ABSTRACT
WRKYs represent an important family of transcription factors that are widely involved in plant development, defense regulation and stress response. Transgenic rice that constitutively expressed ZmWRKY114 had shorter plant height and showed less sensitivity to gibberellic acid (GA3). Further investigation proved that transgenic rice accumulated lower levels of bioactive GAs than that in wild-type plants. Application of exogenous GA3 fully rescued the semi-dwarf phenotype of ZmWRKY114 transgenic plants. Transcriptome and qRT-PCR analyses indicated that the expression of OsGA2ox4, encoding the repressor of GA biosynthesis, was markedly increased. Electrophoretic mobility shift assay and dual-luciferase reporter assay indicated that ZmWRKY114 directly binds to a W-box motif in the OsGA2ox4 promoter. Taken together, these results confirm that ZmWRKY114 is a GA-responsive gene and is participated in the regulation of plant height in rice.
Introduction
Plant height is an important agronomic trait that significantly affect crop yield. Crop with a suitable height often showed improved lodging resistance in unfavorable environments and increased total biomass attributing to enhanced nitrogen-use efficiency.Citation1
Gibberellic acids (GAs), a large class of tetracyclic diterpenoid phytohormone, were proved to play vital role in the regulation of plant height.Citation2 Usually, plants with a dwarf or semi-dwarf phenotype are deficient in GA synthesis. For example, the reduced internode length was often considered to be the most prominent phenotype of GA biosynthesis mutants in Arabidopsis.Citation3 Several of the identified dwarf mutants in rice are also characterized as GA-deficient or GA-insensitive mutants.Citation4–6 Therefore, it is essential for plants to produce and maintain right levels of bioactive GAs (mainly GA1, GA3, GA4, and GA7) to guarantee normal growth and development.Citation7
WRKY transcription factors are one of the biggest families of regulatory proteins in higher plants. In different species, the WRKY family can be classified into three major structural groups, based both on the number of WRKY domains and the types of their zinc finger motifs.Citation8 Recent research has supplied some evidence to prove that WRKY members also play important roles in the regulation of plant height. For example, OsWRKY6-overexpressing rice lines shows a growth-retardation phenotype at the seedling and tillering stages.Citation9 Mutation of rice WRKY11 gene leads to a semi-dwarf and late flowering phenotype.Citation10 OsWRKY78 RNAi lines also exhibited a semi-dwarf and small kernel phenotype because of reduced cell length in these transgenic plants.Citation11 In cotton, silence of WRKY42 gene in VIGS (virus-induced gene silencing) plants significantly reduced plant height.Citation12
Our previous results showed that ZmWRKY114 encoding a WRKY transcription factor involved in plant response to salt stress.Citation13 In this study, we surveyed the functions of this gene in rice growth and development. Our data proved that ZmWRKY114 is participated in regulating plant height through GA signaling.
Materials and methods
Plant materials and growth conditions
The ZmWRKY114-overexpressing transgenic rice lines reported in our previous article were used in this study.Citation13 Zhong Hua 11 (ZH11) rice was used as the control plant for phenotypic examination. The transgenic and wild type (WT) plants were planted in a blue plastic basin which contained field soil and grown in a greenhouse with controlled environmental conditions (28°C, 60% relative humidity and 16 h-light /8 h-dark photoperiod).
Phenotypic analysis
The heights of at least 10 plants were measured at the heading stage in the field. For paraffin section, the penultimate internodes were collected from WT and two transgenic lines and fixed in FAA solution (50% alcohol: formalin (37–40% formaldehyde): glacial acetic acid;18:1:1). Next, the internodes were dehydrated in a series of ethanol solutions (70% (v/v) ethanol, 80% (v/v) ethanol, 85% (v/v) ethanol, 90% (v/v) ethanol, 95% (v/v) ethanol, and anhydrous ethanol) and detained in a series of xylene solutions (3:1 ethanol: xylene, 1:1 ethanol: xylene, 1:3 ethanol: xylene, and pure xylene). The samples were dissected and then observed by light microscope (Leica). Parts of these experiments were performed by Servicebio company (Servicebio technology CO, Wuhan, China).
The GA sensitivity assay of transgenic seedlings
In order to investigate the effect of ZmWRKY114 on plant growth, GA sensitivity assay was firstly performed. The surfaced-sterilized rice seeds of WT and two transgenic lines (OE8 and OE6) were soaked in sterile water at 28°C for 2 days under darkness. Seeds with the same size were inoculated into a sterile box containing 0.5 × MS medium supplemented with 0, 0.1, 1, 10 or 100 μM GA3 + 200 μM Paclobutrazol (PAC, a GA biosynthetic inhibitor, to exogenous GA3 treatments) and 200 μM PAC alone. After grown in a growth chamber for 14 days, the seedling height and root length of 30 seedlings for each sample were measured. Each treatment was repeated three times.
