ABSTRACT
The plant U-box (PUB) gene family, one of the major ubiquitin ligase families in plants, plays important roles in multiple cellular processes including environmental stress responses and resistance. The function of U-box genes has been well characterized in Arabidopsis and other plants. However, little is known about the tea plant (Camellia sinensis) PUB genes. Here, 89 U-box proteins were identified from the chromosome-scale referenced genome of tea plant. According to the domain organization and phylogenetic analysis, the tea plant PUB family were classified into ten classes, named Class I to X, respectively. Using previously released stress-related RNA-seq data in tea plant, we identified 34 stress-inducible CsPUB genes. Specifically, eight CsPUB genes were expressed differentially under both anthracnose pathogen and drought stresses. Moreover, six of the eight CsPUBs were upregulated in response to these two stresses. Expression profiling performed by qRT-PCR was consistent with the RNA-seq analysis, and stress-related cis-acting elements were identified in the promoter regions of the six upregulated CsPUB genes. These results strongly implied the putative functions of U-box ligase genes in response to biotic and abiotic stresses in tea plant.
KEYWORDS:
Introduction
Ubiquitination is one of the major types of post-translational modification (PTM) of proteins in eukaryotes. The ubiquitination pathway, involving the ubiquitin bond to a lysine residue in the target protein, requires three types of enzymes. The E1 activating enzyme is required to recruit and activate ubiquitin, and then the activated ubiquitin is delivered to the E2 conjugating enzyme, and finally the ubiquitin is transferred to the target protein for ubiquitination in presence of the E3 ubiquitin ligase.Citation1 Among these three types of enzymes, the E3 ligases are the largest family and serve as the central players for specifically recognizing the target substrates.Citation2,Citation3 Based on domain organization and function, plant E3 ligases are mainly classified into three families, namely HECT (homologous to E6-associated protein C terminus) ligases,Citation4 RING (really interesting new gene) ligasesCitation5 and U-box ligases.Citation6
The U-box ligases are a class of family with a conserved U-box domain containing approximately 70 amino acids that were first identified in the yeast UFD2 (ubiquitin fusion degradation protein-2).Citation7 The U-box shares great structural similarities with RING domain, while the major difference between these two domains is that the U-box lacks the classical zinc-binding ligand.Citation8 The U-box-containing genes are widely distributed in eukaryotic species. However, compared to the fewer genes in yeast and animals, the plant U-box (PUB) genes are evolutionarily expanded to a larger family with more members. There are only 2 and 11 U-box genes in yeastCitation7,Citation9 and mouse,Citation10 respectively, while 63 PUBs identified in Arabidopsis thaliana,Citation11,Citation12 67 in barley (Hordeum vulgare),Citation13 77 in rice,Citation14 91 in banana (Musa acuminata),Citation15 99 in Brassica oleraceaCitation16 and 125 in soybean.Citation17 The large expansion of U-box proteins in plants implies that PUBs may function in a wide range of cellular processes unique to plants. It is supported by emerging evidences that PUBs act as positive or negative regulators in diverse biological processes including plant response and adaptation to biotic and abiotic stresses. Rice OsPUB44 positively regulates PAMP-triggered immunity, and its E3 ligase activity is restrained by its direct interactor XopPXoo, one of the effectors of rice pathogen Xanthomonas oryzae pv. oryzae.Citation18 Similarly, potato StUBK, a U-box ligase with kinase domain, is targeted by the late blight pathogen effector PiSFI3, resulting in the inhibition of immune responses.Citation19 Another potato U-box ligase, StPUB17, positively regulates plant immunity by degradation of the negative regulator StKH17.Citation20,Citation21 Arabidopsis AtPUB25 and AtPUB26 negatively regulate plant immunity and disease resistance by degrading the inactivated immune kinase Botrytis-INDUCED KINASE 1 (BIK1).Citation22 Conversely, AtPUB25 and AtPUB26 act as positive players conferring freezing tolerance by targeting the cold signaling negative regulator AtMYB15 for degradation.Citation23 Additionally, Arabidopsis AtPUB46,Citation24,Citation25 rice OsPUB67Citation26 and wheat TaPUB1Citation27 are involved in promoting drought tolerance, whereas Arabidopsis AtPUB19,Citation28 AtPUB22 and AtPUB23,Citation29,Citation30 and wheat TaPUB26Citation31 serve as negative regulators in ABA dependent or independent abiotic stress responses.
The tea plant (Camellia sinensis) is an economically important woody crop that is extensively cultivated in tropical and subtropical areas including South China to produce the world’s most popular tea beverage.Citation32 Tea plant throughout the entire life cycle is frequently challenged by diverse abiotic and biotic stresses, such as droughtCitation33,Citation34 and anthracnose pathogens,Citation35,Citation36 resulting in serious quality and yield losses. However, the underlying molecular mechanisms on stress responses and resistance in tea plant remain poorly understood. Recently, several editions of the genomes of two varieties of tea plant, C. sinensis var. assamica (CSA)Citation37 and C. sinensis var. sinensis (CSS)Citation38–41 have been released successively, providing references for functional genomics research on response and adaptation to environmental stresses in tea plant and other Camellia species. Here, we describe the genome-wide identification, evolutionary relationship and expression analysis of tea plant U-box genes (CsPUBs) in response to the anthracnose pathogen Colletotrichum fructicola and drought stresses based on one of the chromosome-scale reference genomes of CSS, hopefully providing insights into U-box genes function on stress responses and breeding for resistance.
Materials and methods
Plant materials
The cultivar “Suchazao” of CSS (CSS-SCZ) was maintained in the Camellia Germplasm Resources Center of Guangxi Forestry Research Institute (GFRI). Two-year-old plants were grown in a growth chamber at 28°C with a photoperiod of 14 h and 80% humidity and used for pathogen and drought treatments in the present study.
Anthracnose pathogen infection
The anthracnose pathogen, C. fructicola, was isolated from diseased leaves of CSS-SCZ. The healthy and unwounded leaves (first-third leaves) were inoculated with conidial suspensions of C. fructicola (106 spores/mL) as described previously.Citation35 The leaf tissues were sampled at 0 h (before inoculation), 24 h, 48 h and 72 h after inoculation (hai), immediately frozen in liquid nitrogen and stored at −80°C for RNA isolation.
Drought treatment
For drought stress, plants were treated by gradually withholding water for 14 d and then re-watering for 3 d. The leaf tissues (first-third leaves) were sampled at the withholding point 0 d, 3 d, 7 d, 14 d and after rewatering for 3 d, respectively.
