ABSTRACT
Aquaporins (AQPs) are channel proteins involved in transporting a variety of substrates. It has been proposed that the constriction regions in the central pores of the AQP channels play a crucial role in determining transport substrates and activities of AQPs. Our previous results suggest that AtNIP1;2, a member of the AQP superfamily in Arabidopsis, facilitates aluminum transport across the plasma membrane. However, the functions of the constriction regions in AtNIP1;2-mediated transport activities are unclear. This study reports that residue substitutions of the constriction regions affect AtNIP1;2-mediated aluminum uptake, demonstrating the critical roles of the constriction regions for transport activities. Furthermore, a constriction region that partially or wholly mimics AtNIP5;1, a demonstrated boric-acid transporter, could not render the boric-acid transport activity to AtNIP1;2. Therefore, besides the constriction regions, other structural features are also involved in determining the nature of AtNIP1;2’s transport activities.
Abbreviations: AIAR: alanine-isoleucine-alanine-arginine; AIGR: alanine-isoleucine-glycine- arginine; AQP: aquaporin; Al-Mal: aluminum-malate; ar/R: aromatic/arginine; AVAR: alanine-valine-alanine-arginine; CK: control; H: helical domain; ICP-MS: inductively coupled plasma mass spectrometry; LA - LE: inter-helical loops A to E; NIP: nodulin 26-like intrinsic protein; NPA: asparagine-proline-alanine; NPG: asparagine-proline- glycine; NPS: asparagine-proline-Serine; NPV: asparagine-proline-valine; ORF: open reading frame; PIP: plasma membrane intrinsic proteins; SIP: small basic intrinsic proteins; TM: transmembrane helices; WIAR: tryptophan-isoleucine-alanine-arginine; WVAR: tryptophan-valine-alanine-arginine; WVGR: tryptophan-valine-glycine- arginine.
Introduction
Aquaporins (AQPs) are channel proteins generally believed to facilitate the permeation of water and small uncharged solutes across the plasma and intracellular membranes.Citation1 However, increasing evidence implicates that some AQP members potentially transport ions in plants.Citation2
Aquaporins exhibit highly conserved structural features.Citation3 Firstly, four AQP monomers form a biologically active tetramer embedded in cell membranes.Citation4, 5 Secondly, each monomer contains an active pore region surrounded by six transmembrane helical domains (H1-H6) connected by five inter-helical loops, i.e., loop A to loop E (LA-LE).Citation5 Thirdly, two major constrictions in the pore are thought to play critical roles in the functional specialization of AQPs.Citation6
The pore’s first constriction comprises two highly conserved asparagine-proline-alanine (NPA) motifs in the hydrophobic LB and LE.Citation6–8 Structurally, the asparagine (N) residues in the two NPA motifs fold back into the core of the protein to form one of the significant constrictions.Citation3,Citation7,Citation9 The second constriction is located near the extracellular end of the pore, designated as an ar/R (aromatic/arginine) region due to the high prevalence of aromatic and basic residues.Citation7 The ar/R region is comprised of four residues, one each from the helix 2 (H2) and helix 5 (H5) and two from the loop E (LE1 and LE2).Citation7,Citation10,Citation11
It has been postulated that the NPA motifs function as a primary filter against protons and other positive ions.Citation6,Citation7,Citation12,Citation13 Structural and functional studies indicated that the NPA motif’s polar asparagine (N) residues are involved in hydrogen-bonding interactions with transport substrates.Citation7,Citation14 For instance, the hydrogen-bonding interactions between the N residues of the NPA motifs and water molecules are essential for maintaining the connectivity of water flow in the pore of the AQP-1 water channel.Citation14 Hence, replacing the N residue with hydrophobic residues caused completely broken aqueous pathways of the AQP-1 channel.Citation14
In contrast, the ar/R constriction is proposed as the primary filter for substrate selectivity.Citation8,Citation10,Citation11,Citation15 Furthermore, the highly conserved Arg (R) residue at the H2 position forms a hydrophilic surface and facilitates hydrogen bonding with the transport substrates.Citation8,Citation16 Thus, the R residue of the ar/R tetrad is critical for substrate selectivity in some AQP members.Citation17,Citation18 For instance, an exchange of the R residue altered substrate selectivity for some TIP members.Citation19