Quantification of endogenous GAs
The 4-week-old seedlings of transgenic and WT plants were sampled for quantitative GA analysis. The samples were frozen in liquid nitrogen and ground into fine powder. Hormones were extracted by 10 mL methanol containing 20% water (v/v) for 12 h at 4°C. After centrifugation at 10,000 × g (4°C) for 5 min, the extracts were collected and passed through a C18 SPE cartridge, then dried in N2. The HPLC-MS/MS analysis was performed using an AB SCIEX QTRAP 6500 System (AB SCIEX, USA). The quantification of endogenous GA1 and GA3 was performed as described by.Citation14
Growth chemical treatments
The 4-week-old transgenic seedlings showing a semi-dwarf phenotype were sprayed with 20 mL of 10 μM GA3 (in water). The treatments were repeated every 15 days for 2 times. Untreated WT and transgenic plants were used as the control. The plant height was measured at 30 days of treatment.
RNA sequencing analysis
Total RNA was extracted from 4-week-old seedlings of WT and two transgenic lines. The cDNA library construction and transcriptome sequencing were performed by Geneseeq company (Nanjing, China) using an Illumina Hiseq 2000 platform. Gene expression levels were estimated based on fragments per kilobase of exon per million fragments mapped (FPKM) using the Cuffinks software (version: 2.2.1). According to the FPKM value, differentially expressed genes (DEGs) between WT and transgenic line were identified. FDR ≦0.05 and ∣log2 (FPKM-transgenic/FPKM-WT)∣≧1 and FPKM ≧2 were regarded as thresholds to determine the significance of differences in gene expression.
Quantitative real-time PCR (qRT-PCR) analysis
Four plant height related genes were selected to verify the RNA-seq analysis using qRT-PCR. qRT-PCR was performed using the ABI 7300 real-time PCR system (Applied Biosystems), and the actin gene was used as an internal control. The PCR reaction system and thermal cycle conditions were referred to previous study.Citation15 The primers used for qRT-PCR analysis are listed in Table S1, and the relative expression level was calculated using the 2−ΔΔCT method.Citation16 Each PCR reaction was performed with three biological replicates and three technical repeats.
Electrophoretic mobility shift assay (EMSA)
Based on RNA-seq and qRT-PCR data, biotin labeled probes containing W-boxes existed in the promotor of OsGA2ox4 were synthesized (Sangon Biotech, Shanghai, China). The EMSA assays were conducted using Thermo Fisher Chemiluminescent EMSA Kits, according to the manufacturer’s instructions (Thermo Fisher Scientific). In brief, probes and proteins were mixed and transferred to Hybond nylon membranes using a sandwich method, and they were cross-linked to the membrane by a UV-light cross-linker. Then, the membranes were transferred to blocking solutions for 15 min with gentle shaking. The membranes were then incubated in another blocking solution with 1:2000 diluted streptavidin-HRP conjugate antigens at RT (15 min) with gentle shaking, and the membranes were washed 4 times with a washing buffer. Finally, 1 mL Luminol/Enhancer Solution and 1 mL Stable Peroxide Solution were added to the membrane and the signals were detected using the Gel Doc EZ gel image analysis system (Bio-Rad, Hercules, CA, USA).
Dual-luciferase reporter assay
The full-length coding sequence of ZmWRKY114 cDNA was inserted into the constructed pGreenII-62-SK vector as the effector, driven by CaMV 35S promoter, and the empty vector was used as a control. The 256 bp OsGA2ox4 promoter fragment containing W-box was ligated into the transient expression pGreenII0800-LUC vector. The primers W1-F, W1-R, W2-F and W2-R are listed in Table S1. The fusion vectors and pSoup helper plasmid were co-introduced into A.tumefaciens strain GV3101. The infiltrated leaves were sprayed with luciferin at 2–3 days post-infiltration and coexpression proteins were monitored using an In Vivo Plant Imaging System (Berthold Technologies, Germany). Cells were lysed and luciferase activity was measured using the dual-luciferase reporter assay kit (TransGen Biotech). The promoter activities were indicated by the LUC/REN ratio.