Sequence retrieval and identification of CsPUB proteins
One edition of the chromosome-scale reference genome of CSS was used hereCitation40 and downloaded from Tea Plant Information Achieve (TPIA, http://tpia.teaplant.org/).Citation42 To identify the CsPUB genes, the Hidden Markov Model (HMM) profile for U-box domains (PF04564) were downloaded from Pfam database (http://pfam.xfam.org/).Citation43 HMM search in HMMER 3.3.2 (http://hmmer.org/download.html) against the CSS genome was performed with an expected value (e-value) threshold of <0.001. Meanwhile, protein blast against CSS genome database on TPIA with Arabidopsis PUB protein sequences downloaded from The Arabidopsis Information Resource (TAIR, https://www.arabidopsis.org/) was performed (e-value = 0.001). Taken these two results together, the final members of CsPUBs were obtained and confirmed by PfamScan (e-value = 0.001, https://www.ebi.ac.uk/Tools/pfa/pfamscan/)Citation44 and SMART (e-value = 0.001, http://smart.embl-heidelberg.de/).Citation45 The basic information of CsPUBs including intron number, intron length, chromosome localization was determined based on the CSS genome database.Citation40 The subcellular localization of CsPUB proteins was predicted using the TargetP 2.0 server (https://services.healthtech.dtu.dk/service.php?TargetP).Citation46
Classification and phylogenetic analysis of PUB genes
The conserved domains or motifs of CSS and Arabidopsis PUB proteins were identified using the online tools PfamScan (e-value = 0.001)Citation44 and SMART (e-value = 0.001).Citation45 According to the extra domain organization, the classification of CsPUB proteins was determined. Meanwhile, the phylogenetic trees were constructed by MEGA7 (www.megasoftware.net) based on Neighbor-Joining method with 1000 bootstrap replications and partial deletion.Citation47 A motif-based sequence analysis tool, the MEME suite tool (http://meme-suite.org/),Citation48 was used to identify extra conserved motifs in Class V PUB proteins. Gene duplication events of U-box genes were analyzed by MCScanX (http://chibba.pgml.uga.edu/mcscan2/)Citation49 and visualized by Cirsco software (http://www.circos.ca/software/).Citation50
Stress-related RNA-Seq data analysis
Two previously released stress-related RNA-seq data-sets of tea plantCitation35,Citation51 were used in the present study to analyze the expression of CSS U-box genes. Expression levels of tea plant genes response to the anthracnose pathogen C. fructicola (accession numbers: SRX3142216, SRX3142220, SRX3142221, SRX3142226, SRX3142227, SRX3142228, SRX3142230, SRX3142231, SRX3142232)Citation35 and drought (accession numbers: SRX2234716, SRX2234790, SRX2234791, SRX2234821, SRX2234822, SRX2234823)Citation51 were available from NCBI SRA (https://www.ncbi.nlm.nih.gov/sra). These two RNA-Seq data were remapped back to the CSS genome used here.Citation40 The differentially expressed genes (DEGs) were defined under the criteria of fold change| (FC) ≥ 1.5. Heatmaps and the Venn diagram were constructed using Excel and PowerPoint of Office 365, respectively.
Quantitative real-time PCR assays
Total RNAs were extracted from the stress-treated samples described above using TRIzol kit (Invitrogen, http://www.invitrogen.com). Quantitative real-time PCR (qRT-PCR) assays were performed as described previouslyCitation35 using an ABI PRISM 7500 Real-time PCR System (Applied Biosystems) and CsPUB specific primers (Table S1) following instructions of the supplier. CsGAPDH1 (CSS0032053) was served as an internal control.
Cis-acting elements analysis
The 1500 bp promoter sequences upstream of the ATG start codon of CsPUB genes were retrieved from TPIA.Citation42 The cis-acting regulatory elements of CsPUB genes were predicted using PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).Citation52
Results
Genome-wide identification of PUB genes in tea plant
To retrieve the PUBs genes from the tea plant, one of the chromosome level reference genome sequences of CSSCitation40 was used in the present study. The hidden Markov model (HMM) profile for U-box domain (PF04564) was obtained from Pfam database.Citation43 HMM search against the CSS genome was performed, and 86 PUB genes were identified from the tea plant. Furthermore, protein blast against the CSS genome database on TPIACitation42 with Arabidopsis PUB protein sequences was performed, and three more CsPUBs were identified. Overall, there are 75 CsPUBs located in all 15 chromosomes, and other 14 CsPUBs found in Contigs. According to the position on the chromosomes, these PUBs of tea plant were named as CsPUB1 to CsPUB89 in order. The basic information of AtPUBs and CsPUBs including the gene name, chromosome position, the number and average length of intron and protein length were listed in Table S2 and Table S3, respectively. Sixty-seven of 89 CsPUB genes in tea plant contained more than one intron, and the intron length was sharply divergent among the tea plant PUB gene family. The average intron length of CsPUBs was 948 bp, which is much larger than that of AtPUBs. The average length of introns of 17 CsPUB genes was larger than 1kb, whereas the average length of introns of all Arabidopsis PUB genes was shorter than 300 bp (Figure S1). Meanwhile, the length of CsPUB protein sequences ranged from 90 to 1066 amino acids, and the average protein length between CsPUBs and AtPUBs was of no significant difference in spite of the large divergence on intron length. Additionally, the PUB proteins of tea plant were predicted to be localized throughout the cell, including nuclear, cytoplasmic, plasma membrane, mitochondrial, chloroplast, and Golgi locations, and the majority of CsPUBs contained putative nuclear localization signal (Table S3).
Classification and phylogenetic analysis of the tea plant PUB proteins
The protein sequences of CsPUBs were used to search with PfamScanCitation44 and SMARTCitation45 tools to identify the extra domains or motifs. The PUB proteins of Arabidopsis were analyzed and reconfirmed in the current study as well. As AtPUB62 (At3g49065) was removed by TAIR (https://www.arabidopsis.org/), 63 AtPUBs were introduced here. Various other domains/motifs were detected in PUB proteins of tea plant and Arabidopsis in addition to U-box domain. According to the presence of the extra domains/motifs, tea plant PUB proteins were grouped into ten classes, following and incorporating the previous defined classes in Arabidopsis (). The phylogenetic tree constructed with Arabidopsis and CSS PUB proteins supported the classification ().
Table 1. Domain organizations of PUB proteins of tea plant and Arabidopsis.
Figure 1. Phylogenetic analysis of the PUB proteins from Arabidopsis and tea plant. The tree was constructed by MEGA7 based on Neighbor-Joining method with 1000 bootstrap replications and partial deletion.Citation47 The PUB proteins of Arabidopsis (AtPUBs in red circles) and CSS (CsPUBs in blue triangles) were classified into ten classes represented in different colors.