Based on their sequence similarity, plant AQPs can be classified into four subfamilies: plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPs), nodulin26-like intrinsic proteins (NIPs), and small basic intrinsic proteins (SIPs).Citation20 The NIP subfamily is unique to plants, and its members facilitate transporting a diversity of substrates, including water, glycerol,Citation21,Citation22 lactic acid,Citation23 urea,Citation24 formamide,Citation24 silicic acid,Citation25 selenite,Citation26 aluminum,Citation27 and metalloids such as arsenite (As(III)), antimonite (Sb(III)), and boron (B).Citation28–33
Arabidopsis has nine NIP members that can be further divided into two subgroups, i.e., NIP-I and NIP-II, based on their different ar/R tetrad patterns.Citation8,Citation9,Citation34 The NIP-I subgroup has six members, i.e., NIP1;1, NIP1;2, NIP2;1, NIP3;1, NIP4;1, and NIP4;2, with a conserved tetrad pattern of Trp (W) at H2, Val/Ile (V/I) at H5, Ala (A) at LE1, and Arg (A) at LE2. Moreover, the NPA-I members have an invariant NPA triad for the NPA1 motif, but variant NPA, NPG, or NPV triads in the NPA2 motif.Citation34
In contrast, three NIP-II members, i.e., NIP5;1, NIP6;1, and NIP7;1, have a similar tetrad pattern except that a smaller Ala (A) residue replaces the bulkier Trp (W) in the first residue of the ar/R tetrad, which results in a broader ar/R constriction that accommodates for transporting larger substrates.Citation8 Furthermore, it has been functionally demonstrated that the NPA-I members could transport formamide and glycerol besides water. However, NPA-IIs show very low water permeability but can transport urea and metalloids.Citation24,Citation35
Our recent studies suggest that AtNIP1;2, an NPA-I member, mediates the permeation of aluminum (Al), possibly in the form of aluminum-malate (Al-Mal) complexes, across the plasma membrane (PM) in Arabidopsis.Citation27 AtNIP1;2 facilitates Al removal from the cell wall into the cytosol in the root-tip region and subsequent root-to-shoot translocation, which are critical steps for an internal Al-resistance mechanism in plants.Citation27,Citation36,Citation37 Furthermore, we have demonstrated that the Al-activated and AtALMT1-mediated malate release into the root cell wallCitation38–40 is a prerequisite for the AtNIP1;2-mediated function and resistance in Arabidopsis.Citation41 However, the roles of the NPA motifs and ar/R selectivity filter in AtNIP1;2’s functions are still unclear.
AtNIP5;1 is the best-characterized NIP-II member that encodes a channel protein responsible for permeation of boric acid (B) into roots under B limitation.Citation33,Citation42 In this study, we tested the roles of the NPA and ar/R constriction regions in determining substrate selectivity and transport activity of AtNIP1;2. Through a site-directed mutagenesis approach, critical residues in the constriction regions of AtNIP1;2 were subject to chemical nature changes, e.g., polar to nonpolar, or conversions to AtNIP5;1-like patterns. Evaluation of substrate selectivity and transport activity suggested that the constriction regions play critical roles in AtNIP1;2-mediated Al selectivity and transport. Furthermore, AtNIP5;1-like NPA and ar/R constriction regions are insufficient to render B transport activities to AtNIP1;2.
Materials and methods
Site-directed mutagenesis
The open reading frame (ORF) of AtNIP1;2 was amplified by primers NIP1;2-F, 5ʹ-CTACggatccAAAATGGCGGAGATCTCGGGAAA-3ʹ and NIP1;2-R, 5ʹ-ATCCgcggccgcACGAGAGCTACCGTTTCGCA-3ʹ (the underlined sequences are restriction enzyme sites for BamH I and Not I, respectively). The PCR product was cut with restriction enzymes BamH I and Not I and cloned into the yeast expression vector, pYES2, to create a pYES2-AtNIP1;2 plasmid. AtNIP1;2 mutants were generated by site-directed mutagenesis using the following synthetic oligonucleotide primers. The lower case letters in the primer sequences represent mismatched nucleotides that introduced single amino-acid substitutions in the translated AtNIP1;2 proteins.
