ABSTRACT
WRKY is one of the largest families of transcription factors in plants. It not only regulates plant growth and development but also participates in the regulation of plant defense against biological and abiotic stresses. In this study, research was aimed to overexpress WRKY39 gene of P. trichocarpa (PtWRKY39) and to identify its important role played in drought and saline-alkali tolerance in tobacco model plant. Under the control of CaMV35S promoter, the overexpression of PtWRKY39 gene was increased to more than 10 times in T3 generation of transgenic tobacco plant. The drought resistance and saline-alkali tolerance were evidenced in overexpressed PtWRKY39 transgenic lines at germination/seedling stage. The overall germination rate, fresh weight, and chlorophyll contents of overexpressed lines were significantly higher while the level of malondialdehyde was significantly lower in PtWRKY39 transgenic lines than that of wild type (WT) lines. The content of H2O2 in leaves was detected by the 3, 3-Diaminobenzidine method showed that the overexpression of PtWRKY39 gene could reduce the accumulation of ROS (mainly H2O2) and enhance salt-alkali tolerance. Phenotypic analysis at 7-leaf pot transgenic seedlings stage treated with the saline-alkali soil extract and salt NaCl under root irrigation stress, revealed growth of the transgenic line was significantly higher than that of WT. This work concludes that overexpression of PtWRKY39 gene can improve the regulation of drought resistance and saline-alkali tolerance of transgenic plants during seed germination and vegetative growth.
1. Introduction
Transcription factor (TF) is a protein that regulates the gene expression by binding to specific DNA sequences in the promoter. WRKY is one of the largest transcription factor families in plants. In 1994, WRKY transcription factor was first discovered in sweet potato by Ishiguro et al.Citation1 and named as SPF1 (sweet potato factor 1). Since then, more WRKY factors have been found in Arabidopsis thaliana and other plants. WRKY factor is named for its special heptapeptide conserved sequence WRKYGOK, which has a zinc finger motifCitation2,Citation3 at the C-terminal. According to the types and number of zinc finger motifs, WRKY transcription factors can be divided into the following three families: type I family zinc finger structure is C2H2 (CX4-5-C-X22-23-H-X1-H), with two WRKY domains. The zinc finger structure of type II family is C2H2 (CX4-5-C-X22-23-H-X1-H) type, which has only one WRKY domain. The second family can be divided into IIa, IIb, IIc, IId and IIe5 subfamilies, while the zinc finger structure of type III family is C2HC (C-X7-C-X23-HX) type and only contains a single WRKY domain.Citation4 The specific-binding promoter sequence of WRKY protein is [(T)TGAC(C/T)].
As a forward or reverse transcriptional regulator, it plays a central role in the regulation of plant growth and development, stress defense and secondary metabolism.Citation5 Current studies have shown that the expression of WRKY transcription factor is significantly induced by drought, high temperature and exogenous abscisic acid (ABA) treatment, and reduce the content of reactive oxygen species (ROS) in transgenic plants under drought stress by enhancing the activities of Superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT).Citation6 From the perspective of application, WRKY gene has the potential value in stress resistance, abiotic and biological stress responses.Citation7 In recent years, studies have shown that overexpression or inhibition of transcription factor genes have an important impact on plant stress resistance. Transcriptional activity analysis showed that GhWRKY9 protein in cotton could activate the transcription of Saccharomyces cerevisiae, which enhanced the drought tolerance, delayed leaf senescence under drought stress, enhanced expression of stress-related genes and decreased expression of senescence-related genes (SAGs).Citation8 Salt-responsive WRKY114, isolated from maize, regulates the expression of related genes. Transgenic rice with a higher level of abscisic acid (ABA) than the wild-type rice, showed decreased salt stress tolerance and ABA sensitivity in transformed rice.Citation9 OsWRKY42 transcription factors in rice can be induced by cell wall damage, which can lead to increased callose deposition in rice and A. thaliana plant. Overexpression of OsWRKY42 gene in A. thaliana can enhance the tolerance to methyl jasmonate growth inhibition, but not to bacterial infection.Citation10
The study on the resistance of the WRKY genes to stress in different plants lays a foundation for the study of plant stress resistance in the future. Poplar is the most widely distributed and adaptable tree species in the world. The function of the WRKY transcription factors has been reported in poplar plant species. The WRKY70 gene of poplar was down-regulated under salt and drought stress, but inhibiting the expression of WRKY70 gene could improve the salt and drought tolerance of poplar.Citation11 After stress treatment, transgenic poplar lines overexpressing PagWRKY75 gene were more sensitive to salt and osmotic stress.Citation12 Although there are many reports about poplar WRKY gene responding to biological stress in recent years, the transgenic research on poplar clone WRKY genes is still limited. In order to cultivate poplar in arid and saline-alkali areas of northeast Songnen Plain of China, this research work aim is to find and analyze the drought and saline-alkali tolerant WRKY39 gene of P. trichocarpa through overexpressing in tobacco. This work will be of great significance to cultivate better resistant poplar varieties with improved germplasm, which is more resistant to drought and saline-alkali land.
2. Materials and methods
2.1. Plant materials
The plant of P. trichocarpa (Torr. & Gray) was donated by Yang Chengjun Research Group of Forest Genetics and Breeding Laboratory of Northeast Forestry University, China. The leaves and stems of P. tomentosa cultivated in the basin were frozen in liquid nitrogen and stored in the freezer at −80°C. The Nicotiana benthamiana seeds of Benji tobacco were preserved in our laboratory. Tobacco seeds were sterilized with 75% alcohol and 1% sodium hypochlorite, germinated on one-half MS solid medium, and the aseptic bottle seedlings were transformed and subsequently cultured.