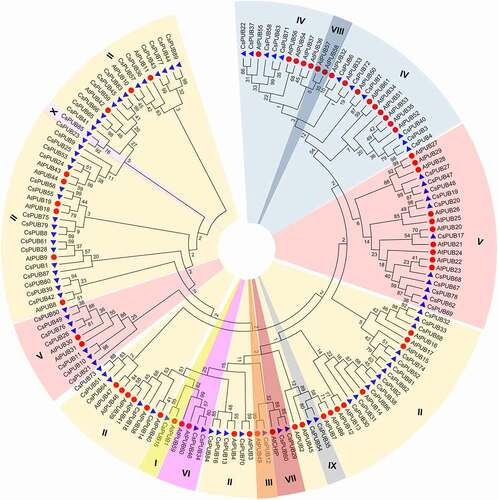
Similar to Arabidopsis, Class I consists of a single PUB protein, CsUFD2 (CsPUB15), which shares 23, 74% and 61% sequence identities with yeast UFD2, Arabidopsis AtUFD2 (AtPUB1) and rice OsUFD2, respectively. Class II with 45 CsPUBs and 28 AtPUBs containing the armadillo (ARM) domain was the largest class in CSS and Arabidopsis. The ARM repeat domain with approximately 40 amino acids was initially identified in Drosophila melanogasterCitation53 and found in all eukaryotes.Citation54 It was involved in protein–protein interaction and functioned in diverse cellular processes including self-incompatibility, hormone signaling and immunity in higher plants.Citation54–56
Class III was originally defined as the second largest class in Arabidopsis and rice PUB families.Citation14 Recent analysis based on newly updated databasesCitation44,Citation45 showed that class III was composed of the PUB proteins with cyclophilin-type peptidyl-prolyl isomerase (PPIase) domain. Our analysis was consistent with the results described previously in barleyCitation13 and other plants.Citation57,Citation58
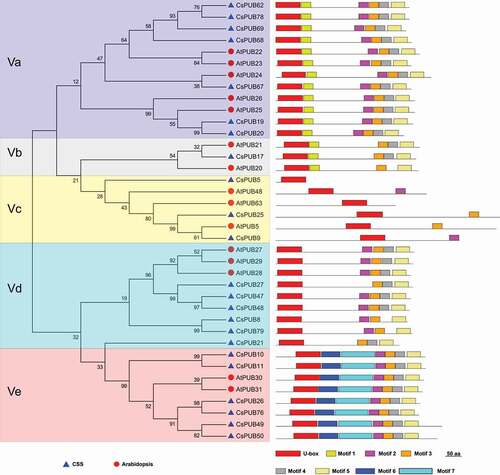
The exact second largest class was Class V with 25 CsPUBs and 15 AtPUBs termed by containing U-box only without any other well-known or defined domains/motifs. To get more information about the structure of Class V PUB proteins, the motif structure of these 38 PUB proteins was analyzed. In addition to the U-box domain, seven extra undefined conserved motifs were identified (). According to the motif organization, the Class V PUBs were categorized into five subclasses (Va – Ve). Class Va, Class Vb and Class Vd contained 3 to 5 extra motifs, while only one or none extra motifs were found in Class Vc. Two extra motifs, motif 6 and motif 7, specially existed in Class Ve. It may provide positive references for the functional study of these Class V U-box genes.
Figure 2. Motif structures and phylogenetic tree of Class V PUB proteins of Arabidopsis and tea plant. Seven extra conserved unknown motifs were identified from the Class V PUB proteins of Arabidopsis and CSS, which hence were further categorized into five subclasses according to the motif structures.
The third largest class, Class IV with 11 CsPUBs and 14 AtPUBs, harbored a kinase domain at the N-terminal region. Phosphorylation by protein kinases was another major type of PTM that controls diverse signaling pathways in eukaryotic cells.Citation59 This class PUB proteins with putative functions of both ligase and kinase might play significant roles in multiple cellular processes in higher plants.
Besides the classes mentioned above, there were several other smaller classes, including Class VI with WD40 repeats domain and Class VII with tetratrico peptide repeat (TPR) domain (). These two domains were documented to be involved in protein interactions.Citation60,Citation61 WD40 repeats containing PUB proteins, CsPUB34, CsPUB46, AtPUB59 and AtPUB60, were homologous to yeast and human Prp19, which was involved in pre-mRNA splicing and DNA repair.Citation62,Citation63 Additionally, it existed another CsPUB with WD40 domain, CsPUB73, which was clustered with Class II PUB proteins.
Moreover, similar to rice and barley, there was no MIFG domain containing PUBs found in CSS. Conversely, there were two smaller classes, Class IX with KAP domain and Class X with PPR repeat domain, identified only in CSS compared to Arabidopsis. KAP (kinesin-associated protein) domain was associated with motor function,Citation64 and PPR (Pentatricopeptide repeat) domain was involved in organelle biogenesis and mRNA processing.Citation65,Citation66
Gene duplication analysis
Gene duplication events are of significant importance for gene family expansion. Here, the duplication events of the PUB genes in CSS genome were analyzed. There were two pairs of tandem repeats, CsPUB10/11 and CsPUB81/82, and 17 pairs of segmental genes between CSS chromosomes (, red lines; Table S4). These results showed that both tandem and segmental gene duplications contributed to the expansion of the PUB gene family in CSS. Specially, it was indicated that the segmental duplication was the major driving force of the CsPUB gene expansion.
Figure 3. Duplication analysis of PUB genes of Arabidopsis and tea plant. The PUB genes in 5 chromosomes of Arabidopsis (in orange) and 15 chromosomes of CSS (in cyan) were introduced here. The orthologous pairs between AtPUBs and CsPUBs were highlighted in blue. The gene pairs of AtPUBs were colored in yellow, while the gene pairs of CsPUBs were colored in red. The gray lines marked the rest genes pairs in Arabidopsis and tea plant.
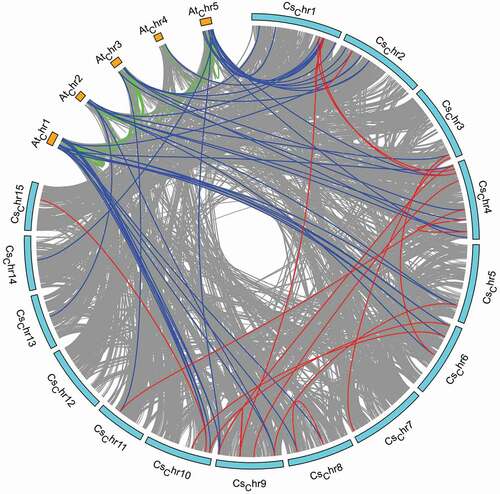
Furthermore, the collinear relationship between CSS and A. thaliana was completed (, blue lines; Table S4). There were 49 orthologous gene pairs between CsPUBs and AtPUBs, of which 12 AtPUBs had two orthologous copies in CSS. It was implied that these orthologous genes might share similar function in these two species.