For NPA modification of AtNIP1;2:
N111LF,5ʹ-CGGTGCTCATTTCctTCCGGCCGTCACAATCGC-3ʹ
N111LR,5ʹ-CGATTGTGACGGCCGGAagGAAATGAGCACCGG-3ʹ
A113SF,5ʹ-GCTCATTTCAATCCGtCCGTCACAATCGCATTCGC-3ʹ;
A113SR,5ʹ-GCGAATGCGATTGTGACGGaCGGATTGAAATGAGC-3ʹ;
N230LF,5ʹ- GGGAGCATCGATGctTCCAGGACGAAGTTTAGG-3ʹ
N230LR,5ʹ-CTAAACTTCGTCCTGGAagCATCGATGCTCCCG-3ʹ
G232VF,5ʹ-GCATCGATGAATCCAGtACGAAGTTTAGGACCTGC-3ʹ;
G232VR,5ʹ-GCAGGTCCTAAACTTCGTaCTGGATTCATCGATGC-3ʹ.
For ar/R modification of AtNIP1;2:
W91AF,5ʹ-CAGGGATCGCCATCGTTgcGGGACTTACCGTCATG-3ʹ;
W91A R, 5ʹ-CATGACGGTAAGTCCCgcAACGATGGCGATCCCTG-3ʹ
V218I F, 5ʹ-CAACAGTGCTACTTAACaTcATAATTGCCGGGCCG-3ʹ
V218I R, 5ʹ-CGGCCCGGCAATTATgAtGTTAAGTAGCACTGTTG-3ʹ
A227G F, 5ʹ-GGCCGGTATCGGGAGgATCGATGAATCCAGGAC-3ʹ
A227G R, 5ʹ-GTCCTGGATTCATCGATcCTCCCGATACCGGCC-3ʹ
R233G F, 5ʹ-ATCGATGAATCCAGGAgGAAGTTTAGGACCTGC-3ʹ
R233G R, 5ʹ- CAGGTCCTAAACTTCcTCCTGGATTCATCGATG-3ʹ
In brief, high-fidelity PCR was performed to amplify the pYES2-AtNIP1;2 plasmid with Pfu DNA polymerase and primer pairs listed above. The resulting PCR products were checked by agarose gel electrophoresis. PCR products with the right size of 6.7 kb were digested with DpnI at 37°C for 3 h, which cut the methylated pYES2-AtNIP1;2 template into small fragments but left the non-methylated and circular PCR products intact. After DpnI digestion, the PCR products were transformed into competent E. coli strain TOP10. The purified plasmids from the transformed TOP10 cells were verified by sequencing. Additional runs of PCR-based site-directed mutagenesis were performed to generate multiple mutations.
Yeast Al sensitivity and uptake analysis
For Al sensitivity evaluation, the pYES2 empty vector or pYES2 carrying wild-type or mutant AtNIP1;2 open reading frames (ORFs) were transformed into the yeast (Sacchromyces cerevisea) strain BY4741. The resultant lines were first cultured in a liquid SD-Ura medium to the stationary phase. Cells were collected by centrifuge at 5,000 g for 5 min, followed by wash 3 times with ddH2O and 3 times with a low pH, low magnesium (LPM) medium, buffered with 5 mM Succinic acid to pH 4.2. The LPM medium contained the following macronutrients in mM: (NH4)2SO4, 40; KCl, 5; NaCl, 2; CaCl2, 0.1; KH2P04, 0.01; MgSO4, 0.25; the following micronutrients in μM: FeCl3,1; H3BO3,10; KI, 0.5; MnSO4, 2.5; Na2MoO4, 1; ZnSO4, 1.5; the following amino acids in mg/l: tyrosine, 0.03; glutamic acid, 0.075; adenine, 0.02; uracil, 0.02; phenylalanine, 0.05; valine, 0.15; serine, 0.4; leucine, 0.03; isoleucine, 0.03; lysine, 0.03; tryptophan, 0.02; arginine, 0.02; histidine, 0.02; methionine, 0.02; aspartic acid, 0.0625; threonine, 0.2; the following vitamins in ng/l: folic acid, 0.2; biotin, 0.2; p-aminobenzoic acid, 20; riboflavin, 20; calcium pantothenate, 40; niacin, 40; pyridoxine hydrochloride, 40; thiamine hydrochloride, 40; inositol, 200; and 2% galactose.