2.2. Carriers, strains and reagents
PMD18-T vector was purchased from Dalian Bao Biology Co., Ltd. Plant expression vectors pQBV3 and pGWB18 were donated by Bu Qingyun Research Group of Chinese Academy of Sciences. Agrobacterium tumefaciens (EHA105) strain was preserved in our laboratory. Competent cells were prepared by our laboratory.Citation13 In this study, saline-alkali stress treatment solution was obtained according to the preparation method described in literature for soda saline-alkali soil extractCitation14
2.3. Extraction of total RNA and cloning of PtWRKY39 gene from P. tomentosa
In this study, kit for one-step separation of plant total RNA (Kangwei Century Biotechnology Co., Ltd. China) was used to lyse cells with a single-phase lysate containing Tris-saturated phenol and isothiocyanate. Genomic DNA and histone proteins were extracted in the organic phase, and then dissolved in chloroform. Total RNA was dissolved in the upper phase, and then it was obtained by purification.Citation15 Using the specific primer PtF1:5ʹ-CCTCATTTGGGTAGGTGCCA-3ʹ, PtR2: 5ʹ-GTTCCATCACATCCCATCTCAC-3ʹ synthesized the total cDNA from the total RNA (target mRNA sequence XM_002319046 registered by GenBank) of leaves and stems of P. tomentosa in cooperation of Kumei Biological Company, China. The target gene fragments were amplified by PCR under the action of Blend-TaqDNA polymerase. The DNA was recovered by agarose gel electrophoresis and inserted into the pMD18-T vector for sample sequencing.
2.4. Bioinformatics analysis of proteins encoded by PtWRKY39 gene
The target plasmid cloned into T-vector was sent to Kumei Biotechnology Co., Ltd (China) for sequencing. NCBI network station (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) was used to analyze the ORF (Open reading frame) region of the gene sequence, and deduced the amino acid sequence. The structure and homology of the amino acid encoded by the target gene were further analyzed and compared. Online ProtParam software (http://web.sxpasy.org/computepi/) calculated the theoretical isoelectric point and molecular weight of PtWRKY39 protein. The secondary structure of the protein was predicted on-line, and the multiple sequences were compared by DNAMAN and ClustalX software.Citation16 The conserved domain and interaction network of the protein were analyzed by SMART (http://smart.embl-heidelberg.de). The phylogeny of PtWRKY39 gene was analyzed by MEGA 6.0 software.
RNA extraction and real-time quantitative PCR analysis of P. tomentosa under drought and salt stress
Approximately, 30 cm twigs with parietal leaves of the same thickness were cultured in sterilized Huo’s nutrient solution for 3 days. About 15 branches were soaked in 20% PEG6000 and 25% volume ratio of heavy saline-alkali soil, respectively, and the treatment time was 0, 12, 24, 48 and 72 hours. According to the method of TOYOBO reverse transcription kit, the total RNA extracted from the P. tomentosa leaves was used as the template to detect the expression characteristics of PtWRKY39 gene under drought and saline-alkali stress. The specific quantitative primer (PtWF3: 5ʹ- cctcatttgggtaggtgcca-3ʹ) was used to extract the corresponding RNA, from the leaves of P. tomentosa by Kangwei Century Co., Ltd. (China). The relative expression of PtWRKY39 gene was detected by MxPro3000 qRT-PCR using primers of PtWR4 gene: 5ʹ-gcataatttgcacagggggc-3ʹ) and internal reference Actin2 gene (Actin2-F: 5ʹ- ttctacaagtgctttgatggtgagttc-3ʹ genes; Actin2-R: 5ʹ- ctattcgatacatagaagatcagaatgttc-3ʹ).
2.6. Subcellular localization of PtWRKY39 protein
The prediction was made by the Network Bioinformatics Software (http://psort.hgc.jp/form.html), and then verified by experimental research. The ORF of PtWRKY39 gene was amplified by PCR with specific primers PtWF5 and PtWR6. The fragment was fused into the N-terminal pBI121-PsWRKY39-GFP of green fluorescent protein (GFP) under the control of (CaMV) 35 s super promoter of Cauliflower Mosaic Virus. For protein fluorescence expression, the recombinant control plasmid pBI121-PtWRKY39-GFP was transformed into onion epidermal cells by particle bombardment. After bombardment, the tissue was incubated on filter paper infiltrated with distilled water at 25°C for 16 h, and onion epidermis cells were pressed under a fluorescence microscope (DAPY Olympus, Tokyo, Japan).
2.7. Construction of plant expression vector and genetic transformation
The ORF region of PtWRKY39 gene was amplified by PCR (primer PtWF5: 5-atggaggaggtt gaggaggt-3ʹ; PtWR6:5ʹ- tcatgtatgcgcagactgtg-3ʹ), and the clone was inserted into the pQBV3 entry vector. The positive inserted clone was detected by PCR, and the correct plasmid DNA was seamlessly linked with the pGWB18 vector to construct the CaM35S promoter and NOS Terminator plant expression vector pGWB18- PtWRKY39. The DNA of plasmid pGWB18-PtWRKY39 was electrocuted into A. tumefaciens EHA105, and the clones were screened on YEP+50 mg/L Kana+100 mg/L Rif plate. Then, the YEP (Yeast Extract Peptone) medium with Kana antibiotics was activated and cultured on the bacterial solution ODλ600 = 0.3 ~ 0.6. The antibiotics and culture medium were removed by centrifugation, and the heavy suspension of AAM was used as the working bacterial liquid to infect the tobacco leaves of 0.4 cm × 0.4 cm. The infection time was 0 ~ 15 min. The stained leaves were co-cultured on MS+0.5 mg/L 6-BA+0.1 mg/L NAA+100 μmol/L AS medium for 72 hours. The resistant buds were transferred to the differentiation medium of MS+0.5 mg/L 6-BA+0.1 mg/L NAA+50 mg/L Hyg for screening, and the differentiated resistant buds were inoculated on MS medium to take root. After ventilation, the seedlings were transferred to flower pots to grow, and the T1 generation-resistant transformants were harvested. Detection of tobacco lines with overexpression of PtWRKY39 transgenic T1 lines with Hyg resistance were screened for up to T3 generations of transformed tobacco. The seeds of T3 lines (T3 color plants) and wild-type (WT) were sown and germinated on the medium containing 50 mg/L Hyg, respectively, The resistant plants were cultured to 5-leaf stage in flower pots. After this, 2–3 fresh and tender heart leaves of transgenic T3 lines and wild types were cut and fully ground under liquid nitrogen. The genomic DNA and total RNA of transgenic leaves were extracted. The method of reverse recording of 1 µg total RNA into cDNA is the same as above. The DNA diluted 50 times was used as template for PCR identification.