Results
Overexpression of ZmWRKY114 led to a semi-dwarf phenotype in rice
Our previous study has proved that transgenic plants overexpressing a maize WRKY114 gene displayed a semi-dwarf phenotype at the seedling stage.Citation13 In fact, the transgenic plants were shorter than the WT almost in the whole developmental process of rice ( and S1). At the heading stage, the average plant height of WT was 91.46 cm; the average height of two ZmWRKY114 overexpression lines, OE8 and OE6 was 63.67 cm and 57.87 cm respectively (). In addition, as shown in and d, OE8 and OE6 were significant shorter than WT in top three internodes.
Figure 1. Phenotypes characterization of the WT and ZmWRKY114-overexpressing plants. (a) WT and ZmWRKY114-overexpressing plants at the heading stage. Bars = 20 cm. (b) Plant height at heading stage. Data are mean ± SD from at least 10 plants. (c) Flag leaves, panicles and internode phenotypes of WT and transgenic plants. fl., flag leaf; P, panicle, I to Ⅳ indicate internodes from top to bottom. Bar, 5 cm. (d) Panicle and internode lengths of WT and transgenic plants. Values are means ± SD of 15 plants. (e) Longitudinal view of parenchyma cells in penultimate internodes of WT and transgenic plants. Bar, 50 μm. (f-h) cell numbers, Cell lengths and cell widths of penultimate internodes in WT and transgenic plants. Values are means ± SD (n = 3). Significant difference was checked by Student’s t-test (* P < .05, **P < .01)
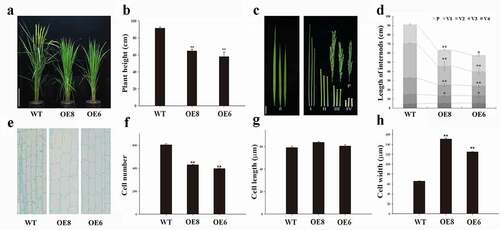
In order to determine the main reason for the semi-dwarf phenotype of ZmWRKY114 transgenic lines, the penultimate internodes of WT and transgenic lines were used for paraffin section observation. Compared to WT, the cell number of OE8 and OE6 was decreased for 34.2% and 28.8%, and the cell width was increased for 52.9% and 44.2% respectively. In addition, there was no significant differences in cell length between WT and transgenic lines (-h). These results showed that a decrease in cell number is the major cause of the dwarfism phenotype in ZmWRKY114 transgenic plants.
ZmWRKY114 overexpression reduced sensitivity to GA3 in rice
To further determine the factors of height reduction in transgenic rice, GA3 and PAC treatments were performed on WT and transgenic rice. There was no difference in seedling heights between transgenic lines and WT plants when treated with 200 μM PAC, but OE8 and OE6 were shorter than WT plants when 0.1, 1, and 10 μM GA3 were added (-c). These results indicated that overexpression of ZmWRKY114 could reduce the GA sensitivity of transgenic plants.
Figure 2. Effects of GA3 on plant height development of ZmWRKY114 transgenic plants. (a) The phenotype of seedlings treated with different concentrations of GA3 and 200 μM PAC. (b and c) Comparison of plant height and root length between WT and two transgenic lines. Data are means ± SD from 30 seedlings. (d) WT and two transgenic lines seedlings aged 4 weeks before GA3 treatment. (e) WT and two transgenic line seedlings were sprayed with 20 mL water for 30 days. (f) WT seedings were sprayed with 20 mL water while two transgenic line seedlings were sprayed with 20 ml 10 μM GA3 for 30 days. Bar, 3.5 cm. (g) Measurement of plant height before and after GA3 treatment. Data are mean ± SD from 15 plants. DAT, day(s) after treatment. (h) GA contents of the 4-week-old seedlings of WT and transgenic plants. Data are means ± SD from two biological samples. Asterisks mean a signifiant difference between WT plants and different transgenic lines by Student’ t-test, * P < .05, **P < .01
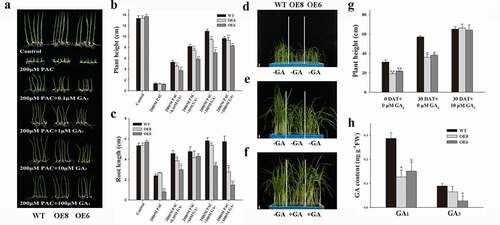
Exogenous GA3 treatment rescued the dwarfism phenotype of ZmWRKY114 transgenic plants
To test whether the semi-dwarf phenotype of ZmWRKY114 transgenic plants was the result of GA deficiency, the 4-week-old transgenic plants were subjected to GA3 treatments by spraying with 10 μM GA3 for 30 days and measuring their heights. As shown in , significant difference in plant height was observed between WT and transgenic plants before GA3 treatment. After 30 days, the average plant height of the wild plants was 1.5 times higher than that of the untreated ZmWRKY114 transgenic strain ( and g). However, after treatment with 10 μM GA3, the ZmWRKY114 overexpression plant height was fully restored and was the same as the average plant height of the wild type ( and g). This result indicated that GA synthesis is possibly repressed in ZmWRKY114-OE plants.