For the drop assay, 5 μl of 10-fold serially diluted cell suspensions were spotted onto solid LPM plates (pH to 4.2), containing 0, 100, or 200 μM Al-Mal (1:2) and 2% galactose for induction of the GAL promoter. The plates were photographed after incubation at 30°C for 4 days.
For determining Al and B contents, yeast cells were cultured in an LPM liquid medium (-Ura, +2% galactose and 1% raffinose, pH 4.2) to a mid-exponential phase. Cells were harvested by centrifuge at 5,000 g for 5 min, followed by 3 time washes with an LPM medium (-Ura, +2% galactose, 1% raffinose). After washing, the cells were transferred to a new LPM medium (2% galactose, pH 4.2 or 7.0 adjusted by 5 mM succinic acid) to an OD600 value at 3.0. Next, Al-malate or H3BO3 was added to the cell culture to a final concentration of 50 μM at pH 4.2 or 7.0, respectively. After 2 h incubation with gentle shaking, cells were harvested by centrifuge at 5000 × g for 5 min and washed 3 times with deionized water (ddH2O) (MilliQ; Millipore), dried, and then digested with 2 N HCl. The Al and B contents of each digested sample were determined by inductively coupled plasma mass spectrometry (ICP-MS) using an Agilent 7500 Series ICP mass spectrometer. Three biological replicates were conducted.
Results and discussion
Sequence comparison of the NPA and ar/R constriction regions between AtNIP1;2 and AtNIP5;1
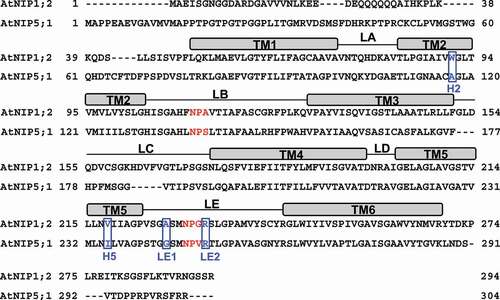
Sequencing alignment indicated that AtNIP1;2, a NIP-I member, has an NPA (Asn-Pro-Ala) and an NPG (Asn-Pro-Gly) sequence of NPA1 and NPA2 motifs, respectively. In contrast, the NIP-II member AtNIP5;1 has NPS (Asn-Pro-Ser) and NPV (Asn-Pro-Val) for the NPA1 and NPA2 motifs, respectively ().Citation24 Therefore, the third residue of both NPA motifs differs between AtNIP1;2 and AtNIP5;1 (). Furthermore, AtNIP1;2 has a WVAR sequence of the ar/R region versus AIGR in AtNIP5;1. Thus, the ar/R tetrad sequences of AtNIP1;2 and AtNIP5;1 differ at the H2, H5, and LE1 positions ().
Figure 1. Sequence alignments of AtNIP1;2 and AtNIP5;1. Red letters define the NPA1 (in LB) and NPA2 (in LE) motifs. Blue boxed letters refer to the tetrad residues of the ar/R constriction region. Rectangle boxes above the amino-acid sequences delineate the transmembrane helices (TM). Thin lines between TMs indicate the inter-helical loops. H, helix; LA-LE, inter-helical loops A-E.
Influence of the NPA and ar/R constrictions on AtNIP1;2-mediated aluminum sensitivity in yeast
The asparagine (N) residue in the NPA1 and NPA2 motifs and the arginine (R) residue in the ar/R selective filter are conserved in the NIP subfamily.Citation34 To investigate the role of these conserved residues in AtNIP1;2-mediated Al-uptake activities, we constructed three AtNIP1;2 single mutants, designated as N111L, N230L, and R233G. In these mutants, the polar N residue of NPA1 and NPA2 was replaced by a nonpolar leucine (L), while the R residue of the ar/R tetrad was replaced by glycine (G).
Aluminum uptake mediated by the wild-type and mutant AtNIP1;2 proteins were investigated by assessing yeast [S. cerevisiae (BY4741)] sensitivity to Al toxicity. Yeast lines harboring an empty pYES2 vector (control, CK) or expressing AtNIP1;2 and its mutants were subjected to Al treatment on low pH, low magnesium (LPM) agar plates (pH 4.2) ().
