suppresses those of CCA1 and ELF3, while PRR7 and TOC1 AS events are accumulated at both low and high temperaturesCitation23,Citation24 (). The photoperiod, light quality, and light intensity also influence the AS of TOC1, ELF3, CCA1, and LHYCitation24–27 (). Plants use circadian clock to monitor the daylength in response to seasonal changes.Citation28 The AS of TOC1 and ELF3 was suppressed during short days,Citation24 indicating the photoperiod influences AS patterns of clock genes. In the case of LHY, an inclusion of exon5a introduces a PTC and is predicted to reduce the amount of functional LHY at low temperature, and the intron retention in 5ʹUTR is robustly elevated when exposed to red light.Citation26 For CCA1, the fourth intron retention variant CCA1-β decreased at low temperature but accumulated under strong light intensity,Citation25,Citation29 but the CCA1-β protein was induced to accumulated in the cytoplasm under high temperature.Citation30
Figure 2. Alternative splicing control of the circadian clock. (a) Alternative splicing of circadian clock genes, including CCA1, LHY, PRR3, PRR5, PRR7, TOC1, and ELF3. Reported alternative splicing events are shown below the gene structure. The black box and bar represent UTRs; the yellow box and bar indicate the exons; the gray box and bar denote the introns. The figure at the top of the gene structure indicates the order of the exon. The alternative splicing events are named according to the alternative splicing types and positions. (b) and (c) Alternative splicing events occurring under different conditions, such as temperatures (b) and lights (c). The arrow indicates the up- or down-regulated alternative splicing event at the indicated conditions. The gray and yellow box represent the intron and exon subjected to degradation via nonsense mediated decay pathway, respectively. (d) The identified splicing factors and regulators in spliceosome conferring circadian clock regulation via alternative splicing of clock genes. The same color indicates the corresponding splicing factor and its specific target as reported, and the other splicing factors affect the alternative splicing of all the clock genes
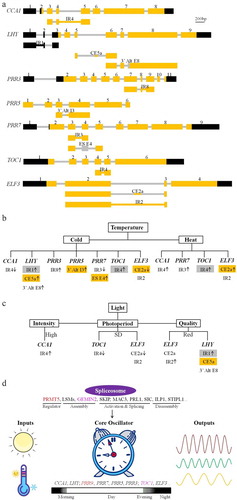
CCA1-α is necessary for proper biological rhythm regulation, while the CCA1-β protein contains a dimerization domain, but lacks a Myb DNA-binding domain at the N terminus. CCA1-β competitively interacts with LHY to antagonistically attenuate the association with CCA1-α, thus abolishing the ability of DNA binding activity.Citation31 In addition to the antagonistic roles of AS in circadian control, multiple abnormal mRNAs produced by circadian clock genes, including PRR5, PRR7, TOC1, LHY, and ELF3, were identified to be targeted by the NMD pathway when exposed to environmental factors, such as temperature fluctuations and photoperiod changesCitation22,Citation23 (, 2c). Despite the fact that the dominant roles in circadian clock control for CCA1 and LHY at different temperatures can be explained at least partly explained by antagonistically AS,Citation32 the functional modes of the considerably validated AS events of circadian clock genes are poorly understood. However, the AS regulation of circadian clock-regulated genes GLYCINE-RICH RNA BINDING PROTEINS 7 (GRP7) and GRP8 was well studied.Citation33,Citation34 Both GRP7 and GRP8 are negatively auto-regulated and reciprocally regulated by directly binding to their own and mutual mRNAs to promote the alternative usage of a cryptic intronic 5ʹ splice site at the expense of their fully spliced transcripts. The yielding of alternative transcripts with PTC, which are linked to NMD, leads to a decrease in the abundance of GRP7 and GRP8 .Citation35,Citation36
During the last decade, increasing evidences have indicated that spliceosome components are implicated in the circadian rhythm by regulating the AS of core circadian clock genes at the posttranscriptional level (). Mutations in PROTEIN ARGININE METHYLTRANSFERASE 5 (PRMT5), which methylates arginine in Sm and LSM spliceosomal proteins, were first identified to lengthen the clock period by the disturbed AS of PRR7 and PRR9.Citation37 However, the lengthened clock in lsm4 and lsm5 might be associated with the AS of other clock genes, because the splicing of PRR9 is normal in lsm4 and lsm5.Citation38 Subsequently, mutants in several spliceosome-associated genes, including SKI-INTERACTING PROTEIN (SKIP), SPLICEOSOME TIMEKEEPER LOCUS 1 (STIPL1), INCREASED LEVEL OF POLYPLOIDY1–1D (ILP1), GEMIN2, SIC, PLEIOTROPIC REGULATORY LOCUS 1 (PRL1), and MODIFIER OF snc1 4 (MOS4)-ASSOCIATED COMPLEX 3 (MAC3), were identified. Interestingly, skip, stipl1, ilp1, sic, prl1, and mac3 confer a long period by altering the AS of multiple circadian clock genes, while the gemin2 mutant shortened the period partly by altering the AS of TOC1. Moreover, SKIP, SIC, and GEMIN2 control the circadian clock by AS in a temperature responsive manner. Mutations in SKIP confer long periods at low temperatures, but not at high temperatures.Citation39 Deficiency in SIC impaired temperature compensation.Citation40 In contrast, the short period resulting from the loss of function of GEMIN2 is most evident at high temperatures.Citation31 Notably, a recent study on PRE-mRNA PROCESSING FACTOR 40 (PRP40) suggests that not all the spliceosome components are involved in circadian clock control.Citation41 Overall, the Arabidopsis circadian clock requires spliceosome activity to mediate the AS of circadian clock transcripts to establish the period, albeit by regulating the AS of different clock genes.
3.3 Alternative splicing in photomorphogenesis
PIF3 acts as a light response repressor in Arabidopsis,Citation42 of which expression regulation has been well studied in the transcriptional and translational level. Recently, the AS of PIF3 at the posttranscriptional level, which results in an intron retention in 5ʹUTR, was identified to be phyB-dependent under a short-day light cycle.Citation43 The AS of UTR influences the coding capacity of the genome and translational efficiency, especially 5ʹUTR, by producing mRNA variants with divergent upstream open reading frame (uORF) or riboswitches.Citation44 Further analysis indicated that the AS in 5ʹUTR inhibits PIF3 protein translation in vitro and in vivo. Consistently, a complementation assay indicated that the intron retention variant failed to rescue the elongated hypocotyl phenotype under the red light.Citation43 Moreover, a considerable number of genes were regulated by uORF-mediated translation inhibition with respect to the light response,Citation45 albeit with an unknown biological function. PIF3 is also demonstrated to confer freezing tolerance by stabilizing the phyB thermosensor,Citation46 but the role of the AS in PIF3 in freezing tolerance remains uncovered.