2.8. Drought and saline stress tolerance analysis of tobacco plant overexpressing PtWRKY39 gene
2.8.1. Detection of germination characteristics under mannitol stress
Around 300 full seeds of each strain (PtWRKY39:T3-#1, #3, #5, and WT seeds of T3 lines) were sterilized with alcohol and sodium hypochlorite, and sown on 1/2 MS+0, 150, 300 mM Mannitol medium, with 1/2 MS medium as control. The experiment was repeated three times. First, vernalization carried out at 4°C for 12 h and then transferred the plant to the incubator. Statistics began on the 4th day, and the average germination rate was calculated on the 14th day.
2.8.2. Analysis of tolerance of tobacco to salt (NaCl) stress
The salt-alkali tolerance of tobacco lines was analyzed for overexpressing T3 transgenic PtWRKY39 gene. The seeds of T3 lines (PtWRKY39:T3-#1, #3, #5) and WT were sterilized and germinated. The 4-leaf seedlings with the same germination were transplanted into the flower pot of 5 cm × 5 cm. The seedlings were cultured in an artificial oxygen waiting room at room temperature of 25 ± 2°C, light intensity of 2600 LX, light/dark time for 16/8 h. Root stressed with salt (NaCl) began at 7-leaf stage. The salt stress treatment concentration was 150 mM, 200 mM, and 225 mM. After 14 days of stress treatment, the phenotype was observed, beside also measured the fresh weight, chlorophyll content,Citation17 and malondialdehyde (MDA) contents.Citation18
2.8.3. Tolerance analysis of growth at seedling stage stressed with saline-alkali soil extract
The tolerance to saline-alkali stress was analyzed PtWRKY39 gene overexpressing T3 tobacco transgenic lines. The seeds of T3 lines (PtWRKY39:T3-#1, #3, #5) and WT were sterilized and germinated, and the three-leaf seedlings with the same germination were transplanted into the flower pot of 5 cm × 5 cm. The seedlings were cultured in an artificial oxygen waiting room at room temperature of 25 ± 2°C, light intensity of 2600 LX, light/dark time for 16/8 h, and root stress treatment of saline-alkali soil extract after one month of culture. The treatment concentration was H2O+SAE:100 ml+0 ml; 70 ml+30 ml; 50 ml+50 ml; 30 ml+70 ml (). The phenotype was investigated after 14 days of stress treatment. The changes in the level of H2O2 at the third leaf stage were detected by 3,3´-Diaminobenzidine (DAB).Citation19,Citation20 For H2O2 staining, the leaves treated with saline-alkali soil extract at different time points were incubated in DAB solution (1 mg/ml, pH 3.8) in the dark at 25°C for 24 hours. After dyeing, the leaves were soaked in 95% ethanol to remove chlorophyll directly.Citation21
Table 1. The leachate’s essential characteristics
2.8.4. Detection of changes in the expression levels of reactive oxygen pathway-related genes after saline-alkali stress at seedling stage
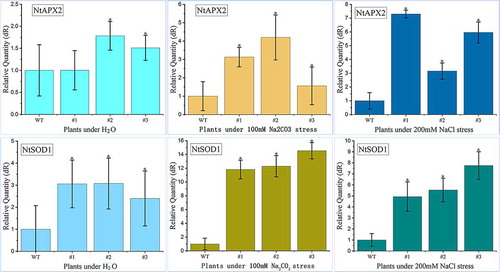
Analysis of the expression levels of reactive oxygen pathway-related genes was carried out after saline-alkali stress in transgenic tobacco lines of T3 generation. The three leaf seedlings T3 strains (PtWRKY39: T3-#1, #3, #5) and WT strain were transplanted to 5 cm × 5 cm pot culture in artificial oxygen room, at 25 ± 2°C, 2600 LX, light/dark time 16/8 h. At stage of 5 leaf period, the roots were irrigated with NaCl (200 mM) and Na2CO3(100 mM) and water irrigated roots used as experimental control group. After 24 hours, RNA was extracted from the experimental group and control group. Using Nt18S RNA (LOC107821161) as internal reference, qRT-PCR was used to analyze the expression levels of oxidative stress response genes NtAPX2 (LOC107759703) and NtCu/Zn SOD1 (LOC107790449).Citation15 Procedures for RNA extraction and qRT-PCR were similar to those described and listed in .
Table 2. The primers used in gene clone and qRT-PCR
3. Results
3.1. Cloning of PtWRKY39 gene
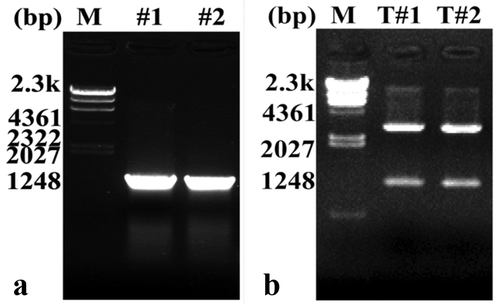
In this study, the total RNA of leaves and stems of P. tomentosa were reverse transcribed to cDNA and the size of the target gene of 1200 bp amplified by qRT-PCR with specific primers (). The plasmid DNA was extracted and then identified the insert by EcoRI/HindIII restriction endonuclease electrophoresis (). Electrophoregram showed that the DNA fragment was of equal in size to the expected target gene.