Figure 2. Mutations in the NPA motifs and ar/R selectivity filter eliminate AtNIP1;2- mediated aluminum sensitivity in yeast cells. Yeast (BY4741) lines carrying the empty vector pYES2, or constructs expressing the wild-type (WT) AtNIP1;2 (NPA/NPG/AIGR) or AtNIP1;2 mutants N111L (LPA/NPG/AIGR), N230L (NPA/LPG/AIGR), or R233G (NPA/NPG/AIGG) were subjected to Al sensitivity tests. Letters in parenthesis refer to the NPA1, NPA2, and ar/R constriction regions, respectively. Red letters indicate exchanged residues. Aliquots (5 μl) of 10-fold serial dilutions of re-suspended cells were spotted onto LPM plates (pH 4.2, 2% galactose) supplemented without (no stress) or with 100 and 200 μM AlCl3 buffered with 200 and 400 μM malate, respectively. The LPM plates were placed in a 30°C incubator for 3 d.
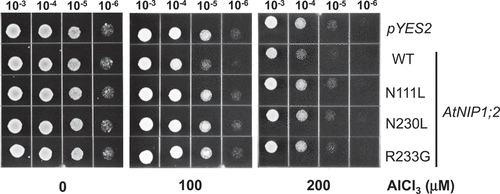
The growth of yeast cells transformed with the empty pYES2 vector or pYES2 containing the wild-type or mutant AtNIP1;2 could not be distinguished on the LPM plates that were not supplemented with Al (). This result indicated that heterologous expression of AtNIP1;2 or its mutants has no harmful effect on yeast growth under standard conditions.
However, yeast cells that expressed wild-type AtNIP1;2 showed significantly reduced growth compared to those carrying the empty vector when exposed to Al stresses by adding 100 or 200 μM Al-malate (Al-Mal) to the growth medium (). The growth inhibition was presumed to be due to AtNIP1;2-mediated Al uptake and accumulation, as Wang et al. (2017) reported previously.
In contrast, N111L, N230L, and R233G grew similarly to the empty vector control (CK) line under – and + Al treatments (). These results indicated mutations in the conserved N and R residues on the NPA motifs and the ar/R region, respectively, abolished AtNIP1;2-mediated Al sensitivity in yeast (), suggesting critical roles of these residues in AtNIP1;2-mediated Al-Mal uptake and accumulation.
Importance of the NPA and ar/R constriction regions for AtNIP1;2-mediated Al uptake
To confirm the differences in Al sensitivity were associated with Al uptake and accumulation in the yeast cells, short-term (2 h) Al uptake was evaluated for individual yeast lines. As indicated in , the yeast line harboring the native AtNIP1;2 construct had an ~3-fold higher Al uptake rate than the control (CK) line. This result is consistent with our previous observation that AtNIP1;2 facilitates across-PM Al uptake in yeast.Citation27
Figure 3. Influence of N/L substitution in the NPA motifs and R/G substitution in the ar/R selectivity filter on AtNIP1;2-mediated Al uptake. Yeast (BY4741) lines carrying the empty control (CK) vector pYES2 and expressing the native or mutated (i.e., N111L, N230L, and R233G) AtNIP1;2 were subject to short-term (2 h) Al uptake assays. Data are means ± SD (n = 3). Different letters above the columns indicate statistically significant differences at P < .05 by Tukey’s test.
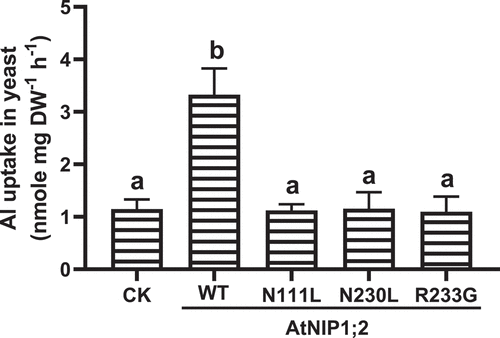
In contrast, the Al uptake in the yeast lines expressing AtNIP1;2 mutants (N111L, N230L, or R233G) was comparable to those of the CK line but not the native AtNIP1;2 line (), indicating that that the mutations caused a loss of AtNIP1;2-mediated Al-Mal uptake in yeast. Thus, the asparagine (N) residue in the NPA motifs and the arginine (R) residue of the ar/R selective filter are critical for AtNIP1;2-mediated across-membrane Al uptake.