3.4 Alternative splicing in flowering time control
The appropriate timing of flowering is crucial for plants’ reproductive success. Arabidopsis accessions exhibit different flowering time, partly by the various usage of alternative splice sites of FLOWERING LOCUS C (FLC),Citation47 which is the central floral repressor integrating the endogenous and environmental pathways in the flowering time control network. In Col-0 accession, FLC is alternatively spliced,Citation48 but the significance of the AS variants in flowering is unknown. However, FLC is the best-characterized gene which is involved in the cross-regulation of the sense and antisense transcript.Citation49 The FLC antisense transcript COOLAIR extends beyond the end of the sense transcript, which functionally repressed FLC expression at the early stage of cold inductionCitation50,Citation51 (). Due to the AS plus with alternative 3ʹpolyadenylation, the antisense transcript produces two main classes of COOLAIR isoforms: the proximally polyadenylated class I (terminating at the sixth intron of FLC gene locus) and the distally polyadenylated class II (terminating at the promoter of FLC gene locus)Citation50,Citation51 (). Vernalization influences COOLAIR splicing with a consequence of an increase of class I/class II ratioCitation51 (), implying their different effects on FLC expression level. Recently, much progress has been achieved in the identification of splicing factors (), including RZ-1B and RZ-1 C, BAD RESPONSE TO REFRIGERATION 2a (BRR2a), SC35 and SC35-like, U2AF65, KHZs, which target FLC sense transcript to favor the splicing efficiency, but with the usage of unnatural splice sites. Interestingly, SKIP mediates FLC expression by the splicing of SERRATED LEAVES AND EARLY FLOWERING (SEF) pre-mRNA to control flowering time.Citation52 Another evolutionarily conserved splicing factor PRP8 (), in which mutations lead to flowering time delay, was identified to repress FLC expression by favoring class I isoform production,Citation53 suggesting that splicing of COOLAIR is functionally important. In agreement, loss of GRP7 reduces class I/class II ratio but upregulates FLC expressionCitation54 (). In contrast, class I isoform is increased in nsra mutant (), resulting in an early flowering phenotype.Citation55 Taken together, these results suggest that class I is associated with FLC repression, but class II is linked with high FLC expression levels.
Figure 3. AS events and their regulators involved in flowering time control. (a) Alternative splicing of flowering-related genes, including FLC, COOLAIR, FLM, MAF2, and CO. Reported alternative splicing events are shown. The yellow box indicates the exon; the gray bar denotes the intron; the black box represent UTR or long non-coding RNA. (b) Alternative splicing events occurring under different temperatures. The arrow indicates the up- or down-regulated alternative splicing event at the indicated conditions. The yellow box represent the variants subjected to degradation by NMD. (c) The identified splicing factors and regulators in spliceosome conferring flowering time control via alternative splicing of flowering-related genes. The same color or the star indicate the corresponding splicing factor and its target as reported, and the splicing factors involved in the alternative splicing of MAF2 and CO are unknown
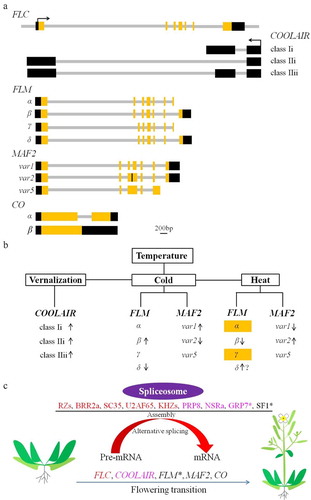
The flowering time is regulated by endogenous factors and environmental signals. Among environmental cues, little is known about how flowering is controlled in response to ambient temperature.Citation56 FLOWERING LOCUS M (FLM), an FLC-clade member, interacts with floral repressor SHORT VEGETATIVE PHASE (SVP) to repress flowering.Citation40,Citation57 FLM undergoes AS, yielding four AS isoforms, namely FLM-α, -β, -γ, and -δCitation58 (). FLM-β and FLM-δ were the dominant isoforms by selectively skipping the second exon and third exon, respectively, which encodes part of the MIKC intervening (I) region for protein–protein interaction.Citation58 FLM-β dominates in cool temperature (16°C) and FLM-δ accumulates in warm temperature (27°C) (). Initially, it was widely accepted that FLM functions in a temperature-dependent manner by the ratio of FLM-β/FLM-δ, because SVP-FLM-δ heterodimer abolished the DNA binding activity compared with SVP-FLM-β, thereby releasing the downstream floral activator to promote flowering at a warm temperature.Citation40,Citation57 Most recently, several lines of evidences have suggested that a high temperature accelerates flowering, predominantly by the down-regulation of FLM-β expression via producing the NMD-sensitive variants (), which is supported by the fact that the thermal induction of flowering is not abolished in flm-δ, but in upf mutants.Citation59,Citation60 Collectively, early flowering under an elevated temperature is only caused by the repression of FLM-β, rather than the decrease of the FLM-β/FLM-δ ratio.Citation40,Citation57,Citation59,Citation60 FLM AS is regulated by splicing factors (), for example, inactivation of SF1 decreased the ratio of FLM-β/FLM-δ,Citation61 and overexpression of GRP7 selectively favors the formation of FLM-β.Citation62
Similarly, another FLC-clade member called MADS-AFFECTING FLOWERING 2 (MAF2), which functions redundantly with FLM,Citation63 is also subjected to AS in a temperature-dependent manner.Citation64,Citation65 Three main transcripts are generated from the MAF2 gene locus, namely MAF2var1, -var2, and -var5Citation64,Citation65 (). MAF2var1, acting as a repressor in flowering by interacting with SVP to regulate downstream gene expression, is induced by low temperature (16°C) (). When the temperature reaches 27°C, the MAF2var2 isoform predominates (). MAF2var2 lacks part of the K-domain and all of the C-domain by introducing the third intron, which disrupts the interaction with SVP. Further genetic analysis indicates overexpression of var2 flowers at the same time with wild type.Citation64 Therefore, at low temperatures MAF2 represses flowering in parallel with FLM by interacting with SVP, providing an additional input for the control of the ambient temperature-mediated flowering. Although ectopic expression of the third isoform MAF2var5 leads to early flowering, the endogenous MAF2var5 exhibits a very low expression level at all tested temperatures in vivo, suggesting a weak contribution to the flowering time in Col.Citation65 However, the biological significance of the rare variants of FLM and MAF2 is required to be determined in specific accessions.