Expression analysis of CsPUB genes under anthracnose pathogen and drought stresses
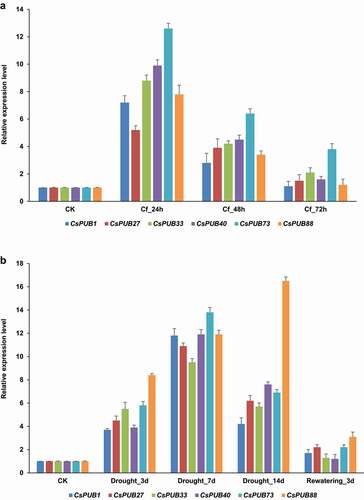
Tea plant is frequently challenged by various environmental stresses, including anthracnose pathogens and drought. In order to investigate the expression patterns of CsPUBs under these two stresses, two recently published stress-related RNA-seq libraries were used in the current study. There were 14 and 28 CsPUB genes differentially expressed in response to anthracnose pathogen () and Table S5) and drought ( and Table S6), respectively. Among them, eight CsPUB genes shared overlapping roles responding to both stresses ()). Furthermore, six of the eight CsPUBs, CsPUB1, CsPUB27, CsPUB33, CsPUB40, CsPUB73 and CsPUB88, were significantly upregulated under these two stresses. The expression levels of these six CsPUBs under pathogen infection and drought treatment were confirmed by qRT-PCR (), which were consistent with the result of RNA-seq analysis.
Figure 4. Expression profiles of CsPUB genes under C. fructicola infection and drought treatment. (a) 14 CsPUB genes were differentially expressed responding to anthracnose pathogen; (b) 28 CsPUB genes were differentially expressed responding to drought stress; (c) Venn diagram analysis of the 34 total differentially expressed CsPUBs under these two stresses. Eight CsPUBs were expressed differentially under both stresses, while six of the eight CsPUBs (in bold) were upregulated under both stresses.
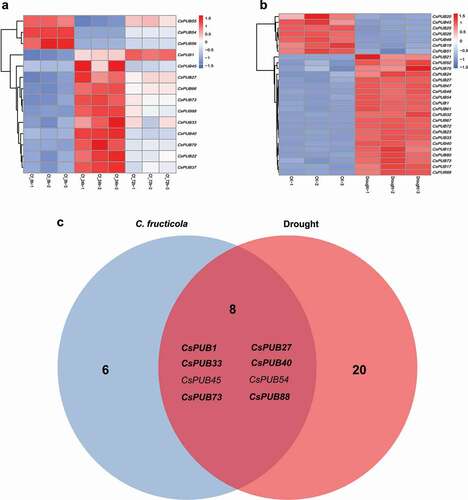
Figure 5. Expression patterns of the six CsPUB genes confirmed by qRT-PCR. (a) Expression levels of the six upregulated CsPUBs under C. fructicola attack; (b) Expression levels of the six upregulated CsPUBs under drought treatment. Three independent experiments were performed. CsGAPDH1 acted as the internal control gene to normalize the expression data.
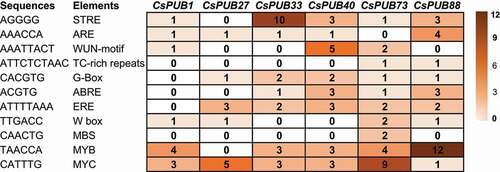
Stress-associated cis-acting elements in the six CsPUB promoters
To get more evidences for the six upregulated CsPUB genes on stress responses, the cis-acting regulatory elements in the promoters of the six CsPUBs were predicted by PlantCARE.Citation52 There were 11 well-known stress-related elements identified (). STRE (stress response element; AGGGG) was involved in rapid response to abiotic stresses such as heat and osmotic stresses,Citation67,Citation68 while the ARE (antioxidant response element; AAACCA) was related to the anaerobic and oxidative stress induction.Citation69 WUN-motif (wound-responsive element; AAATTACT), TC-rich repeats (ATTCTCTAAC),Citation70 and G-box (CACGTG)Citation71 were three types of elements involved in biotic stress response and defense. ABRE (abscisic acid response element; ACGTG) and ERE (ethylene response elements; ATTTTAAA) were related to the major stress-related hormones ABA and ethylene, respectively. W box (WRKY binding site; TTGACC), MBS (MYB binding site; CAACTG), MYB (TAACCA) and MYC (CATTTG) were associated with the transcription factors WRKY, MYB and MYC, respectively, which were involved in environmental stresses response and adaptation.Citation72,Citation73 These results further provided auxiliary evidence for the six up-regulated CsPUB genes in response to abiotic and biotic stresses.
Figure 6. Stress-related cis-acting elements in the six upregulated CsPUB genes. The cis-acting regulatory elements in the promoters of the six CsPUBs were predicted by PlantCARE.Citation52 Eleven well-known stress-related elements were identified, which were reported to be involved in environmental stress response and adaptation. The degree of red represents the number of the elements in the CsPUB promoters.
Discussion
The PUB gene family, one of the largest ubiquitin ligase families in plants, was known to play significant roles in plant response to biotic and abiotic stresses. More and more evidences were reported about the functional characterization of PUBs in Arabidopsis,Citation22,Citation23 riceCitation18 and other plants.Citation19–21,Citation31 However, the function of tea plant PUB genes on stress response is largely unknown. Here, we identified 89 PUB genes from tea plant genome. There were 14 pathogen-induced CsPUBs and 28 drought-induced CsPUBs identified from two recently published stress-related RNA-seq libraries, respectively. Among them, the expression levels of six CsPUB genes were significantly upregulated in response to both anthracnose pathogen and drought stresses.