Influence of the NPA and ar/R constrictions on substrate selectivity of AtNIP1;2
Except for the conserved 1st residues of N in NPA1 and NPA2 motifs and the 4th residue of R in the ar/R region, other residues show more diversity between the NIP-I and NIP-II subgroups.Citation34 For instance, AtNIP1;2, a NIP-I member, and AtNIP5;1, a NIP-II member, differ in the 3rd residues of the NPA motifs and the first three residues of the ar/R tetrad ().
To investigate the impact of the identity of pore constriction regions on uptake activities and substrate selectivity, the NPA motifs and ar/R selective filter of AtNIP1;2 were exchanged to partially or entirely mimic those in AtNIP5;1, a demonstrated B transporter of the NIP-II subgroup.Citation33
As indicated in , a single alanine-to-serine (A113S) exchange at the 3rd residue of the NPA1 motif (NPA to NPS) abolished the AtNIP1;2-mediated Al-Mal transport activity (). This result indicates that the third residue of S is as critical as the first residue of N in the NPA1 motif for the AtNIP1;2-facilitated Al uptake (). In contrast, a glycine-to-valine (G232V) exchange at the 3rd residue of the NPA2 motif had a much weaker impact on Al uptake: Al uptake activity decreased ~40% compared with the native AtNIP1;2 (). Furthermore, the double AtNIP1;2 mutant (A113S/G232V), carrying NPA1/NPA2 motifs mimicked to those of AtNIP5;1 (NPS/NPV), also showed no increased Al transport activities (). This result indicates that the effect of A113S overrides that of G232V. Similarly, a single AVAR mutant, which contained a tryptophan-to-alanine (W91A) substitution that mimicks the first residue of the ar/R tetrad in AtNIP5;1, did not show any Al-uptake activity compared with the control line ().
Figure 4. Impacts of third residue substitutions in the NPA motifs and residue substitutions in the ar/R selectivity filter on AtNIP1;2-mediated Al uptake. Yeast (BY4741) lines carrying the control (CK) empty vector pYES2 and the native (NPA/NPG/WVAR) or mutated AtNIP1;2 were subject to short-term (2 h) Al-uptake assays. (a) Red letters indicate substitutions of the third residue in the NPA1 and NPA2 motifs. (b) Red letters indicate residue substitutions of the ar/R tetrad. Data are means ± SD (n = 3). Different letters above the columns indicate statistically significant differences among groups at P < .05 by Tukey’s test.
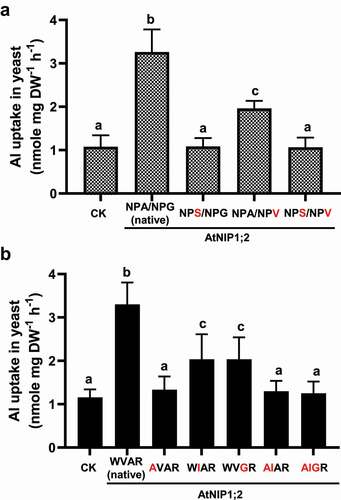
The strong effects of the A-to-S in NPA1 and W-to-A in the ar/R tetrad on AtNIP1;2-mediated Al transport could be because of the serine’s hydroxyl group and tryptophan indole group affecting the folding and orientation of the channel protein. Or, the A-to-S substitution at NPA1 may affect post-translational modification, e.g., phosphorylation.Citation43,Citation44
Aluminum-uptake phenotypes differed in the other two single mutant lines, i.e., WIAR and WVGR (). The WIAR and WVGR lines contained valine-to-isoleucine (V218I) and alanine-to-glycine (A227G) exchanges that mimicked the 2nd and 3rd residues of the ar/R tetrad in AtNIP5;1. As shown in , Al uptake decreased ~38% in the WIAR and WVGR mutant lines than the wild-type (native) AtNIP1;2 (). These results suggest that the 2nd and 3rd residues have fewer impacts on the AtNIP1;2-mediated Al uptake than the 1st () and 4th () residues of the ar/R tetrad.