Apart from the central flowering repressors mentioned above, CONSTANS (CO), the flowering activator in the photoperiod pathway,Citation66 also undergoes alternative splicing. CO produces two protein variants: the full-length COα that is equivalent to the well-known CO protein for flowering induction,Citation67 and the truncated COβ lacking the DNA-binding domain (). COβ interacts with COα to attenuate its function by competitively compromising the formation of COα-HAP5a dimers,Citation68 thereby affecting the DNA-binding activity on FT promoter. Furthermore, COβ, resistant to E3-mediated protein degradation, induces the interactions between CO-α and CO-destabilizing E3 ligase but inhibits the association between CO-α and CO-stabilizing E3 ligase, ensuring its diurnal accumulation dynamics during photoperiodic flowering.Citation69 In spite of the identifications of these AS events involved in flowering time control, the regulators of these AS events are poorly understood, and not all splicing factors regulate flowering by splicing the same target (). Therefore, the characterizations of splicing regulators or factors will provide a new layer to establish the flowering regulation network.
3.5 Alternative splicing in floral organs regulation
SR proteins are crucial regulators in constitutive and alternative splicing and other aspects of mRNA metabolism.Citation70 More interestingly, most SRs are subjected to alternative splicing, although the biological functions are largely unknown.Citation71 SR45, encoding a unique plant SR protein, was identified to yield the two transcripts of SR45.1 and SR45.2, which have distinct roles during normal plant development and salt stress.Citation72,Citation73 Compared with SR45.1, SR45.2 replaces eight amino acids (TSPQRKTG) with a single arginine by the alternative usage of 21 nucleotides in 3ʹ splice site. Loss-of-function mutant sr45–1, which affects both isoforms, shows several developmental defects, including defects in petal development, root growth, and salt tolerance. Genetic analysis revealed that SR45.1-GFP complements the flower petal and salt-sensitive phenotypes, but not the root growth.Citation72,Citation73 Conversely, SR45.2-GFP only rescues root growth, and not others.Citation73 Interestingly, both isoforms are able to rescue the hypersensitivity to glucose and ABA.Citation74 Collectively, the conclusion can be drawn that both isoforms are functional, albeit with diverse roles.
Auxin response factors (ARFs) mediate multiple cellular responses to auxin by activating or repressing downstream developmental genes.Citation75 Different splice variants have been identified for several ARFs. In the case of ARF8, the full-length ARF8.1 contains 14 exons and ARF8.2 is alternatively spliced, leading to a premature stop codon four nucleotides ‘GTAA’ downstream of the 3ʹ end of exon 13. ARF8.3, which lacks exon1 and 34 nucleotides of exon 2, was annotated in Araport11 without function annotation.Citation76 Recently, a third splice variant ARF8.4, the eighth in-frame intron retention version compared with ARF8.2, was identified to be specifically expressed during late stages of stamen development. Genetic and molecular analysis revealed that ARF8.4 controls filament elongation and endothecium lignification by directly regulating the expression of AUX/IAA19 and MYB26.Citation77 In contrast, ARF8.1 and ARF8.2 have no or a minor effect on stamen elongation. In addition, ARF8.2 and ARF8.4 collaboratively regulate anther dehiscence, while ARF8.1 is not involved.Citation77
3.6 Alternative splicing in gametogenesis
ARF5, also known as MP, is an important transcription factor in auxin signaling and activate downstream gene expression with elevated auxin levels to regulate diverse developmental processes, including embryo development, primary and lateral root formation, vasculature patterning, shoot apical meristem maintenance and floral and ovule initiation. Most recently, MP is identified to be expressed in ovule primordia with auxin minima to activate the expression of direct target genes by AS.Citation78 MP is subject to AS with the 11th intron retention mainly in inflorescences, which is designated as MP11ir and only accounts for approximately 6% of total transcripts. MP11ir encodes MP isoforms lacking C-terminal PB1 domain, which is required for the interaction between ARFs and AUX/IAA proteins,Citation79 thereby functioning independently of auxin levels and AUX/IAA. Complementation analysis indicated that MP11ir is not essential for the complementation of lateral branching, flower and pistil morphology. In terms of ovule development, both MP and MP11ir partially restores mp mutants, but MP11ir is more efficient,Citation78 suggesting MP11ir is tissue- and developmental-specific in ovule and both MP isoforms synergistically act in Arabidopsis development.
Besides the roles for AS of MP in ovule, AS of CYCLIN-DEPENDENT KINASE G1(CDKG1) is implicated in pollen development in response to ambient temperature.Citation80 Two AS events resulted from CDKG1 via the selective intron mainly encodes two protein isoforms CDKG1L (long) and CDKG1S (short). The long isoform exclusively localized in the nuclear, but the short isoform lacking two out of four SR domains and NLS localized in both the nuclear and cytoplasm. At low temperature, mostly the long isoform is produced, while at high temperature, both long and short sioforms are produced. Therefore, the ratio between the long and short isoforms is regulated by temperature.Citation81 Moreover, inactivation of CDKG1 results in male sterility by meiosis and pollen wall defects at high temperature, but is fertile at low temperature,Citation82,Citation83 suggesting the long isoform is required for pollen development at high temperature. In agreement, the long isoform could restore the cdkg1 mutant defect in terms of the male sterility at high temperature, while the short isoform could not.Citation80 The above mentioned results indicate CDKG1L and CDKG1S is regulated by temperature to antagonistically control pollen development. However, on the other hand, CDKG1L and CDKG1S synergistically mediate the correct splicing of U2AF65A in response to fluctuating ambient temperatures.Citation81