Figure 1. Cloning and identification of PtWRKY39 gene (a) The cDNA of PtWRKY39 gene of #1 leaves, #2 leaves, amplified by PCR. M: Molecular marker; # 1 and # 2: PCR products of PtWRKY39 gene in stems of P. tomentosa, respectively. (b) pMD18-T- PtWRKY39 digested by double enzymes M: Molecular marker; T#1&T#2: pMD18-T-PtWRKY39 digested by double enzymes
3.2. Bioinformatics analysis of PtWRKY39 gene
In order to detect the correct nucleotides of the cloned gene, the DNA sequence of the pMD18-T-plasmid was tested by NCBI network (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Analysis showed that the ORF of 1065 bp of the gene encodes 354 amino acids (), which was consistent with the size of the registered PtWRKY39 gene. ExPaSy analysis showed that expected protein has the formula C1732H2816N528O518S24, with relative molecular weight of 40.1 Kd. The theoretical isoelectric point (pI) was 9.79, and the hydrophilic/hydrophobic GRAVY value of −0.719 indicates the hydrophobic nature of the protein.
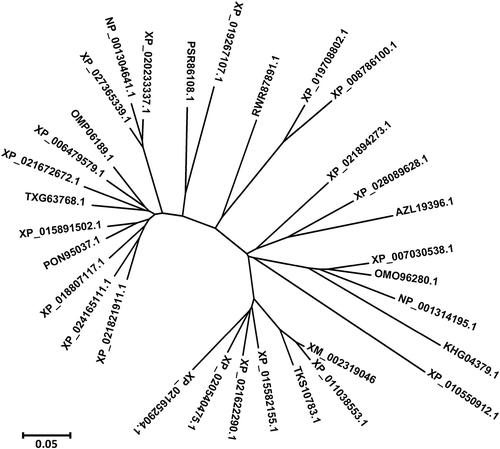
Online analysis tool PSIPREDV4.0 predicts the secondary structure of PtWRKY39 protein (). It was found that there were nine coils and multiple crimped structures. SWISS-MODEL online analysis tool analyzed the tertiary structure; it was found that the existence of WRKY structure was consistent with the predicted results of secondary structure ().The SMART analysis of chloro acid sequence derived by PtWRKY39 showed that the similarity of the protein with P.euphratica and P.tomentosa was 99.72 and 97.18, respectively (). Protein has the specific domain of WRKY transcription factor family which was located in the zinc finger and WRKYGQKP domains of 238–283 and 285–345 amino acids, respectively (). Phylogenetic analysis based on relationship between P. euphratica and P. trichocarpa, showed belong to the same branch (). One or two copies of WRKY exist in of plant transcription factors’ superfamily. These transcription factors are involved in regulating a variety of plant-specific physiological processes, including pathogen defense, senescence, hair development, and biosynthesis of secondary metabolites.
Figure 3. Prediction of the conserved domains of PtWRKY39 protein (a) Prediction of secondary structure of PtWRKY39, (b) Prediction of PtWRKY39 tertiary structure, (c) Prediction of PtWRKY39 conserved domain
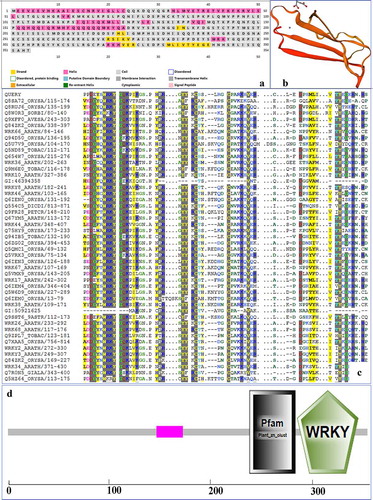
Figure 4. Phylogenetic tree of PtWRKY39 protein
3.3. Expression analysis ofPtWRKY39 gene under drought and saline-alkali stress
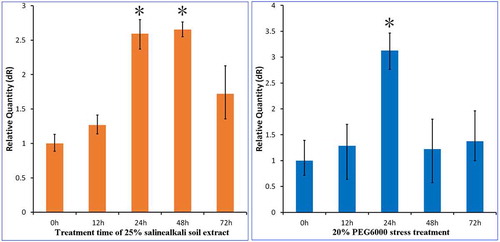
In this study, through the analysis of qRT-PCR quantitative data, it was found that drought and compound saline-alkali stress could induce the mRNA expression of PtWRKY39 gene, which showed a parabolic relationship between expression and induction time. When PEG6000 simulated drought stress, the expression reached the maximum after 24 hours stress. The relative expression increased 3.127 times, and then began to decrease. Soda saline-alkali soil is composed of compound salt and high pH. The extract of severe alkali spot soil was used to simulate the saline-alkali stress in Songnen Plain. The mRNA expression of PtWRKY39 gene increased with the extension of stress time, and the relative expression was 2.653 times higher than that of the control at 48 hours. From the qRT-PCR quantitative data analysis, it was found that the mRNA expression of PtWRKY39 gene increased with the stress time. The change of PtWRKY39 gene response induced by saline-alkali stress was consistent with that of PEG6000 simulated drought (), indicating that its response to drought and compound saline-alkali stress may be related to it.
3.4. Subcellular localization of PtWRKY39 protein in P. tomentosa
In this study, the amino acids encoded by the cloned gene PtWRKY39 protein were entered into the biological software PSORT, to select the subcellular localization of plant protein online. The preliminary prediction results show that it is 30.0% in the nucleus, 30.0% in the microbody, 10% in the mitochondrial matrix, and 0% in the cell membrane table (). Through subcellular prediction, it can be seen that nucleus and microbodies are the most likely sites for localization, which needs to be verified by further experimental study of subcellular localization.