The CsPUB and AtPUB proteins were categorized into ten classes based on the domain organization, which was consistent with the adjusted structure of U-box genes described by Ryu et al.Citation13 There were two CSS special classes of PUB genes identified from tea plant, Class IX with KAP domain (CsPUB35 and CsPUB54) and Class X with PRR repeats (CsPUB85). KAP domain was shown to be involved in motor functionCitation64 and was found in U-box proteins of Medicago truncatula as well.Citation57 PPR domain was reported to be associated with organelle biogenesis and mRNA processing.Citation65,Citation66 Moreover, these three CsPUBs were closely neighbored to the Class II PUB proteins with ARM domain. Hence, the functional characterization of these three CsPUB genes is needed. Compared to Arabidopsis PUB gene family, the CsPUB family was with a notable expansion in total. Obviously, the larger Class II of CsPUBs in tea plant mainly led to the expansion, over a half of CsPUBs belonged to Class II. Furthermore, it was indicated that the tandem and segmental gene duplications contributed to the CsPUB family expansion. Additionally, the number of Class V CsPUB genes also slightly increased in comparison to that of Class V AtPUBs. Class V was ever defined as the U-box proteins without known domains. However, we identified several undefined or unknown conserved motifs from Class V PUB proteins in Arabidopsis and tea plant. The motifs structure of Class V varied widely, and it could be further assorted into five different subclasses. It might be positively helpful for the further investigation of Class V U-box genes.
Based on previously released stress-related RNA-seq data, we identified approximately 40% of CsPUB genes expressed differentially responding to drought or fungal attack by C. fructicola. Among them, eight CsPUB genes were differentially expressed in response to both anthracnose pathogen and drought stresses. Particularly, six of the eight CsPUBs were up-regulated under these two stresses. CsPUB1, CsPUB33 and CsPUB88 all belonged to Class II. CsPUB73, though with WD40 domain, was also clustered with Class II CsPUB proteins with ARM domain. Multiple evidences showed that ARM domain containing proteins were involved in diverse cellular processes including cytoskeletal regulation, flowering, self-incompatibility and disease resistance.Citation54–56 Specially, the closest ortholog of CsPUB33 and CsPUB88, Arabidopsis AtPUB16 and AtPUB17, were reported to function in plant defense.Citation74,Citation75 CsPUB27, the Class Vd U-box protein with undefined conserved motifs, shared high sequence similarity to its closest ortholog AtPUB27, which was immediately activated on infection with a pathogen-derived elicitor.Citation76 CsPUB40 was the only Class IV U-box gene among the six upregulated genes. Class IV CsPUBs harbor a kinase domain with putative kinase function, which play central roles in multiple cellular processes through its phosphorylation.Citation59 The function of this Class U-box genes in Arabidopsis and other plants remains largely unknown. Referentially, a well-characterized gene encoding a protein with both E3 ligase and kinase activities, namely KEEP ON GOING (KEG), plays critical roles in seedling establishment,Citation77 hormone signalingCitation77–79 and stress responses.Citation80,Citation81 These evidences implied that the six CsPUB genes might putatively function on stress responses. However, whether and how these CsPUB genes play dual positive roles in response to both biotic and abiotic stresses, more proofs are required.
In conclusion, there were 89 U-box genes identified from tea plant, while six of them were upregulated under both anthracnose pathogen and drought stresses. Further investigations on these six CsPUB genes are needed to clarify the potential positive roles and underlying molecular mechanisms in biotic and abiotic stress responses.
Author contributions
J. L. M., R. Q. Z. and H. Z. L. conceived and designed the experiments; H. Z. L., W. J. L. and D. X. Z. performed the experiments and analyzed the data; H.Z.L., R. Q. Z. and J. L. M. wrote the paper.
Supplemental Material
Download Zip (450 KB)Acknowledgments
We thank Dr Hang Ye and Ms Yu Wei from GFRI for their assistance in this study. The sweet wife (Y. Y.) and the lovely son (C. Y. L.) of H. Z. L. are acknowledged for their warm support.
Disclosure statement
The authors declare no conflict of interests.
Supplementary Material
Supplemental data for this article can be accessed on the publisher’s website.
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Callis J. The ubiquitination machinery of the ubiquitin system. Arabidopsis Book. 2014;12:1. doi:https://doi.org/10.1199/tab.0174.
- Chen L, Hellmann H. Plant E3 ligases: flexible enzymes in a sessile world. Mol Plant. 2013;6(5):1388–12. doi:https://doi.org/10.1093/mp/sst005.
- Mazzucotelli E, Belloni S, Marone D, De Leonardis A, Guerra D, Di Fonzo N, Cattivelli L, Mastrangelo A. The E3 ubiquitin ligase gene family in plants: regulation by degradation. Current Genomics. 2006;7(8):509–522. doi:https://doi.org/10.2174/138920206779315728.
- Marín I, Zhang Z. Evolution of plant HECT ubiquitin ligases. PLoS ONE. 2013;8(7):e68536–e. doi:https://doi.org/10.1371/journal.pone.0068536.
- Stone SL, HauksdÓttir H, Troy A, Herschleb J, Kraft E, Callis J. Functional analysis of the RING-type ubiquitin ligase family of arabidopsis. Plant Physiol. 2005;137(1):13–30. doi:https://doi.org/10.1104/pp.104.052423.
- Yee D, Goring DR. The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J Exp Bot. 2009;60(4):1109–1121. doi:https://doi.org/10.1093/jxb/ern369.
- Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96(5):635–644. doi:https://doi.org/10.1016/S0092-8674(00)80574-7.
- Aravind L, Koonin EV. The U box is a modified RING finger — a common domain in ubiquitination. Curr Biol. 2000;10(4):R132–R4. doi:https://doi.org/10.1016/S0960-9822(00)00398-5.
- Ohi MD, Vander Kooi CW, Rosenberg JA, Chazin WJ, Gould KL. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat Struct Mol Biol. 2003;10(4):250–255. doi:https://doi.org/10.1038/nsb906.
- Hou X, Zhang W, Xiao Z, Gan H, Lin X, Liao S, Han C. Mining and characterization of ubiquitin E3 ligases expressed in the mouse testis. BMC Genomics. 2012;13(1):495. doi:https://doi.org/10.1186/1471-2164-13-495.
- Azevedo C, Santos-Rosa MJ, Shirasu K. The U-box protein family in plants. Trends Plant Sci. 2001;6(8):354–358. doi:https://doi.org/10.1016/s1360-1385(01)01960-4.
- Wiborg J, O’Shea C, Skriver K. Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochem J. 2008;413(3):447–457. doi:https://doi.org/10.1042/BJ20071568.
- Ryu MY, Cho SK, Hong Y, Kim J, Kim JH, Kim GM, Chen YJ, Knoch E, Møller BL, Kim WT, Lyngkjær MF, Yang SW. Classification of barley U-box E3 ligases and their expression patterns in response to drought and pathogen stresses. BMC Genomics. 2019;20(1):326. doi:https://doi.org/10.1186/s12864-019-5696-z.