3.7 Alternative splicing in phytohormone signaling
As sessile organisms, plants have evolved sophisticated mechanisms including AS to fight against unfavorable stimuli. The ABSCISIC ACID (ABA) signal take vital actions against stress, in which many advances has been achieved in the PYL-PP2C-SnRK2 regulatory module.Citation84 In the absence of ABA, the PP2Cs interact with SnRK2s to inhibit its kinase activity by dephosphorylation, resulting in ABA signaling off. ABA binds to its receptor PYLs to promote the interactions with PP2Cs and inhibit their phosphotase activity, thereby releasing SnRK2s from the PP2C-SnRK2 complex.Citation85,Citation86 The released SnRK2s phosphotase the downstream effectors and activate ABA signaling.Citation85 HYPERSENSITIVE TO ABA 1 (HAB1), a component of Group A PP2C, undergoes AS and yields two major isoforms, namely HAB1.1 and HAB1.2.Citation87,Citation88 HAB1.2 differs from HAB1.1 in terms of retaining the third intron to yield a truncated protein by a premature stop codon which abolishes the phosphatase activity. However, the truncated protein HAB1.2 still physically interacts with, but fails to dephosphorylate SnRK2.6.Citation87 In the presence of ABA, HAB1.2 displays increased expression level and the ratio of HAB1.1 to HAB1.2 is decreased, thereby enhancing ABA signaling. Therefore, HAB1.2 isoform antagonistically functions with HAB1.1 as a dominant-negative regulator in ABA signaling.Citation87 RNA BINDING PROTEIN 25 (RBM25), an evolutionarily conserved splicing factor, has been identified to mediate the AS of HAB1.Citation88 The inactivation of RBM25 increases the ratio of HAB1.2 to HAB1.1, especially in the presence of ABA. Consistently, the rbm25 mutant exhibits substantially increased sensitivity to increasing concentrations of ABA in terms of seed germination and cotyledon greening. Furthermore, solid evidences have indicated that overexpression of HAB1.1 completely complements ABA-sensitive phenotypes of the rbm25 mutant in genetic analysis and RBM25 is capable of binding HAB1 pre-mRNA at a molecular level, suggesting that RBM25 directly regulates the AS of HAB1 to fine-tune ABA signaling.Citation87,Citation88
JASMONATE ZIM-domain (JAZ) transcription factors act as repressors in JASMONATE (JA) signaling, which is induced by abiotic and biotic stresses.Citation89 JAZ proteins contain two highly conserved sequence motifs, including that the Jas domain and the ZIM domain. The Jas domain for interacting with E3 to destabilize the repressors and the ZIM/TIFY domain for JAZs interactions are key features of JAZ proteins.Citation90 JAZ10 was experimentally identified to produce three alternative splicing variants, namely JAZ10.1, JAZ10.3, and JAZ10.4. JA10.3 weakly interacts with the E3 ligase COI1 due to lacking seven amino acids from the C-terminal end of the Jas domain,Citation91 while JAZ10.4 abolishes the entire Jas domain, thereby being resistant to degradation.Citation90 Overexpression of JAZ10.3 confers partial insensitivity to JA,Citation91 and the ectopic expression of JAZ10.4 shows more severe JA-insensitive phenotypes,Citation90 which is consistent with the biochemical results. The Jas intron-dependent AS of JAZ transcription factors was identified to be a general mechanism to desensitize or deactivate JA responses, including JAZ5, JAZ6 and JAZ9. Moreover, recent research suggested that MED25 recruits splicing factors PRP39a and PRP40a to prevent JAZ AS-mediated excessive desensitization of JA signaling.Citation92
Another case is BRI-EMS-SUPPRESSOR1 (BES1), which is the downstream transcription factor in BR signaling. BES1 regulates the expression of thousands of target genes in response to BRs in the nucleus.Citation93 BES1 shows two variants: BES1-S and BES1-L. Compared with the canonical and widely used short BES1-S, BES1-L, with two extra exons and one intron instead of BES1-S 5ʹUTR, encodes an N-terminal bipartite nuclear localization signal domain, which enables BES1-L to exclusively localize in the nucleus.Citation94 In contrast, BES1-S localizes in both the nucleus and cytoplasm, and nuclear localization is accumulated by BR treatment. Interestingly, the nuclear-localized BES1-L promotes the nuclear localization of BES1-S and BZR1 via dimerization. Further phenotypic analysis of the overexpression of BES1 variants indicated that BES1-L is more important in BR signaling in Arabidopsis.Citation94 Intriguingly, BES1-L is a recently evolved isoform because the BES1-L-like transcript is present in the closer relative A. lyrata, but absent in distant relatives Capsella rubella and Thellungiella salsuginea,Citation94 suggesting a special and essential role for BES1-L in Arabidopsis.
3.8 Alternative splicing in abiotic stress response
AS also affects protein subcellular localization. ZINC-INDUCED FACILITATOR-LIKE 1 (ZIFL1) is a paralog of major facilitator superfamily (MFS) transporter ZIF1 that is localized in tonoplasts and required for zinc tolerance.Citation95 The ZIFL1 gene generates three distinct transcripts, namely ZIFL1.1-ZIFL1.3. ZIFL1.1 encodes the full-length protein containing 12 membrane-spanning regions, while ZIFL1.2 uses an alternative start codon in-frame in the fourth exon, resulting in a protein lacking two first N-terminal membrane-spanning regions. The third transcript ZIFL1.3 causes frame shift by an alternative 3ʹ splice site selection in the fourteenth intron, abolishing the two last N-terminal membrane-spanning regions by PTC.Citation96 Genetic and complementation analysis indicated that ZIFL1.1 is specifically required for cellular auxin efflux during shootward auxin transport at the root tip by regulating PINs abundance and ZIFL1.3 exclusively confers drought tolerance by modulating stomatal closure, which can be explained by the divergent subcellular localization, with ZIFL1.1 in the tonoplast membrane of root cells and ZIFL1.3 in plasma membrane of stomatal guard cells.Citation96,Citation97 Interestingly, no roles in auxin transport or drought tolerance for ZIFL1.2 were observed, suggesting that the two first N-terminal membrane-spanning regions are essential.