Table 3. Preliminary forecast of PtWRKY39 Protein
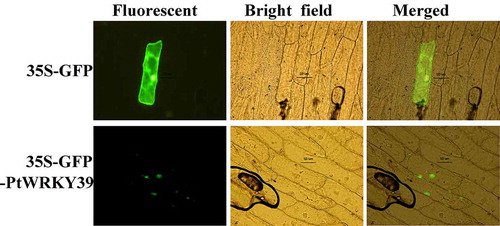
By scanning the transformed onion epidermis cells under confocal microscope (Olympus, Japan), it was found that the green fluorescent protein was expressed in the nucleus at the initial stage after 35S-GFP-PtWRKY39 fusion (), and the nucleus overlapped with the bright field, which indicated that the PtWRKY39 transcriptional protein was in the nucleus. It was consistent with the high probability of subcellular location in the nucleus predicted by the above PSORT software.
3.5. Genetic transformation and molecular identification of PtWRKY39 gene in tobacco
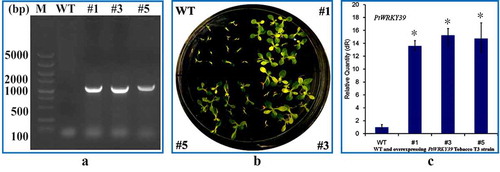
In this study, the plasmid DNA of plant expression vector pGWB18-PtWRKY39 transformed by electric shock was introduced into A. tumefaciens EHA105. Resistant T1 transformants were obtained, and the integration of PtWRKY39 gene was detected by PCR (). The resistant transformants were screened and cultured under 50 mg/L Hyg selective pressure and harvested into T3 generation seeds. The seeds of T3 lines were sterilized and seeded on 50 mg/L Hyg on 1/2 MS solid medium for 15 days. As shown in , although the wild type could germinate but it no longer grew and developed new leaves whereas the transgenic lines continued to grow and developed new leaves. It showed Hyg resistant growth in transgenic lines (). qRT-PCR detection of PtWRKY39 gene expression in leaves of transgenic line # 1,3,5 after 30 days of germination showed that the expression of PtWRKY39 gene in transgenic line # 1,3,5 was overexpressed relative to WT (), which was reserved for subsequent resistance test.
3.6. Drought resistance of tobacco with overexpression of PtWRKY39 gene during germination
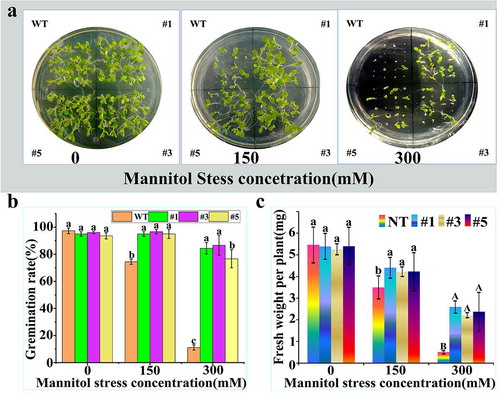
In this study, the drought resistance of tobacco with overexpression of PtWRKY39 gene under the action of CaM35S promoter was detected. The seeds of T3 generation of tobacco # 1, 3, 5 and WT lines were sown on a drought stress plates, and different drought conditions were simulated with 0, 150, 300 mM mannitol. The growth phenotype of WT on control 1/2 MS plate was observed after 14 days of germination. There was no difference in growth between WT and 3 transgenic tobacco lines during the germination period. The growth of tobacco at bud stage was affected by the addition of mannitol. Under the treatment of 150 mM mannitol, only two cotyledons were noticed in WT, while new leaves also grew in the overexpressed lines. Under the treatment of 300 mM mannitol, the number of cotyledons were less and the buds were smaller in WT. It was obvious that the growth of overexpressed lines was superior to that of WT (). In other studies, the germination rate was investigated from the 4th day to the 14th day, and the total germination rate analysis showed that the value of overexpressed lines was significantly higher than that of WT line (). The fresh weight of a single plant was significantly different from that of WT (), which was obviously inhibited by mannitol stress compared with no treatment, but the transgenic tobacco showed more resistance than WT tobacco. The germination experiment simulated by mannitol showed that the overexpression of PtWRKY39 gene enhanced the germination growth of resistant transgenic lines of tobacco plant. Hence, the PtWRKY39 transcription factor was related to its drought resistance.