- Zeng LR, Park CH, Venu RC, Gough J, Wang GL. Classification, expression pattern, and E3 ligase activity assay of rice U-box-containing proteins. Mol Plant. 2008;1(5):800–815. doi:https://doi.org/10.1093/mp/ssn044.
- Hu H, Dong C, Sun D, Hu Y, Xie J. Genome-wide identification and analysis of U-box E3 ubiquitin protein ligase gene family in banana. Int J Mol Sci. 2018;19(12):3874. doi:https://doi.org/10.3390/ijms19123874.
- Hu D, Xie Q, Liu Q, Zuo T, Zhang H, Zhang Y, Lian X, Zhu L. Genome-Wide Distribution, Expression and Function Analysis of the U-Box Gene Family in Brassica oleracea L. Genes. 2019;10(12):1000. doi:https://doi.org/10.3390/genes10121000.
- Wang N, Liu Y, Cong Y, Wang T, Zhong X, Yang S, Li Y, Gai J. Genome-wide identification of soybean U-box E3 ubiquitin ligases and roles of GmPUB8 in negative regulation of drought stress response in Arabidopsis. Plant Cell Physiol. 2016;57(6):1189–1209. doi:https://doi.org/10.1093/pcp/pcw068.
- Ishikawa K, Yamaguchi K, Sakamoto K, Yoshimura S, Inoue K, Tsuge S, Kojima C, Kawasaki T. Bacterial effector modulation of host E3 ligase activity suppresses PAMP-triggered immunity in rice. Nat Commun. 2014;5(1):5430. doi:https://doi.org/10.1038/ncomms6430.
- He Q, McLellan H, Hughes RK, Boevink PC, Armstrong M, Lu Y, Banfield MJ, Tian Z, Birch PRJ. Phytophthora infestans effector SFI 3 targets potato UBK to suppress early immune transcriptional responses. New Phytol. 2019;222(1):438–454. doi:https://doi.org/10.1111/nph.15635.
- He Q, McLellan H, Boevink PC, Sadanandom A, Xie C, Birch PR, Tian Z. U-box E3 ubiquitin ligase PUB17 acts in the nucleus to promote specific immune pathways triggered by Phytophthora infestans. J Exp Bot. 2015;66(11):3189–3199. doi:https://doi.org/10.1093/jxb/erv128.
- McLellan H, Chen K, He Q, Wu X, Boevink PC, Tian Z, Birch PRJ. The ubiquitin E3 ligase PUB17 positively regulates immunity by targeting a negative regulator, KH17, for degradation. Plant Commun. 2020;1(4):100020. doi:https://doi.org/10.1016/j.xplc.2020.100020.
- Wang J, Grubb LE, Wang J, Liang X, Li L, Gao C, Ma M, Feng F, Li M, Li L, et al. A regulatory module controlling homeostasis of a plant immune kinase. Mol Cell. 2018;69(3):493–504. doi:https://doi.org/10.1016/j.molcel.2017.12.026.
- Wang X, Ding Y, Li Z, Shi Y, Wang J, Hua J, Gong Z, Zhou J-M, Yang S. PUB25 and PUB26 promote plant freezing tolerance by degrading the cold signaling negative regulator MYB15. Dev Cell. 2019;51(2):222–235. doi:https://doi.org/10.1016/j.devcel.2019.08.008.
- Adler G, Mishra AK, Maymon T, Raveh D, Bar-Zvi D. Overexpression of Arabidopsis ubiquitin ligase AtPUB46 enhances tolerance to drought and oxidative stress. Plant Sci. 2018;276:220–228. doi:https://doi.org/10.1016/j.plantsci.2018.08.018.
- Adler G, Konrad Z, Zamir L, Mishra AK, Raveh D, Bar-Zvi B-ZD. The Arabidopsis paralogs, PUB46 and PUB48, encoding U-box E3 ubiquitin ligases, are essential for plant response to drought stress. BMC Plant Biol. 2017;17(1):8. doi:https://doi.org/10.1186/s12870-016-0963-5.
- Qin Q, Wang Y, Huang L, Du F, Zhao X, Li Z, Wang W, Fu B. A U-box E3 ubiquitin ligase OsPUB67 is positively involved in drought tolerance in rice. Plant Mol Biol. 2020;102(1–2):89–107. doi:https://doi.org/10.1007/s11103-019-00933-8.
- Zhang G, Zhang M, Zhao Z, Ren Y, Li Q, Wang WW. Wheat TaPUB1 modulates plant drought stress resistance by improving antioxidant capability. Sci Rep. 2017;7(1):7549. doi:https://doi.org/10.1038/s41598-017-08181-w.
- Liu Y-C, Wu Y-R, Huang X-H, Sun J, Xie Q. AtPUB19, a U-Box E3 ubiquitin ligase, negatively regulates abscisic acid and drought responses in Arabidopsis thaliana. Mol Plant. 2011;4(6):938–946. doi:https://doi.org/10.1093/mp/ssr030.
- Zhao J, Zhao L, Zhang M, Zafar SA, Fang J, Li M, Zhang W, Li X. Arabidopsis E3 ubiquitin ligases PUB22 and PUB23 negatively regulate drought tolerance by targeting ABA receptor PYL9 for degradation. Int J Mol Sci. 2017;18(9):1841. doi:https://doi.org/10.3390/ijms18091841.
- Cho SK, Ryu MY, Song C, Kwak JM, Kim WT. Arabidopsis PUB22 and PUB23 are homologous U-box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell. 2008;20(7):1899–1914. doi:https://doi.org/10.1105/tpc.108.060699.
- Wu Y, Wang W, Li Q, Zhang G, Zhao X, Li G, Li Y, Wang Y, Wang W. The wheat E3 ligase TaPUB26 is a negative regulator in response to salt stress in transgenic Brachypodium distachyon. Plant Sci. 2020;294:110441. doi:https://doi.org/10.1016/j.plantsci.2020.110441.
- Xia E-H, Tong W, Wu Q, Wei S, Zhao J, Zhang -Z-Z, Wei C-L, Wan X-C. Tea plant genomics: achievements, challenges and perspectives. Hortic Res. 2020;7(1):7. doi:https://doi.org/10.1038/s41438-019-0225-4.
- Parmar R, Seth R, Singh P, Singh G, Kumar S, Sharma RK. Transcriptional profiling of contrasting genotypes revealed key candidates and nucleotide variations for drought dissection in Camellia sinensis (L.) O. Kuntze. Sci Rep. 2019;9(1):7487. doi:https://doi.org/10.1038/s41598-019-43925-w.