Subsequently, GA1 and GA3 contents of WT and transgenic lines were measured using HPLC-MS/MS. As shown in , both the GA1 content and GA3 content were lower in ZmWRKY114-overexpressing plants compared with WT. This result strongly implies that overexpression of ZmWRKY114 has a negative effect on GA biosynthetic pathway.
ZmWRKY114 overexpression altered the expression of plant height related genes in rice
To further examine whether the semi-dwarf phenotype was caused by ZmWRKY114 overexpression, RNA-seq analysis was conducted for WT and two transgenic lines. Each sample was biologically repeated twice. 5799 DEGs in OE8 and 4828 DEGs in OE6 compared with the WT were found (Table S2 and Table S3). Among these DEGs, 1722 up-regulated and 1661 down-regulated genes had significantly different transcript abundance ( and b), many genes were reported to involve in plant height control through regulating the GA synthesis, such as OsEUI1, OsLOL2, OsYAB1, OsGI and OsGA2ox4 ().
Table 1. Plant height related genes altered in two transgenic lines compared to WT plants
Figure 3. Venn diagram and qRT-PCR analyses. (a) Overlapping of up-regulated genes. (b) Overlapping of down-regulated genes. The overlap of genes that were up- or down-regulated in the different comparisons. (c) qRT-PCR analyses of expression of plant height related genes including OsGA2ox4 (AK107211), OsYAB1 (NM_001065472), OsGhd2 (XM_015767173), OsEUI1 (AK109526) and OsJar3 (NM_001062995) in WT and two transgenic lines. OsActin gene was used as an internal control. Data indicate mean ± SD of three replicates. Asterisks mean a signifiant difference by Student’ t-test, * P < .05, **P < .01
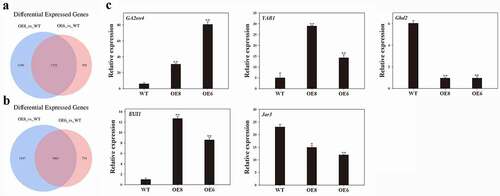
The expression levels of plant height related genes, such as OsEUI1, OsGA2ox4, OsYAB1, OsGhd2 and OsJar3 genes were compared between WT and ZmWRKY114-overexpressing plants using qRT-PCR method. The results showed that ZmWRKY114 overexpression indeed alters the expression of OsGA2ox4, OsYAB1, OsGhd2, OsEUI1 and OsJar3 (). The expression levels of OsEUI1, OsGA2ox4 and OsYAB1 were up-regulated but OsGhd2 and OsJar3 expression was down-regulated.
ZmWRKY114 can bind to the W-box motif in OsGA2ox4 promoter
RNA-seq and qRT-PCR data showed that the OsGA2ox4 expression was altered significantly in the ZmWRKY114-overexpressing plants, suggesting that OsGA2ox4 may be a direct target of ZmWRKY114 transcription factor. To further test whether ZmWRKY114 can bind to the W-box motif in the promoter region of OsGA2ox4, EMSAs were used with ZmWRKY114-MBP protein. We identified W-box motif by examining the promoter region of OsGA2ox4. As shown in , the potential ZmWRKY114-binding W-box site, TTGACT at −1136 to − 1142 bp upstream of the ATG position of OsGA2ox4, was discovered. In , shifted band was detected when probes harboring TTGACT in the OsGA2ox4 promoter region were incubated with ZmWRKY114 protein (lane 2). Conversely, there was no shift band was observed when the solution harboring the probe alone (lane 1). In addition, the ability of ZmWRKY114 binding to the OsGA2ox4 promoter was alleviated along with increased concentrations of competitive probes (lane 3; 4). However, the mPr with mutated W-box elements did not change significantly (lane 5; 6).