Figure 8. Growth of tobacco seeds with overexpression of PtWRKY39 gene under drought treatment. (a) Drought-resistant phenotype of tobacco at germination stage with transgene PtWRKY39 gene. (b) Germination rate of tobacco seeds overexpressing PtWRKY39 gene under mannitol stress. (c) Analysis of fresh weight of over-expressed PtWRKY39 gene in tobacco under mannitol stress
3.7. Salt (NaCl) tolerance of tobacco with overexpression of PtWRKY39 gene at seedling stage
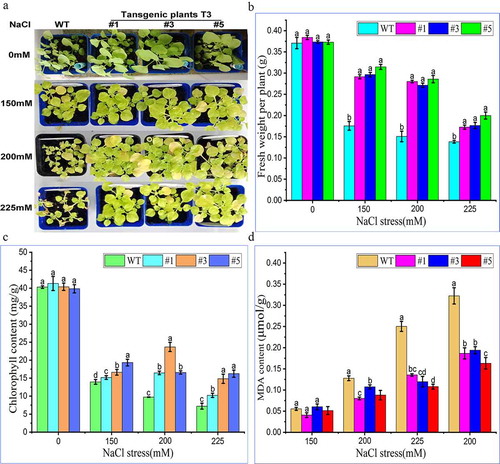
In this study, the 4-leaf seedlings of T3 generation lines and WT seedlings with the same germination were transplanted into flowerpots. After 3 weeks of growth, the 7-leaf potted seedlings’ roots were treated with salt (NaCl) and stressed for 14 days to observe their phenotypes. It can be seen that the growing plants were injured to different degrees compared with the control group without treatment, and the higher the concentration of NaCl treatment, the greater the damage to the plants (). When the concentration of NaCl in root irrigation treatment was 150 mM, the growth of overexpressed plants T3#1, 3, 5 was better than that of WT. When the concentration of NaCl was 200 mM, the leaves of overexpressed plants were significantly larger than that of WT, and the plant height was higher than that of WT. When the concentration of NaCl was 225 mM, the growth of WT stopped and the growth of overexpressed plants was also inhibited, but the growth degree of them was higher than that of wild type and remained in living condition. According to the statistical analysis, the fresh weight of control (non-salt treated) tobacco plant was observed much higher than the fresh weight of overexpressed tobacco plant group (). It indicated that salt stress inhibited the growth of tobacco, but the fresh weight of transgenic plants was better than that of WT, under salt stress. Chlorophyll was extracted from tobacco leaves under salt stress at seedling stage. It was found that the content of chlorophyll in transgenic tobacco was higher than that in WT under salt stress (). Under the same treatment, the content of MDA in WT was much higher than that in transgenic tobacco (). It can be concluded that the overexpression of PtWRKY39 gene can enhance the salt (NaCl) tolerance of tobacco plants.
Figure 9. Salt (NaCl) tolerance of tobacco plant overexpressing PtWRKY39 gene. Fig. A Salt (NaCl) tolerance in tobacco plants with overexpression of PtWRKY39 gene. Fig. B Detection and analysis of fresh weight of tobacco plant overexpressing PtWRKY39 gene under Salt (NaCl) stress. Fig. C Analysis of chlorophyll content in tobacco overexpressing PtWRKY39 gene under Salt (NaCl) stress. Fig. D Detection of MDA content in tobacco overexpressing PtWRKY39 gene under salt (NaCl) stress
3.8. Salt and alkaline tolerance of tobacco seedlings with overexpression of PtWRKY39 gene
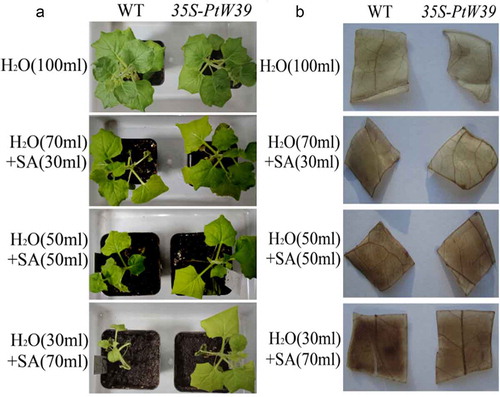
In order to analyze the saline-alkali tolerance of PtWRKY39 gene, three-leaf seedlings with the same germination of T3 line (T3#1) and WT were transplanted into flowerpots. After 3 weeks of growth, the phenotypes of 7-leaf potted seedlings treated with saline-alkali extract were investigated. Compared with the control, the growing plants were treated and observed more the amount of saline-alkali soil extract, greater the growth inhibition (). Under the treatment of adding 30 ml extract, the leaf color of overexpressed plants was darker than that of WT. Under the treatment of adding 50 ml extract, the leaves of overexpressed plants were larger than WT leaves. The leaf yellowing degree was lower and WT no longer grew in response to 70 ml extract. The growth of overexpressed plants was significantly more than that of WT, which showed salt-alkali tolerance. Hydrogen peroxide in leaves of the plant was detected by DAB method. The results showed that the staining degree of leaves was gradually deepened with the increase of the amount of treatment solution (). But the staining of leaves of overexpressed plants showed corresponding differences and the brownish-red color lower than WT. This observation indirectly indicated that the content of H2O2 in leaves of overexpressed plants was lower than that in WT leaves. After pretreatment with Na2CO3 (100 mM) and NaCl (200 mM) for 24 hours, the expression level of NtSOD1 and NtAPX2 gene was increased by qRT-PCR (), These genes improved the activity of ROS enzyme, which plays an important role in the balance of ROS, which indicated that the overexpression of PtWRKY39 gene could reduce oxidative stress.
4. Discussion
WRKY protein is a kind of plant-specific transcriptional regulatory factors, which is involved in plant development, senescence and stress resistance. More and more attention has been paid to its role in the regulation of plant responses to various biotic and abiotic stresses.Citation22,Citation23 In recent years, most of the studies on the function of WRKY transcription factors have been focused on model plants such as A. thaliana and rice. Compared with the widely studied AtWRKYs and OsWRKYs proteins, there are few studies on the functions related to drought and saline-alkali tolerance in poplar species.
In order to explore the role of PtWRKYs transcription factor in trees, a PtWRKY39 gene was cloned from P. tomentosa. The ORF 1065bp nucleotides encode 354 amino acids. The amino acid homology comparison showed that the similarity was higher than that of P. euphratica and P. tomentosa. The zinc finger domain and WRKYGQKP conserved domain of amino acids at position 238–283 and 285–345 were members of WRKY subgroup IId.Citation7 According to the above characteristics of IId members of the WRKY subgroup, a C region sequence was identified at the N-terminal of the protein, which contained a nuclear targeting sequence. Subcellular detection showed that the PtWRKY39 protein was located in the nucleus. The expression pattern of a gene is usually an indicator of its function. It has been reported that the expression of GhWRKY39 gene is induced by high salt stress, suggesting that it plays a role in defense response and participates in the regulation of stress pathways.Citation24 In this study, through the analysis of qRT-PCR quantitative data, it was found that drought and compound saline-alkali stress could induce the mRNA expression of PtWRKY39 gene. The extract of severe alkali soil and high pH was used to simulate saline-alkali stress in Songnen Plain. The mRNA expression of PtWRKY39 gene increased at first and then decreased. It is suggested that its response to drought and compound saline-alkali stress may be related to the overexpression of PtWRKY39 gene.