- Guo Y, Zhao S, Zhu C, Chang X, Yue C, Wang Z, Lin Y, Lai Z. Identification of drought-responsive miRNAs and physiological characterization of tea plant (Camellia sinensis L.) under drought stress. BMC Plant Biol. 2017;17(1):211. doi:https://doi.org/10.1186/s12870-017-1172-6.
- Wang Y, Hao X, Lu Q, Wang L, Qian W, Li N, Ding C, Wang X, Yang Y. Transcriptional analysis and histochemistry reveal that hypersensitive cell death and H2O2 have crucial roles in the resistance of tea plant (Camellia sinensis (L.) O. Kuntze) to anthracnose. Hortic Res. 2018;5(1):18. doi:https://doi.org/10.1038/s41438-018-0025-2.
- Wang Y-C, Hao X-Y, Wang L, Xiao B, Wang X-C, Yang Y-J. Diverse Colletotrichum species cause anthracnose of tea plants (Camellia sinensis (L.) O. Kuntze) in China. Sci Rep. 2016;6(1):35287. doi:https://doi.org/10.1038/srep35287.
- Xia E-H, Zhang H-B, Sheng J, Li K, Zhang Q-J, Kim C, Zhang Y, Liu Y, Zhu T, Li W, et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol Plant. 2017;10(6):866–877. doi:https://doi.org/10.1016/j.molp.2017.04.002.
- Wei C, Yang H, Wang S, Zhao J, Liu C, Gao L, Xia E, Lu Y, Tai Y, She G, et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc Natl Acad Sci U S A. 2018;115(18):E4151–E8. doi:https://doi.org/10.1073/pnas.1719622115.
- Chen J-D, Zheng C, Ma J-Q, Jiang C-K, Ercisli S, Yao M-Z, Chen L. The chromosome-scale genome reveals the evolution and diversification after the recent tetraploidization event in tea plant. Hortic Res. 2020;7(1):63. doi:https://doi.org/10.1038/s41438-020-0288-2.
- Xia E, Tong W, Hou Y, An Y, Chen L, Wu Q, Liu Y, Yu J, Li F, Li R, et al. The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into its genome evolution and adaptation. Mol Plant. 2020;13(7):1013–1026. doi:https://doi.org/10.1016/j.molp.2020.04.010.
- Zhang Q-J, Li W, Li K, Nan H, Shi C, Zhang Y, Dai Z-Y, Lin Y-L, Yang X-L, Tong Y, et al. The chromosome-level reference genome of tea tree unveils recent bursts of non-autonomous LTR retrotransposons in driving genome size evolution. Mol Plant. 2020;13(7):935–938. doi:https://doi.org/10.1016/j.molp.2020.04.009.
- Xia E-H, Li F-D, Tong W, Li P-H, Wu Q, Zhao H-J, Ge R-H, Li R-P, Li -Y-Y, Zhang -Z-Z, et al. Tea Plant Information Archive: a comprehensive genomics and bioinformatics platform for tea plant. Plant Biotechnology Journal. 2019;17(10):1938–1953. doi:https://doi.org/10.1111/pbi.13111.
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44(D1):D279–D285. doi:https://doi.org/10.1093/nar/gkv1344.
- Madeira F, Park Y, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47(W1):W636–W41. doi:https://doi.org/10.1093/nar/gkz268.
- Letunic I, Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018;46(D1):D493–D6. doi:https://doi.org/10.1093/nar/gkx922.
- Almagro Armenteros JJ, Salvatore M, Emanuelsson O, Winther O, von Heijne G, Elofsson A, Nielsen H. Detecting sequence signals in targeting peptides using deep learning. Life Sci Alliance. 2019;2(5):e201900429. doi:https://doi.org/10.26508/lsa.201900429.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi:https://doi.org/10.1093/molbev/msw054.
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server):W202–8. doi:https://doi.org/10.1093/nar/gkp335.
- Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee T-H, Jin H, Marler B, Guo H, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49. doi:https://doi.org/10.1093/nar/gkr1293.
- Krzywinski MI, Schein JE, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi:https://doi.org/10.1101/gr.092759.109.
- Zheng C, Wang Y, Ding Z, Zhao L. Global transcriptional analysis reveals the complex relationship between tea quality, leaf senescence and the responses to cold-drought combined stress in Camellia sinensis. Front Plant Sci. 2016;7:1858. doi:https://doi.org/10.3389/fpls.2016.01858.
- Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–327. doi:https://doi.org/10.1093/nar/30.1.325.
- Riggleman B, Wieschaus E, Schedl P. Molecular analysis of the armadillo locus: uniformly distributed transcripts and a protein with novel internal repeats are associated with a Drosophila segment polarity gene. Genes Dev. 1989;3(1):96–113. doi:https://doi.org/10.1101/gad.3.1.96.
- Samuel MA, Salt JN, Shiu SH, Goring DR. Multifunctional arm repeat domains in plants. Int Rev Cytol. 2006;253:1–26. doi:https://doi.org/10.1016/s0074-7696(06)53001-3.
- Zhou J, Lu D, Xu G, Finlayson SA, He P, Shan L. The dominant negative ARM domain uncovers multiple functions of PUB13 in Arabidopsis immunity, flowering, and senescence. J Exp Bot. 2015;66(11):3353–3366. doi:https://doi.org/10.1093/jxb/erv148.
- Wang H, Lu Y, Jiang T, Berg H, Li C, Xia XY. The Arabidopsis U-box/ARM repeat E3 ligase AtPUB4 influences growth and degeneration of tapetal cells, and its mutation leads to conditional male sterility. Plant J. 2013;74(3):511–523. doi:https://doi.org/10.1111/tpj.12146.
- Song J, Mo X, Yang H, Yue L, Song J, Mo MB, Yang ZM. The U-box family genes in Medicago truncatula: key elements in response to salt, cold, and drought stresses. PLoS ONE. 2017;12(8):e0182402. doi:https://doi.org/10.1371/journal.pone.0182402.
- Wang C, Duan W, Riquicho AR, Jing Z, Liu T, Hou X, Li Y. Genome-wide survey and expression analysis of the PUB family in Chinese cabbage (Brassica rapa ssp. pekinesis). Mol Genet Genomics. 2015;290(6):2241–2260. doi:https://doi.org/10.1007/s00438-015-1075-x.
- Liang X, Zhou J-M. Receptor-like cytoplasmic kinases: central players in plant receptor kinase–mediated signaling. Annu Rev Plant Biol. 2018;69(1):267–299. doi:https://doi.org/10.1146/annurev-arplant-042817-040540.