Figure 4. EMSA analysis showing binding of recombinant ZmWRKY114 protein to the promoter of OsGA2ox4. (a) The W-box was predicted in the promoter region of OsGA2ox4. (b) The black arrow indicates the binding of ZmWRKY114 and the biotin-labeled W-box probe. Pr: TTGACTTTGACTTTGACT; mPr: TTTACTTTTACTTTTACT. The + and − signs represent the presence and absence of corresponding components, respectively. (c) Schematic diagrams of the effector and reporter constructs for dual luciferase assay. REN, Renilla luciferase; LUC, firefly luciferase. (d) Luciferase assay of the OsGA2ox4 promoter. proOsGA2ox4::LUC reporter constructs were transiently expressed in leaves of N.benthamiana together with either the empty vector or 35S::ZmWRKY114 effector, and the LUC signal was imaged at 72 h post-transfection. (e) The LUC/REN ratio as determined using a dual luciferase assay. Data are means (±SD) of at least three biological replicates. Statistical significance was determined using Student’s t-test: **P < .01
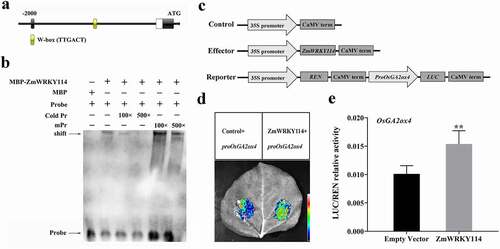
We then performed dual luciferase assays to determine whether ZmWRKY114 was an activator or a repressor of OsGA2ox4 transcription. A 256 bp OsGA2ox4 promoter fragment containing the TTGACT sequence was introduced into a vector to generate a reporter construct, which was then co-transformed with either the effector or the control (). Subsequent analysis showed that the co-expression of 35S::ZmWRKY114 with proOsGA2ox4::LUC led to a 1.5-fold rise in firefly luciferase (LUC) activity compared with control ( and e), suggesting that ZmWRKY114 directly binds to the OsGA2ox4 promoter and acts as a transcriptional activator.
Discussion
Since the important roles of WRKY TFs in plant development and stress response were demonstrated, members of this gene family have been identified from various higher plants.Citation17–22 At present, Wei et al.Citation23 identified 119 WRKY genes from the complete genome of maize B73 inbred line, and only a few members have been functionally characterized. Most of the identified maize WRKY members are involved in plant abiotic stress response, including ZmWRKY17, ZmWRKY58 and ZmWRKY114.Citation24,Citation25 Another maize WRKY protein ZmWRKY79 has been found to play an important role in plant biotic stress response via participation in phytohormone metabolism and ROS scavenging.Citation26
In this study, the reduced plant height of ZmWRKY114 transgenic lines implied this gene could involve in the regulation of plant growth, largely because of its highest expression in stem.Citation13 In particular, the phytohormone GA is an important factor in determining plant height through affecting the length and number of internode cells.Citation27 Light microscope of internode tissues indicated that the semi-dwarf phenotype of transgenic plants was caused by reduced cell numbers (-h). ZmWRKY114-overexpressing plants were more insensitive to GA3 and exogenous GA3 treatment could rescue the dwarfism phenotype of these transgenic plants, suggesting that ZmWRKY114 overexpression affected the homeostasis of GA in two transgenic lines, which was confirmed by the reduced bioactive GA1 and GA3 content in transgenic plants compared with that of WT.
To further uncover the molecular mechanisms underlying the decreased plant height of ZmWRKY114 transgenic lines, RNA-seq and qRT-PCR experiments were performed. The data showed that ZmWRKY114 overexpression could alter the expression of some GA related genes including OsEUI1, OsGA2ox4, OsYAB1 and OsLOL2. OsEUI1 was reported to encode a cytochrome P450 monooxygenase that deactivates bioactive GAs and transgenic plants overexpressing this gene exhibited a severe dwarf phenotype.Citation28 Gibberellin 2-oxidases (GA2oxs) control plant growth by inactivating endogenous bioactive GAs through 2β-hydroxylation.Citation3 Functional studies have testified that increased expression of these GA2oxs leads to reduced levels of bioactive GAs and corresponding dwarf phenotypes.Citation3,Citation7 GA2ox4 has been shown to act exclusively as a C19-GA 2-oxidase.Citation29 Overexpression of OsYAB1 also resulted in a semi-dwarf phenotype by impairing GA-mediated repression of GA3ox2.Citation30 In addition, as a positive regulator in the process of GA biosynthesis, OsLOL2 promotes the accumulation of bioactive GA through activating the expression of OsKS1, a gene encoding for GA biosynthetic enzyme.Citation31 In the light of the significant change of OsGA2ox4 expression in the transgenic plants reflected by RNA-seq and qRT-PCR assays, we scan the promoter region of OsGA2ox4 and a W-box cis-element was found in its promoter. Data from EMSA assay and dual-luciferase reporter assay showed that OsGA2ox4 is a direct target of ZmWRKY114 (). The result further proved that semi-dwarf phenotype in ZmWRKY114-overexpressing plants result from GA deficiency. All these results suggest that overexpression of the maize WRKY114 gene in transgenic rice regulates the expression level of OsGA2ox4 by binding to the W-box element in the promoter, and that increased expression of OsGA2ox4 leads to reduced bioactive GAs levels and a corresponding dwarf phenotype.