In this study, the plants overexpressing PtWRKY39 gene were more resistant to drought stress than wild plants. The overall germination rate of transgenic lines was higher than that of WT (), and the fresh weight per plant was also higher than that of WT (). This shows that PtWRKY39 protein can improve drought stress, which is closely related to the regulation of drought resistance. Compared with the control, overexpression of ZmWRKY101 in A. thaliana enhanced the salt and drought tolerance of transgenic plants, and endowed the transgenic plants with salt and drought tolerance.Citation25 In this study, in order to analyze the salt-alkali tolerance of PtWRKY39 gene, treated the transformed plants and WT pot seedlings, with salt (NaCl) which revealed that the salt-tolerance (NaCl) of transgenic lines (T3# 1, 3, 5) was much better than that of WT (), the fresh weight of transgenic plants under salt stress was higher than that of WT (), and the chlorophyll content was higher than that of WT (). Through the treatment of saline-alkali soil extract on transgenic plants and WT pot seedlings, it was found that the growth was different (). More the amount of saline-alkali soil extract, greater the growth inhibition. When the amount of added extract reached 70 ml treatment, WT no longer grew, and the growth of overexpressed lines was significantly more than that of WT, showed the saline-alkali tolerance. We speculate that the defense mechanism mediated by PtWRKY39 gene may be related to PAMP-triggered immunity. Participating in the defense response of ROS system, the content of H2O2 in leaves was detected by DAB method (). With the increase of the amount of treatment solution, the content of H2O2 in leaves of overexpressed plants was lighter than that in WT, indicating that the content of H2O2 in leaves was lower than that in WT leaves.
In response to 24 hours of NaCl (200 mM) and Na2CO3 (100 mM) stress and reactive oxygen species generate, the expression level of NtSOD1 and NtAPX2 genes was increased obviously. NtSOD1 and NtAPX2 are the most important antioxidant enzyme in plant active oxygen metabolism, especially to remove the key enzymes of H2O2 in the chloroplast. The expression of these two enzymes activity also shows that the overexpression of PtWRKY39 gene can reduce ROS (mainly H2O2) accumulation and effectively remove excessive H2O2 to enhance salt resistance (). This shows that the overexpression of PtWRKY39 gene can reduce the accumulation of ROS (mainly H2O2 and effectively eliminate excessive H2O2 to enhance salt-alkali tolerance).
It has been reported that PAMP-triggered immunity and effector-triggered immunity activate local and systemic defense responses, which are regulated by plant hormones, especially jasmonic acid and salicylic acid.Citation26,Citation27 The role of ROS in defense response and HR activation is mainly related to NADPH oxidase, which catalyzes oxygen reduction.Citation2 Furthermore, superoxide disproportionation oxygen is used to produce the most stable reactive oxygen species and hydrogen peroxide.Citation28 The overexpression of chrysanthemum DgWRKY5 gene enhanced the tolerance of tobacco to salt stress, decreased the accumulation of MDA and hydrogen peroxide, while the activity of antioxidant enzymes in transgenic plants was higher than that in wild plants.Citation29 To sum up, overexpression of PtWRKY39 gene can make plants resistant to drought and complex saline-alkali stress, and participate in the regulation of stress defense. We believe that the molecular mechanism mediated by PtWRKY39 gene is quite complex in the process of defense response, and PtWRKY39 gene may be involved in multiple signal pathways and interact with multiple defense genes, which need to be further studied.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (Grant No. 2016YFC0501203), Natural Science Foundation of Heilongjiang Province (Grant No. C2017009), the HeilongjiangPostdoctoral Scientific Research Developmental Fund (Grant No. LBH-Q15004) and the College Students’ innovation and Entrepreneurship Project. The funders had no role in the design of the study and collection, analysis, interpretation of data and in writing the manuscript.
Additional information
Funding
References
- Eulgem T, Rushton PJ, Schmelzer E, Hahlbrock K, Somssich IE. Early nuclear events in plant defence signalling: rapid gene activation by WRKY transcription factors. Embo J. 1999;18(17):1–12. doi:10.1093/emboj/18.17.4689.
- Yongjuan R, Dongjiao W, Yachun S, Ling W, Xu Z, Weihua S, Youxiong Q. Plant WRKY transcription factors: structure, classification, evolution and function. J Agric Biotechnol. 2021;29:105–124.
- Caihong J, Zhuo W, Jianbin Z, Jingyi W, Hongxia M, Juhua L, Zhiqiang J, Biyu X. Cloning and expression analysis of 8 WRKY transcription factors in banana. J Trop Crops. 2018;39:2193–2199.
- Maeo K, Hayashi S, Kojima-Suzuki H, Morikami A, Nakamura K. Role of conserved residues of the WRKY domain in the DNA-binding of tobacco WRKY family proteins. Biosci Biotechnol Biochem. 2001;65(11):2428–2436. doi:10.1271/bbb.65.2428.
- Li J, Xiong Y, Li Y, Ye S, Yin Q, Gao S, Yang D, Yang M, Palva ET, Deng X. Comprehensive Analysis and Functional Studies of WRKY Transcription Factors in Nelumbo nucifera. Int J Mol Sci. 2019;20(20):5006. doi:10.3390/ijms20205006.
- Wang CT, Ru JN, Liu YW, Li M, Zhao D, Yang JF, Fu JD, Xu ZS. Maize WRKY Transcription Factor ZmWRKY106 Confers Drought and Heat Tolerance in Transgenic Plants. Int J Mol Sci. 2018;19(10):3046. doi:10.3390/ijms19103046.