HOT SHOCK TRANSCRIPTION FACTOR A2 (HSFA2) is a key regulator in both basal and aquired themotolerance.Citation98,Citation99 The HSFA2 locus consists of two exons and one intron. The alternative splicing of HSFA2 is temperature-dependent.Citation100 At 22°C, HSFA2 generates the full-length transcript. At 37°C, another nonfunctional variant named HSFA2-II, which contains a premature termination codon by retaining a 31 bp cryptic miniexon from the intron, is generated and subsequently degraded by nonsense-mediated pathway. When the temperature reaches 42°C, HSFA2 will generate a third transcript HSFA2-III, which retains the upstream region of the intron splited by the miniexon to encodes the short HsfA2 protein (referred to as S-HsfA2) with 129 amino acids in length. As the temperature increases to 45°C, HSFA2-II disappears, but HSFA2-III accumulates. S-HSFA2, which is derived from HSFA2-III, directly binds to its own promoter to activate the fully spliced HASFA2 expression to confer thermotolerance.Citation100 Notably, not only HSFA2 but also other heat shock transcription factors, including HSFA7b, HSFB1, HSFB2a and HSFA4c, undergo AS at 42°C,Citation100 indicating that AS for HSFs is a general mechanism underlying heat stress. Interestingly, loss of function of SF1 promotes the production of productive HSFA2, leading to increased resistance to heat stress.Citation61
ZINC-INDUCED FACILITATOR 2 (ZIF2) encodes a functional transporter that is Zn-induced. 5ʹUTR of ZIF2 generates two splice variants, namely ZIF2.1 and ZIF2.2, which encode the same protein. Compared with ZIF2.1, ZIF2.2 is 139 nucleotides longer in length due to retaining the first intron in the 5ʹUTR to favor translation,Citation101 which is a conserved but poorly understood mechanism. The regulation of translation efficiency driven by ZIF2.2 5ʹUTR largely depends on a predicted stable stem loop upstream of the start codon which is excised in the ZIF2.1. Therefore, ZIF2.2 confers greater Zn tolerance under excess Zn than ZIF2.1 in a Zn-responsive manner by efficiently enhancing protein translation.Citation101
4 Prospects
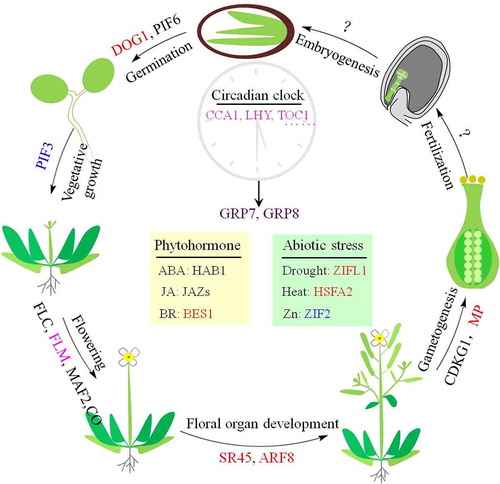
In the last decades, many AS events that are developmentally regulated or stress induced and their corresponding biological functions with different underlying mechanisms have been well studied in the model plant Arabidopsis (). However, AS events functioning in fertilization and embryogenesis have not been uncovered yet. It will be of great interest to intensively explore novel AS events that play vital roles during these processes, thereby establishing elaborate AS-regulatory mechanisms during Arabidopsis life cycle from seed germination to embryogenesis.
Figure 4. Development-regulated and stress-induced alternative splicing events in Arabidopsis. Alternative splicing is a general mechanism in which gene expression is subtly regulated. During Arabidopsis life cycle, the well-studied alternatively spliced genes that regulate plant development and confer stress tolerance are shown at the corresponding positions. The question mark represents the unknown alternative splicing events. The genes that generate alternative splicing variants playing potentially antagonistic or synergistic roles, altered translation efficiency, and linked to NMD are represented black, red, blue and purple, respectively. The genes in magenta indicate the alternative splicing variants are proceeded by combined functional modes
Additionally, extensive studies have uncovered numerous alternative splicing events, especially in unfavorable conditions via developed sequencing technology in crops in recent years. However, compared with those from Arabidopsis, only several genes in crops have been characterized. Whether AS event regulation in polyploidy crops, such as wheat and oilseed rape, shares the same mechanism as that in Arabidopsis remains unclear. The characterization and functional analysis of the AS events for specific tissues, time or conditions will help us to better understand the mechanisms of plant development and adaptation to the fluctuating environments, thereby promoting application for genetic improvements in crops.
Conflicts of interest
The authors declare no conflicts of interest.
Availability of data and material
Not applicable.
Code availability
Not applicable.
Authors contribution
Not applicable.
Ethics approval
Not applicable.
Consent to participate
All listed authors are consent to participate.
Consent for publication
All listed authors are consent for publication.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (31700274).
Additional information
Funding
References
- Chen W, Moore MJ. Spliceosomes. Current Biology. 2015;25:1–12.
- Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136(4):701–718. doi:10.1016/j.cell.2009.02.009.
- Kalyna M, Simpson CG, Syed NH, Lewandowska D, Marquez Y, Kusenda B, Marshall J, Fuller J, Cardle L, McNicol J, et al. Alternative splicing and NMD modulate expression. Nucleic Acids Res. 2012;40:2454–2469.
- Carvalho RF, Feijão CV, Duque P. On the physiological significance of alternative splicing events. Protoplasma. 2012;250:639-650.
- Leff SE, Rosenfeld MG. Complex transcriptional units: diversity in gene expression by alternative RNA splicing. Annu Rev Biochem. 1986;55(1):1091–1117. doi:10.1146/annurev.bi.55.070186.005303.
- Werneke JM, Chatfield JM, Ogren WL. Alternative mRNA splicing generates the two ribulose bisphosphate carboxylase/oxygenase activase polypeptides in Spinach and Arabidopsis. Plant Cell. 1989;1(8):815–825. doi:10.1105/tpc.1.8.815.
- Marquez Y, Brown JW, Simpson C, Barta A, Kalyna M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 2012;22(6):1184–1195. doi:10.1101/gr.134106.111.
- Shang X, Cao Y, Ma L. Alternative splicing in plant genes: a means of regulating the environmental fitness of plants. Int J Mol Sci. 2017;18:432.
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415.
- Chamala S, Feng G, Chavarro C, Barbazuk WB. Genome-wide identification of evolutionarily conserved alternative splicing events in flowering plants. Front Bioeng Biotechnol. 2015;3:33. doi:10.3389/fbioe.2015.00033.
- Käufer F, Potashkin J. The splicing machinery comparision among budding yeast, fission yeast and human. Nucleic Acids Res. 2000;28(16):3003–3010. doi:10.1093/nar/28.16.3003.
- Koncz C, deJong F, Villacorta N, Szakonyi D, Koncz Z. The spliceosome-activating complex: molecular mechanisms underlying the function of a pleiotropic regulator. Front Plant Sci. 2012;3. doi:10.3389/fpls.2012.00009.
- Meyer K, Koester T, Staiger D. Pre-mRNA splicing in plants: in vivo functions of RNA-binding proteins implicated in the splicing process. Biomolecules. 2015;5(3):1717–1740. doi:10.3390/biom5031717.
- Bentsink L, Jowett J, Hanhart CJ, Koornneef M (2006). Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA 103, 17042–17047.
- Finch-Savage WE, Footitt S. Seed dormancy cycling and the regulation of dormancy mechanisms to time germination in variable field environments. J Exp Bot. 2017;68(4):843–856. doi:10.1093/jxb/erw477.
- Nakabayashi K, Bartsch M, Ding J, Soppe WJ. Seed dormancy in Arabidopsis requires self-binding ability of DOG1 protein and the presence of multiple isoforms generated by alternative splicing. PLoS Genet. 2015;11(12):e1005737. doi:10.1371/journal.pgen.1005737.
- Kowalczyk J, Palusinska M, Wroblewska-Swiniarska A, Pietras Z, Szewc L, Dolata J, Jarmolowski A, Swiezewski S. Alternative polyadenylation of the sense transcript controls antisense transcription of DELAY OF GERMINATION 1 in Arabidopsis. Mol Plant. 2017;10(10):1349–1352. doi:10.1016/j.molp.2017.07.011.