In addition, A phylogenetic analysis was carried out to evaluate the evolutionary relationships between ZmWRKY114 and other WRKY proteins. As shown in Fig. S2, ZmWRKY114 displayed a close relationship with OsWRKY51 and OsWRKY42 in rice. It has been reported that OsWRKY51 and OsWRKY71 physically interact in the nuclei to synergistically suppress the induction of Amy32b by GA.Citation32 Ectopic expression of OsWRKY42 results in reduced expression of cell wall damage and salt stress induced jasmonic acid biosynthesis and response genes.Citation33 It is worth mentioning that we found that the expression level of Ghd2, a novel CO-like gene which has been reported to be a regulator in the control of grain number, heading date, and plant height, was decreased approximately 4-fold in transgenic plants compared to WT.Citation34 Yeast two-hybrid and pull-down assays confirmed that OsGhd2 could interact with ZmWRKY114 (Fig. S3). Previous study showed that OsGhd2 also specifically interacts with three 14-3-3 proteins,Citation34 which have been reported to function as the WRKY-interacting partners.Citation35 The results implied that ZmWRKY114 involved in the control of plant height through a complex regulatory network. In addition, the decreased expression of OsGhd2 might affect the GA biosynthesis through altering the expression level of GA2-oxidase genes (OsGA2ox4 in this study), just like its homolog OsGhd7Citation36 to influence the plant height.
In conclusion, this study indicates that ZmWRKY114 functions as a negative regulator in GA biosynthesis during growth and development in rice.
Author contributions
RC and QM conceived and designed the research; XF, CB, MW, HY, WL and HC performed the experiments; XF and RC wrote the manuscript. All authors had read and approved the final manuscript.
Supplemental Material
Download Zip (6.3 MB)Acknowledgments
This work was supported by the China Postdoctoral Science Foundation (2018M642506) and the National Natural Science Foundation of China (31801365 and 31971895), the Natural Science Foundation of Anhui Province (1808085QC88).
Disclosure statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
Additional information
Funding
References
- Cho SH, Kang K, Lee SH, Lee IJ, Paek NC. OsWOX3A is involved in negative feedback regulation of the gibberellic acid biosynthetic pathway in rice (Oryza sativa). J Exp Bot. 2016;67(6):1–7. doi:https://doi.org/10.1093/jxb/erv559.
- Chu YL, Xu N, Wu Q, Yu B, Li XX, Chen RR, Huang JL. Rice transcription factor OsMADS57 regulates plant height by modulating gibberellin catabolism. Rice. 2019;12(1):38. doi:https://doi.org/10.1186/s12284-019-0298-6.
- Schomburg FM, Bizzell CM, Lee DJ, Zeevaart JAD, Amasino RM. Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell. 2003;15(1):151–163. doi:https://doi.org/10.1105/tpc.005975.
- Asano K, Hirano K, Ueguchi-Tanaka M, Angeles-Shim RB, Komura T, Satoh H, Kitano H, Matsuoka M, Ashikari M. Isolation and characterization of dominant dwarf mutants, Slr1-d, in rice. Mol Genet Genomics. 2009;281(2):223–231. doi:https://doi.org/10.1016/j.jchromb.2011.03.003.
- Li JA, Jiang JF, Qian QA, Xu YY, Zhang C, Xiao J, Du C, Luo W, Zou GX, Chen ML, et al. Mutation of rice BC12/GDD1, which encodes a kinesin-like protein that binds to a GA biosynthesis gene promoter, leads to dwarfism with impaired cell elongation. Plant Cell. 2011;23(2):628–640. doi:https://doi.org/10.1105/tpc.110.081901.
- Zhang J, Liu XQ, Li SY, Cheng ZK, Li CY. The rice semi-dwarf mutant sd37, caused by a mutation in CYP96B4, plays an important role in the fine-tuning of plant growth. PloS One. 2014:9. doi:https://doi.org/10.1371/journal.pone.0088068.
- Lo S-F, Yang S-Y, Chen K-T, Hsing Y-I, Zeevaart JAD, Chen L-J, Yu S-M. A novel class of gibberellin 2-Oxidases control semidwarfism, tillering, and root development in rice. Plant Cell. 2008;20(10):2603–2618. doi:https://doi.org/10.1105/tpc.108.060913.