- Jing Z, Meiyan Z. Identification of WRKY gene family and analysis of drought stress response model of poplar. Chin J Cell Biol. 2019;541:2160–2173.
- Gu L, Ma Q, Zhang C, Wang C, Wei H, Wang H, Yu S. The Cotton GhWRKY91 Transcription Factor Mediates Leaf Senescence and Responses to Drought Stress in Transgenic Arabidopsis thaliana. Front Plant Sci. 2019;29(10):1352. doi:10.3389/fpls.2019.01352.
- Bo C, Chen H, Luo G, Li W, Zhang X, Ma Q, Cheng B, Cai R. Maize WRKY114 gene negatively regulates salt-stress tolerance in transgenic rice. Plant Cell Rep. 2020;39(1):135–148. doi:10.1007/s00299-019-02481-3.
- Pillai SE, Kumar C, Patel HK, Sonti RV. Overexpression of a cell wall damage induced transcription factor, OsWRKY42, leads to enhanced callose deposition and tolerance to salt stress but does not enhance tolerance to bacterial infection. BMC Plant Biol. 2018;18(1):177. doi:10.1186/s12870-018-1391-5.
- Zhao H, Jiang J, Li K, Liu G. Populus simonii × Populus nigra WRKY70 is involved in salt stress and leaf blight disease responses. Tree Physiol. 2017;37(6):827–844. doi:10.1093/treephys/tpx020.
- Zhao K, Zhang D, Lv K, Zhang X, Cheng Z, Li R, Zhou B, Jiang T. Functional characterization of poplar WRKY75 in salt and osmotic tolerance. Plant Sci. 2019;289:110259. doi:10.1016/j.plantsci.2019.110259.
- Prescott D. Molecular cloning: a laboratory manual. In: Sambrook J, Fritsch EF, Maniatis T editors. Analytical Biochemistry. 1990. Vol. 3. 2nd. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1989. p. 182–183.
- Wang H, Takano T, Liu S. Screening and Evaluation of Saline–Alkaline Tolerant Germplasm of Rice (Oryza sativa L.) in Soda Saline–Alkali Soil. Agronomy. 2018;8(10):205. doi:10.3390/agronomy8100205.
- Guan QJ, Ma HY, Wang ZJ, Wang ZY, Bu QY, Liu SK. A rice LSD1-like-type ZFP gene OsLOL5 enhances saline-alkaline tolerance in transgenic Arabidopsis thaliana, yeast and rice. BMC Genomics. 2016;17(1):142. doi:10.1186/s12864-016-2460-5.
- Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinf. 2002;1:1-22
- Nianwei Q, Xiushun W, Fabin Y, Xiaogang Y, Wen Y, Runjie D, Xiu W, Jing C, Feng Z. Rapid extraction and precise determination of chlorophyll. Acta Botanica Sinica. 2016;51:667–678.
- Zifang L, Xidong W. Experimental design scheme for determination of malondialdehyde content in plants. Tianjin Agric Sci. 2016;22:49–51.
- Mahalingam R, Jambunathan N, Gunjan SK, Faustin E, Weng H, Ayoubi P. Analysis of oxidative signalling induced by ozone in Arabidopsis thaliana. Plant Cell Environ. 2006;29(7):1357–1371. doi:10.1111/j.1365-3040.2006.01516.x.
- Xiaoli Z, Pengcheng W, Chunpeng S. Method for determination of hydrogen peroxide in plant cells. J Bot Sci. 2009;44:103–106.
- Hans TC, Ziguo Z, Yangdou W, David BC. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley—powdery mildew interaction. Plant J. 1997;11(6):1187–1194. doi:10.1046/j.1365-313X.1997.11061187.x.
- Li W, Pang S, Lu Z, Jin B. Function and Mechanism of WRKY Transcription Factors in Abiotic Stress Responses of Plants. Plants. 2020;9(11):1515. doi:10.3390/plants9111515.
- Chen C, Chen X, Han J, Lu W, Ren Z. Genome-wide analysis of the WRKY gene family in the cucumber genome and transcriptome-wide identification of WRKY transcription factors that respond to biotic and abiotic stresses. BMC Plant Biol. 2020;20(1):443. doi:10.1186/s12870-020-02625-8.
- Weina S, Dongdong L, Lili H, Changai W, Xingqi G, Han L. GhWRKY39, a member of the WRKY transcription factor family in cotton, has a positive role in disease resistance and salt stress tolerance. Plant Cell Tissue Organ Cult. 2014;118(1):17–32. doi:10.1007/s11240-014-0458-8.
- Yumin G, Yunhua Z, Tao J, Xiaoping Z. Overexpression of maize transcription factor ZmWRKY101 gene improves salt tolerance of Arabidopsis thaliana. J Plant Physiol. 2020;56:1921–1932.
- Bostock RM. Signal crosstalk and induced resistance: straddling the line between cost and benefit. Annu Rev Phytopathol. 2005;43(1):545–580. doi:10.1146/annurev.phyto.41.052002.095505.
- Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol. 2004;42(1):185–209. doi:10.1146/annurev.phyto.42.040803.140421.
- Lamb C, Dixon RA. THE OXIDATIVE BURST IN PLANT DISEASE RESISTANCE. Annu Rev Plant Physiol Plant Mol Biol. 1997;48(1):251–275. doi:10.1146/annurev.arplant.48.1.251.
- Liang QY, Wu YH, Wang K, Bai Z-Y, Liu Q-L, Pan Y-Z, Zhang L, Jiang -B-B. Chrysanthemum WRKY gene DgWRKY5 enhances tolerance to salt stress in transgenic chrysanthemum. Sci Rep. 2017;7(1):4799. doi:10.1038/s41598-017-05170-x.