- Cyrek M, Fedak H, Ciesielski A, Guo Y, Sliwa A, Brzezniak L, Krzyczmonik K, Pietras Z, Kaczanowski S, Liu F, et al. Seed dormancy in Arabidopsis is controlled by alternative polyadenylation of DOG1. Plant Physiol. 2016;170(2):947–955. doi:10.1104/pp.15.01483.
- Penfield S, Josse EM, Halliday KJ. A role for an alternative splice variant of PIF6 in the control of Arabidopsis primary seed dormancy. Plant Mol Biol. 2010;73(1–2):89–95. doi:10.1007/s11103-009-9571-1.
- Yan Q, Xia X, Sun Z, Fang Y. Depletion of Arabidopsis SC35 and SC35-like serine/arginine-rich proteins affects the transcription and splicing of a subset of genes. PLoS Genet. 2017;13(3):e1006663. doi:10.1371/journal.pgen.1006663.
- Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009;60(1):357–377. doi:10.1146/annurev.arplant.043008.092054.
- Mateos JL, De Leone MJ, Torchio J, Reichel M, Staiger D. Beyond transcription: fine-tuning of circadian timekeeping by post-transcriptional regulation. Genes. 2018;9:616.
- James AB, Syed NH, Bordage S, Marshall J, Nimmo GA, Jenkins GI, Herzyk P, Brown JW, Nimmo HG. Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell. 2012;24(3):961–981. doi:10.1105/tpc.111.093948.
- Kwon YJ, Park MJ, Kim SG, Baldwin IT, Park CM. Alternative splicing and nonsense-mediated decay of circadian clock genes under environmental stress conditions in Arabidopsis. BMC Plant Biol. 2014;14(1):136. doi:10.1186/1471-2229-14-136.
- Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010;20(1):45–58. doi:10.1101/gr.093302.109.
- Shikata H, Hanada K, Ushijima T, Nakashima M, Suzuki Y, Matsushita T (2014). Phytochrome controls alternative splicing to mediate light responses in Arabidopsis. Proc Natl Acad Sci USA 111, 18781–18786.
- Xin R, Zhu L, Salome PA, Mancini E, Marshall CM, Harmon FG, Yanovsky MJ, Weigel D, Huq E (2017). SPF45-related splicing factor for phytochrome signaling promotes photomorphogenesis by regulating pre-mRNA splicing in Arabidopsis. Proc Natl Acad Sci USA 114, E7018–E7027.
- Imaizumi T. Arabidopsis circadian clock and photoperiodism: time to think about location. Curr Opin Plant Biol. 2010;13(1):83–89. doi:10.1016/j.pbi.2009.09.007.
- Filichkin SA, Cumbie JS, Dharmawardhana P, Jaiswal P, Chang JH, Palusa SG, Reddy AS, Megraw M, Mockler TC. Environmental stresses modulate abundance and timing of alternatively spliced circadian transcripts in Arabidopsis. Mol Plant. 2015;8(2):207–227. doi:10.1016/j.molp.2014.10.011.
- Zhang S, Liu H, Yuan L, Li X, Wang L, Xu X, Xie Q. Recognition of CCA1 alternative protein isoforms during temperature acclimation. Plant Cell Rep. 2021;40(2):421–432. doi:10.1007/s00299-020-02644-7.
- Seo PJ, Park MJ, Lim MH, Kim SG, Lee M, Baldwin IT, Park CM. A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell. 2012;24(6):2427–2442. doi:10.1105/tpc.112.098723.
- Gould PD, Locke JC, Larue C, Southern MM, Davis SJ, Hanano S, Moyle R, Milich R, Putterill J, Millar AJ, et al. The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell. 2006;18(5):1177–1187. doi:10.1105/tpc.105.039990.
- Heintzen C, Nater M, Apel K, Staiger D (1997). AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc Natl Acad Sci USA 94, 8515–8520.
- Carpenter CD, Kreps JA, Simon AE. Genes encoding glycine-rich Arabidopsis thaliana proteins with RNA-binding motifs are influenced by cold treatment and an endogenous circadian rhythm. Plant Physiol. 1994;104:1015–1025.
- Staiger D, Zecca L, Wieczorek Kirk DA, Apel K, Eckstein L. The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J. 2003;33(2):361–371. doi:10.1046/j.1365-313X.2003.01629.x.
- Schoning JC, Streitner C, Meyer IM, Gao Y, Staiger D. Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic Acids Res. 2008;36(22):6977–6987. doi:10.1093/nar/gkn847.
- Sanchez SE, Petrillo E, Beckwith EJ, Zhang X, Rugnone ML, Hernando CE, Cuevas JC, Godoy Herz MA, Depetris-Chauvin A, Simpson CG, et al. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature. 2010;468(7320):112–116. doi:10.1038/nature09470.
- Perez-Santangelo S, Mancini E, Francey LJ, Schlaen RG, Chernomoretz A, Hogenesch JB, Yanovsky MJ (2014). Role for LSM genes in the regulation of circadian rhythms. Proc Natl Acad Sci USA 111, 15166–15171.
- Wang X, Wu F, Xie Q, Wang H, Wang Y, Yue Y, Gahura O, Ma S, Liu L, Cao Y, et al. SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell. 2012;24(8):3278–3295. doi:10.1105/tpc.112.100081.
- Pose D, Verhage L, Ott F, Yant L, Mathieu J, Angenent GC, Immink RG, Schmid M. Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature. 2013;503(7476):414–417. doi:10.1038/nature12633.
- Hernando CE, Garcia Hourquet M, De Leone MJ, Careno D, Iserte J, Mora Garcia S, Yanovsky MJ. A role for Pre-mRNA-PROCESSING PROTEIN 40C in the control of growth, development, and stress tolerance in Arabidopsis thaliana. Front Plant Sci. 2019;10:1019. doi:10.3389/fpls.2019.01019.
- Pham VN, Kathare PK, Huq E. Phytochromes and phytochrome interacting factors. Plant Physiol. 2018;176(2):1025–1038. doi:10.1104/pp.17.01384.
- Dong J, Chen H, Deng XW, Irish VF, Wei N. Phytochrome B induces intron retention and translational inhibition of PHYTOCHROME-INTERACTING FACTOR3. Plant Physiol. 2020;182(1):159–166. doi:10.1104/pp.19.00835.
- Roy B, Von Arnim AG. Translational regulation of cytoplasmic mRNAs. The Arabidopsis Book. 2013;11:e0165. doi:10.1199/tab.0165.