- Rushton PJ, Somssich IE, Ringler P, Shen QXJ. WRKY transcription factors. Trends Plant Sci. 2010;15(5):247–258. doi:https://doi.org/10.1016/j.tplants.2010.02.006.
- Choi C, Hwang SH, Fang IR, Il Kwon S, Park SR, Ahn I, Kim JB, Hwang DJ. Molecular characterization of Oryza sativa WRKY 6, which binds to W-box-like element 1 of the Oryza sativa pathogenesis-related (PR) 10a promoter and confers reduced susceptibility to pathogens. New Phytol. 2015;208(3):846–859. doi:https://doi.org/10.1093/dnares/dsr048.
- Cai Y, Chen X, Xie K, Xing Q, Wu Y, Li J, Du C, Sun Z, Guo Z, Zhang JSJPO. Dlf1, a WRKY transcription factor, is involved in the control of flowering time and plant height in rice. PLoS One. 2014;9(7):e102529. doi:https://doi.org/10.1371/journal.pone.0102529.
- Zhang CQ, Xu Y, Lu Y, Yu HX, Gu MH, Liu QQ. The WRKY transcription factor OsWRKY78 regulates stem elongation and seed development in rice. Planta. 2011;234(3):541–554. doi:https://doi.org/10.1007/s00425-011-1423-y.
- Gu L, Wei H, Wang H, Su J, Yu SJBG. Characterization and functional analysis of GhWRKY42, a group IId WRKY gene, in upland cotton (Gossypium hirsutum L.). BMC Genet. 2018;19(1):48. doi:https://doi.org/10.1186/s12863-018-0653-4.
- Bo C, Chen HW, Luo GW, Li W, Zhang XG, Ma Q, Cheng BJ, Cai RH. Maize WRKY114 gene negatively regulates salt-stress tolerance in transgenic rice. Plant Cell Rep. 2020;39(1):135–148. doi:https://doi.org/10.1007/s00299-019-02481-3.
- Chen ML, Huang YQ, Liu JQ, Yuan BF, Feng YQ. Highly sensitive profiling assay of acidic plant hormones using a novel mass probe by capillary electrophoresis-time of flight-mass spectrometry. J Chromatogr B. 2011;879(13–14):938–944. doi:https://doi.org/10.1016/j.jchromb.2011.03.003.
- Han SH, Yoo SC, Lee BD, An G, Paek NC. Rice flavin-binding, kelch repeat, F-BOX 1 (OsFKF1) promotes flowering independent of photoperiod. Plant Cell Environ. 2015;38(12):2527–2540. doi:https://doi.org/10.1111/pce.12549.
- Garrido JMG, Morcillo RJL, Rodriguez JAM, Bole JAO. Variations in the mycorrhization characteristics in roots of wild-type and ABA-deficient tomato are accompanied by specific transcriptomic alterations. Mol Plant Microbe In. 2010;23(5):651–664. doi:https://doi.org/10.1094/mpmi-23-5-0651.
- Pandey SP, Somssich IE. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009;150(4):1648–1655. doi:https://doi.org/10.1104/pp.109.138990.
- Ulker B, Somssich IE. WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol. 2004;7(5):491–498. doi:https://doi.org/10.1016/j.pbi.2004.07.012.
- Tripathi P, Rabara RC, Langum TJ, Boken AK, Rushton DL, Boomsma DD, Rinerson CI, Rabara J, Reese RN, Chen XF, et al. The WRKY transcription factor family in Brachypodium distachyon. BMC Genomics. 2012:13. doi:https://doi.org/10.1186/1471-2164-13-270.
- Ling J, Jiang WJ, Zhang Y, Yu HJ, Mao ZC, Gu XF, Huang SW, and Xie BY. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genomics. 2011:12. doi:https://doi.org/10.1186/1471-2164-12-471.
- Tang J, Wang F, Hou XL, Wang Z, Huang ZN. Genome-Wide fractionation and identification of WRKY transcription factors in Chinese Cabbage (Brassica rapa ssp pekinensis) reveals collinearity and their expression patterns under abiotic and biotic stresses. Plant Mol Biol Rep. 2014;32(4):781–795. doi:https://doi.org/10.1007/s11105-013-0672-2.
- Bencke-Malato M, Cabreira C, Wiebke-Strohm B, Bucker-Neto L, Mancini E, Osorio MB, Homrich MS, Turchetto-Zolet AC, Mccg DC, Stolf R, et al. Genome-wide annotation of the soybean WRKY family and functional characterization of genes involved in response to Phakopsora pachyrhizi infection. BMC Plant Biol. 2014:14. doi:https://doi.org/10.1186/S12870-014-0236-0.