- Xu C, Min J. Structure and function of WD40 domain proteins. Protein Cell. 2011;2(3):202–214. doi:https://doi.org/10.1007/s13238-011-1018-1.
- Zeytuni N, Zarivach R. Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure. 2012;20(3):397–405. doi:https://doi.org/10.1016/j.str.2012.01.006.
- Kuraoka I, Ito S, Wada T, Hayashida M, Lee L, Saijo M, Nakatsu Y, Matsumoto M, Matsunaga T, Handa H, et al. Isolation of XAB2 complex involved in Pre-mRNA splicing, transcription, and transcription-coupled repair. J Biol Chem. 2008;283(2):940–950. doi:https://doi.org/10.1074/jbc.M706647200.
- Tarn WY, Lee KR, Cheng SC. Yeast precursor mRNA processing protein PRP19 associates with the spliceosome concomitant with or just after dissociation of U4 small nuclear RNA. Proc Natl Acad Sci U S A. 1993;90(22):10821–10825. doi:https://doi.org/10.1073/pnas.90.22.10821.
- Lu L, Lee YRJ, Pan R, Maloof JN, Liu B. An internal motor kinesin is associated with the golgi apparatus and plays a role in trichome morphogenesis in Arabidopsis. Mol Biol Cell. 2005;16(2):811–823. doi:https://doi.org/10.1091/mbc.e04-05-0400.
- Lurin C, Andreés C, Aubourg S, Bellaoui M, Bitton F, Bruyére C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. Genome-Wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis[W]. Plant Cell. 2004;16(8):2089–2103. doi:https://doi.org/10.1105/tpc.104.022236.
- O’Toole N, Hattori M, Andres C, Iida K, Lurin C, Schmitz-Linneweber C, Sugita M, Small I. On the expansion of the pentatricopeptide repeat gene family in plants. Mol Biol Evol. 2008;25(6):1120–1128. doi:https://doi.org/10.1093/molbev/msn057.
- Kobayashi N, McEntee K. Identification of cis and trans components of a novel heat shock stress regulatory pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:248–256. doi:https://doi.org/10.1128/mcb.13.1.248-256.1993.
- Schuller C, Brewster JL, Alexander MR, Gustin MC, Ruis RH. The HOG pathway controls osmotic regulation of transcription via the stress response element (STRE) of the Saccharomyces cerevisiae CTT1 gene. EMBO J. 1994;13(18):4382–4389. doi:https://doi.org/10.1002/j.1460-2075.1994.tb06758.x.
- Fan F, Liu L, Zhang Z, Zou Z, Jiang J, HuangJ, Fan L, Zhang Z, Deng X, Ge Q, Gong W, Li J, Gong J, Shi Y, Lei K, Zhang S, Jia T, Zhang L, Yuan Y, Shang H. Genome-wide identification and expression analysis of the Metacaspase gene family in Gossypium species. Genes. 2019;10(7):527. doi:https://doi.org/10.3390/genes10070527.
- Wen Z, Yao L, Wan R, Li Z, Liu C, Wang X. Ectopic expression in Arabidopsis thaliana of an NB-ARC encoding putative disease resistance gene from wild Chinese Vitis pseudoreticulata enhances resistance to Phytopathogenic Fungi and Bacteria. Front Plant Sci. 2015;6:1087. doi:https://doi.org/10.3389/fpls.2015.01087.
- Chen S-P, Kuo C-H, Lu -H-H, Lo H-S, Yeh K-W, Coaker G. The sweet potato NAC-domain transcription factor IbNAC1 is dynamically coordinated by the activator IbbHLH3 and the repressor IbbHLH4 to reprogram the defense mechanism against wounding. PLoS Genet. 2016;12(10):e1006397. doi:https://doi.org/10.1371/journal.pgen.1006397.
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell. 1997;9:1859–1868. doi:https://doi.org/10.1105/tpc.9.10.1859.
- Rushton DL, Tripathi P, Rabara RC, Lin J, Ringler P, Boken AK, Langum TJ, Smidt L, Boomsma DD, Emme NJ, et al. WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnology Journal. 2012;10(1):2–11. doi:https://doi.org/10.1111/j.1467-7652.2011.00634.x.
- Libault M, Wan J, Czechowski T, Udvardi M, Stacey G. Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Molecular Plant-Microbe Interactions®. 2007;20(8):900–911. doi:https://doi.org/10.1094/mpmi-20-8-0900.
- Yang C-W, González-Lamothe R, Ewan RA, Rowland O, Yoshioka H, Shenton M, Ye H, O’Donnell E, Jones JDG, Sadanandom A, et al. The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. The Plant Cell. 2006;18(4):1084–1098. doi:https://doi.org/10.1105/tpc.105.039198.
- Heise A, Lippok B, Kirsch C, Hahlbrock K. Two immediate-early pathogen-responsive members of the AtCMPG gene family in Arabidopsis thaliana and the W-box-containing elicitor-response element of AtCMPG1. Proceedings of the National Academy of Sciences. 2002;99(13):9049–9054. doi:https://doi.org/10.1073/pnas.132277699.
- Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J. KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. The Plant Cell. 2007;18(12):3415–3428. doi:https://doi.org/10.1105/tpc.106.046532.
- Wawrzynska A, Christiansen KM, Lan Y, Rodibaugh NL, Innes RW. Powdery mildew resistance conferred by loss of the ENHANCED DISEASE RESISTANCE1 protein kinase is suppressed by a missense mutation in KEEP ON GOING, a regulator of abscisic acid signaling. Plant Physiology. 2008;148(3):1510–1522. doi:https://doi.org/10.1104/pp.108.127605.
- Pauwels L, Ritter A, Goossens J, Durand AN, Liu H, Gu Y, Geerinck J, Boter M, Vanden Bossche R, De Clercq R, et al. The RING E3 ligase KEEP ON GOING modulates JASMONATE ZIM-DOMAIN12 stability. Plant Physiol. 2015;169(2):1405–1417. doi:https://doi.org/10.1104/pp.15.00479.
- Gu Y, Innes RW. The KEEP ON GOING Protein of Arabidopsis Regulates Intracellular Protein Trafficking and Is Degraded during Fungal Infection. The Plant Cell. 2012;24(11):4717–4730. doi:https://doi.org/10.1105/tpc.112.105254.
- McNeilly D, Schofield A, Stone SL. Degradation of the stress-responsive enzyme formate dehydrogenase by the RING-type E3 ligase KEEP ON GOING and the ubiquitin 26S proteasome system. Plant Mol Biol. 2018;96(3):265–278. doi:https://doi.org/10.1007/s11103-017-0691-8.