- Kurihara Y, Makita Y, Kawashima M, Fujita T, Iwasaki S, Matsui M (2018). Transcripts from downstream alternative transcription start sites evade uORF-mediated inhibition of gene expression in Arabidopsis. Proc Natl Acad Sci USA 115, 7831–7836.
- Jiang B, Shi Y, Peng Y, Jia Y, Yan Y, Dong X, Li H, Dong J, Li J, Gong Z, et al. Cold-induced CBF-PIF3 interaction enhances freezing tolerance by stabilizing the phyB thermosensor in Arabidopsis. Mol Plant. 2020;13(6):894–906. doi:10.1016/j.molp.2020.04.006.
- Caicedo AL, Stinchcombe JR, Olsen KM, Schmitt J, Purugganan MD (2004). Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc Natl Acad Sci USA 101, 15670–15675.
- Severing EI, Van Dijk AD, Morabito G, Busscher-Lange J, Immink RG, Van Ham RC. Predicting the impact of alternative splicing on plant MADS domain protein function. PLoS One. 2012;7(1):e30524. doi:10.1371/journal.pone.0030524.
- Ietswaart R, Wu Z, Dean C. Flowering time control: another window to the connection between antisense RNA and chromatin. Trends Genet. 2012;28(9):445–453. doi:10.1016/j.tig.2012.06.002.
- Swiezewski S, Liu F, Magusin A, Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis polycomb target. Nature. 2009;462(7274):799–802. doi:10.1038/nature08618.
- Csorba T, Questa JI, Sun Q, Dean C (2014). Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc Natl Acad Sci USA 111, 16160–16165.
- Cui Z, Tong A, Huo Y, Yan Z, Yang W, Yang X, Wang X. SKIP controls flowering time via the alternative splicing of SEF pre-mRNA in Arabidopsis. BMC Biol. 2017;15(1):80. doi:10.1186/s12915-017-0422-2.
- Marquardt S, Raitskin O, Wu Z, Liu F, Sun Q, Dean C. Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol Cell. 2014;54(1):156–165. doi:10.1016/j.molcel.2014.03.026.
- Xiao J, Li C, Xu S, Xing L, Xu Y, Chong K. JACALIN-LECTIN LIKE1 regulates the Nuclear Accumulation of GLYCINE-RICH RNA-BINDING PROTEIN7, influencing the RNA processing of FLOWERING LOCUS C antisense transcripts and flowering time in Arabidopsis. Plant Physiol. 2015;169(3):2102–2117. doi:10.1104/pp.15.00801.
- Bazin J, Romero N, Rigo R, Charon C, Blein T, Ariel F, Crespi M. Nuclear speckle RNA binding proteins remodel alternative splicing and the non-coding Arabidopsis transcriptome to regulate a cross-talk between auxin and immune responses. Front Plant Sci. 2018;9:1209. doi:10.3389/fpls.2018.01209.
- Wigge PA. Ambient temperature signalling in plants. Curr Opin Plant Biol. 2013;16(5):661–666. doi:10.1016/j.pbi.2013.08.004.
- Lee JH, Ryu HS, Chung KS, Pose D, Kim S, Schmid M, Ahn JH. Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science. 2013;342(6158):628–632. doi:10.1126/science.1241097.
- Scortecci KC, Michaels SD, Amasino RM. Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J. 2001;26(2):229–236. doi:10.1046/j.1365-313x.2001.01024.x.
- Sureshkumar S, Dent C, Seleznev A, Tasset C, Balasubramanian S. Nonsense-mediated mRNA decay modulates FLM-dependent thermosensory flowering response in Arabidopsis. Nature Plants. 2016;2(5):16055. doi:10.1038/nplants.2016.55.
- Lutz U, Pose D, Pfeifer M, Gundlach H, Hagmann J, Wang C, Weigel D, Mayer KF, Schmid M, Schwechheimer C. Modulation of ambient temperature-dependent flowering in Arabidopsis thaliana by natural variation of FLOWERING LOCUS M. PLoS Genet. 2015;11(10):e1005588. doi:10.1371/journal.pgen.1005588.
- Lee KC, Jang YH, Kim SK, Park HY, Thu MP, Lee JH, Kim JK. RRM domain of Arabidopsis splicing factor SF1 is important for pre-mRNA splicing of a specific set of genes. Plant Cell Rep. 2017;36(7):1083–1095. doi:10.1007/s00299-017-2140-1.
- Steffen A, Elgner M, Staiger D. Regulation of flowering time by the RNA-Binding proteins AtGRP7 and AtGRP8. Plant Cell Physiol. 2019;60(9):2040–2050. doi:10.1093/pcp/pcz124.
- Ratcliffe OJ, Nadzan GC, Reuber TL, Riechmann JL. Regulation of flowering in Arabidopsis by an LC Homologue. Plant Physiol. 2001;126(1):122–132. doi:10.1104/pp.126.1.122.
- Rosloski SM, Singh A, Jali SS, Balasubramanian S, Weigel D, Grbic V. Functional analysis of splice variant expression of MADS AFFECTING FLOWERING 2 of Arabidopsis thaliana. Plant Mol Biol. 2013;81(1–2):57–69. doi:10.1007/s11103-012-9982-2.
- Airoldi CA, McKay M, Davies B. MAF2 is regulated by temperature-dependent splicing and represses flowering at low temperatures in parallel with FLM. PLoS One. 2015;10(5):e0126516. doi:10.1371/journal.pone.0126516.
- Shim JS, Imaizumi T. Circadian clock and photoperiodic response in Arabidopsis: from seasonal flowering to redox homeostasis. Biochem. 2015;54(2):157–170. doi:10.1021/bi500922q.
- Song YH, Ito S, Imaizumi T. Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 2013;18:575–583.
- Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell. 2006;18(11):2971–2984. doi:10.1105/tpc.106.043299.
- Gil KE, Park MJ, Lee HJ, Park YJ, Han SH, Kwon YJ, Seo PJ, Jung JH, Park CM. Alternative splicing provides a proactive mechanism for the diurnal CONSTANS dynamics in Arabidopsis photoperiodic flowering. Plant J. 2017;89(1):128–140. doi:10.1111/tpj.13351.
- Barta A, Kalyna M, Reddy AS. Implementing a rational and consistent nomenclature for serine/arginine-rich protein splicing factors (SR proteins) in plants. Plant Cell. 2010;22(9):2926–2929. doi:10.1105/tpc.110.078352.
- Reddy AS, Shad Ali G. Plant serine/arginine-rich proteins: roles in precursor messenger RNA splicing, plant development, and stress responses. Wiley Interdiscip Rev RNA. 2011;2(6):875–889. doi:10.1002/wrna.98.