- Wei KF, Chen J, Chen YF, Wu LJ, Xie DX. Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 2012;19(2):153–164. doi:https://doi.org/10.1093/dnares/dsr048.
- Cai RH, Dai W, Zhang CS, Wang Y, Wu M, Zhao Y, Ma Q, Xiang Y, Cheng BJ. The maize WRKY transcription factor ZmWRKY17 negatively regulates salt stress tolerance in transgenic Arabidopsis plants. Planta. 2017;246(6):1215–1231. doi:https://doi.org/10.1007/s00425-017-2766-9.
- Cai RH, Zhao Y, Wang YF, Lin YX, Peng XJ, Li Q, Chang YW, Jiang HY, Xiang Y, Cheng BJ. Overexpression of a maize WRKY58 gene enhances drought and salt tolerance in transgenic rice. Plant Cell Tiss Org. 2014;119(3):565–577. doi:https://doi.org/10.1007/s11240-014-0556-7.
- Fu JY, Liu Q, Wang C, Liang J, Liu LJ, Wang Q. ZmWRKY79 positively regulates maize phytoalexin biosynthetic gene expression and is involved in stress response. J Exp Bot. 2018;69(3):497–510. doi:https://doi.org/10.1093/jxb/erx436.
- Wang L, Yin Y, Wang L-F, Wang ML, Zhao M, Tian Y, Li Y-F. Transcriptome profiling of the elongating internode of Cotton (Gossypium hirsutum L.) seedlings in response to Mepiquat Chloride. Front Plant Sci. 2020:10. doi:https://doi.org/10.3389/Fpls.2019.01751.
- Zhu YY, Nomura T, Xu YH, Zhang YY, Peng Y, Mao BZ, Hanada A, Zhou HC, Wang RX, Li PJ, et al. ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell. 2006;18(2):442–456. doi:https://doi.org/10.1105/tpc.105.038455.
- Rieu I, Eriksson S, Powers SJ, Gong F, Griffiths J, Woolley L, Benlloch R, Nilsson O, Thomas SG, Hedden P, et al. Genetic analysis reveals that C19-GA 2-oxidation is a major gibberellin inactivation pathway in arabidopsis. Plant Cell. 2008;20(9):2420–2436. doi:https://doi.org/10.1105/tpc.108.058818.
- Dai MQ, Zhao Y, Ma QF, Hu Y, Hedden PF, Zhang Q, Zhou DX. The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiol. 2007;144(1):121–133. doi:https://doi.org/10.1104/pp.107.096586.
- Xu CX, He CZ. The rice OsLOL2 gene encodes a zinc finger protein involved in rice growth and disease resistance. Mol Genet Genomic. 2007;278(1):85–94. doi:https://doi.org/10.1007/s00438-007-0232-2.
- Xie Z, Zhang ZL, Zou XL, Yang GX, Komatsu S, Shen QXJ. Interactions of two abscisic-acid induced WRKY genes in repressing gibberellin signaling in aleurone cells. Plant J. 2006;46(2):231–242. doi:https://doi.org/10.1111/j.1365-313X.2006.02694.x.
- Pillai SE, Kumar C, Patel HK, Sonti RV. Overexpression of a cell wall damage induced transcription factor, OsWRKY42, leads to enhanced callose deposition and tolerance to salt stress but does not enhance tolerance to bacterial infection. BMC Plant Biol. 2018:18. doi:https://doi.org/10.1186/s12870-018-1391-5.
- Liu JH, Shen JQ, Xu Y, Li XH, Xiao JH, Xiong LZ. Ghd2, a CONSTANS -like gene, confers drought sensitivity through regulation of senescence in rice. J Exp Bot. 2016;67(19):5785–5798. doi:https://doi.org/10.1093/jxb/erw344.
- Chi YJ, Yang Y, Zhou Y, Zhou J, Fan BF, Yu J-Q, Chen ZX. Protein–Protein interactions in the regulation of WRKY transcription factors. Mol Plant. 2013;6(2):287–300. doi:https://doi.org/10.1093/mp/sst026.
- Weng X, Wang L, Wang J, Hu Y, Du H, Xu C, Xing Y, Li X, Xiao J, Zhang QJPP. Grain number, plant height and heading date7 is a central regulator of growth, development, and stress response. Plant Physiol. 2014;164(2):735–747. doi:https://doi.org/10.1186/s12863-018-0653-4.