- Albaqami M, Laluk K, Reddy ASN. The Arabidopsis splicing regulator SR45 confers salt tolerance in a splice isoform-dependent manner. Plant Mol Biol. 2019;100(4–5):379–390. doi:10.1007/s11103-019-00864-4.
- Zhang XN, Mount SM. Two alternatively spliced isoforms of the Arabidopsis SR45 protein have distinct roles during normal plant development. Plant Physiol. 2009;150(3):1450–1458. doi:10.1104/pp.109.138180.
- Carvalho RF, Carvalho SD, Duque P. The plant-specific SR45 protein negatively regulates glucose and ABA signaling during early seedling development in Arabidopsis. Plant Physiol. 2010;154(2):772–783. doi:10.1104/pp.110.155523.
- Guilfoyle TJ, Hagen G. Auxin response factors. Curr Opin Plant Biol. 2007;10(5):453–460. doi:10.1016/j.pbi.2007.08.014.
- Cheng CY, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, Town CD. Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017;89(4):789–804. doi:10.1111/tpj.13415.
- Ghelli R, Brunetti P, Napoli N, De Paolis A, Cecchetti V, Tsuge T, Serino G, Matsui M, Mele G, Rinaldi G, et al. A newly identified flower-specific splice variant of AUXIN RESPONSE FACTOR8 regulates stamen elongation and endothecium lignification in Arabidopsis. Plant Cell. 2018;30(3):620–637. doi:10.1105/tpc.17.00840.
- Cucinotta M, Cavalleri A, Guazzotti A, Astori C, Manrique S, Bombarely A, Oliveto S, Biffo S, Weijers D, Kater MM, et al. Alternative splicing generates a MONOPTEROS isoform required for ovule development. Curr Biol. 2020;31:892–899.
- Hamann T, Benkova E, Baurle I, Kientz M, Jurgens G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002;16:1610–1615.
- Nibau C, Dadarou D, Kargios N, Mallioura A, Fernandez-Fuentes N, Cavallari N, Doonan JH. A functional kinase is necessary for Cyclin-Dependent Kinase G1 (CDKG1) to maintain fertility at high ambient temperature in Arabidopsis. Front Plant Sci. 2020;11:586870.
- Cavallari N, Nibau C, Fuchs A, Dadarou D, Barta A, Doonan JH. The cyclin-dependent kinase G group defines a thermo-sensitive alternative splicing circuit modulating the expression of Arabidopsis ATU2AF65A. Plant J. 2018;94:1010–1022.
- Nibau C, Lloyd A, Dadarou D, Betekhtin A, Tsilimigka F, Phillips DW, Doonan JH. CDKG1 is required for meiotic and somatic recombination intermediate processing in Arabidopsis. Plant Cell. 2020;32:1308–1322.
- Huang XY, Niu J, Sun MX, Zhu J, Gao JF, Yang J, Zhou Q, Yang ZN. CYCLIN-DEPENDENT KINASE G1 is associated with the spliceosome to regulate CALLOSE SYNTHASE5 splicing and pollen wall formation in Arabidopsis. Plant Cell. 2013;25:637–648.
- Hirayama T, Shinozaki K. Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci. 2007;12:343–351.
- Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev. 2010;24:1695–1708.
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E. ABA perception and signalling. Trends Plant Sci. 2010;15:395–401.
- Wang Z, Ji H, Yuan B, Wang S, Su C, Yao B, Zhao H, Li X. ABA signalling is fine-tuned by antagonistic HAB1 variants. Nat Commun. 2015;6:8138.
- Zhan X, Qian B, Cao F, Wu W, Yang L, Guan Q, Gu X, Wang P, Okusolubo TA, Dunn SL, et al. An Arabidopsis PWI and RRM motif-containing protein is critical for pre-mRNA splicing and ABA responses. Nat Commun. 2015;6:8139.
- Feng G, Yoo MJ, Davenport R, Boatwright JL, Koh J, Chen S, Barbazuk WB. Jasmonate induced alternative splicing responses in Arabidopsis. Plant Direct. 2020;4:e00245.
- Chung HS, Howe GA. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell. 2009;21:131–145.
- Yan Y, Stolz S, Chetelat A, Reymond P, Pagni M, Dubugnon L, Farmer EE. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell. 2007;19:2470–2483.
- Wu F, Deng L, Zhai Q, Zhao J, Chen Q, Li C. Mediator subunit MED25 couples alternative splicing of JAZ genes with fine-tuning of jasmonate signaling. Plant Cell. 2020;32:429–448.
- Yu X, Li L, Zola J, Aluru M, Ye H, Foudree A, Guo H, Anderson S, Aluru S, Liu P, et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 2011;65:634–646.
- Jiang J, Zhang C, Wang X. A recently evolved isoform of the transcription factor BES1 promotes brassinosteroid signaling and development in Arabidopsis thaliana. Plant Cell. 2015;27:361–374.
- Haydon MJ, Cobbett CS. A novel major facilitator superfamily protein at the tonoplast influences zinc tolerance and accumulation in Arabidopsis. Plant Physiol. 2007;143:1705–1719.
- Remy E, Cabrito TR, Baster P, Batista RA, Teixeira MC, Friml J, Sa-Correia I, Duque P. A major facilitator superfamily transporter plays a dual role in polar auxin transport and drought stress tolerance in Arabidopsis. Plant Cell. 2013;25:901–926.
- Remy E, Baster P, Friml J, Duque P. ZIFL1.1 transporter modulates polar auxin transport by stabilizing membrane abundance of multiple PINs in Arabidopsis root tip. Plant Signal Behav. 2013;8. doi:10.4161/psb.25688.
- Nover L, Bharti K, Doring P, Mishra SK, Ganguli A, Scharf KD. Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones. 2001;6:177–189.
- Von Koskull-doring P, Scharf KD, Nover L. The diversity of plant heat stress transcription factors. Trends Plant Sci. 2007;12:452–457.
- Qi X, Sun N, Liu M, Qi X. An autoregulatory loop controlling Arabidopsis HsfA2 expression: role of heat shock-induced alternative splicing. Plant Physiol. 2013;162(1):512–521. doi:10.1104/pp.112.205864.
- Remy E, Cabrito TR, Batista RA, Hussein MA, Teixeira MC, Athanasiadis A, Sa-Correia I, Duque P. Intron retention in the 5ʹUTR of the novel ZIF2 transporter enhances translation to promote zinc tolerance in Arabidopsis. PLoS Genet. 2014;10(5):e1004375. doi:10.1371/journal.pgen.1004375.