Furthermore, a double AIAR mutant and a triple AIGR mutant contained W91A/V218I and W91A/V218I/A227G residue substitutions partially and wholly mimicked the AtNIP5;1 ar/R tetrad. Both AIAR and AIGR lines resembled the AVAR single mutant in Al-uptake activities (). This result indicates that the tryptophan (W) residue at H2 has a dominant effect on influencing AtNIP1;2-mediated Al uptake over the valine (V) and alanine (A) residues at H5 and LE1, respectively.
AtNIP5;1-like constriction regions could not render boron transport activity to AtNIP1;2
To test whether an ar/R tetrad that mimics AtNIP5;1 can render B transport activity to AtNIP1;2, tryptophan (W) at H2, valine (V) at H5, or alanine (A) at LE1 in AtNIP1;2 were changed individually or in combination to alanine (A), isoleucine (I), or glycine (G), respectively (). No B uptake activities were observed for the native AtNIP1;2 and its single, double, or triple ar/R mutants ().
Figure 5. AtNIP5;1-like NPA motifs and ar/R selectivity filter does not render B uptake activity to AtNIP1;2. Yeast (BY4741) lines carrying the control (CK) empty vector pYES2 or expressing AtNIP1;2 containing the native or mutated NPA and ar/R constriction regions were subject to short-term (2 h) B uptake assays. (a) Red letters indicate residue substitutions in the ar/R tetrad. (b) Residue substitutions (red letters) that transform the AtNIP1;2 NPA and ar/R constriction regions to an AtNIP5;1 type. Data are means ± SD (n = 3). Different letters above the columns indicate statistically significant differences at P < .05 by Tukey’s test.
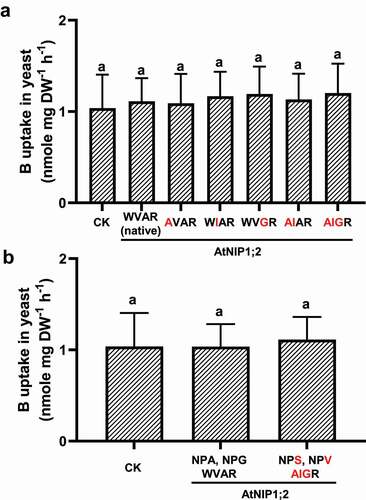
We then generated a quintuple mutant (NPS/NPV/AIGR, the underlines indicate residue substitutions) where the NPA motifs and ar/R selective filter mimics AtNIP5;1. However, as shown in , this AtNIP1;2 quintuple mutant could not transport B also.
These results indicate that 1) boric acid is not a transport substrate for AtNIP1;2 as the native AtNIP1;2 is incapable of transporting boric acid; 2) partial or complete mimics of the NPA motifs and the ar/R tetrad of AtNIP5;1 could not convert AtNIP1;2 from an Al to a B transporter.
In conclusion, the substrate selectivity of AtNIP1;2 is not simply controlled by the NPA motifs and ar/R selectivity filter. Other structural features and post-translational modifications, e.g., phosphorylation,Citation43,Citation44 methylation and acetylation,Citation45 and glycosylation,Citation46 may also be necessary for determining the nature of the substrate specificity and transport activities.
Author’s contributions
J.L. conceived, projected, guided the experiments, and wrote the article; Y.W. designed and performed the experiments, analyzed the data, and wrote the paper; E. X., G.W., Q.B., F.X., X.J., and C.L. performed the experiments and analyzed the data; L. L. analyzed the data and reviewed and edited the article. All authors have read and approved the final manuscript.
Acknowledgments
Not applicable
Disclosure statement
No potential conflict of interest was reported by the author(s).
Availability of data and material
The data supporting this study’s findings are available from the corresponding author, JL, upon reasonable request.
Additional information
Funding
References
- Maurel C, Verdoucq L, Luu D-T, Santoni V. D-T Luu and V Santoni. Plant aquaporins: membrane channels with multiple integrated functions. Annual Review of Plant Biology. 2008;59(1):1–7. doi:https://doi.org/10.1146/annurev.arplant.59.032607.092734.
- Tyerman SD, McGaughey SA, Qiu J. AJ Yool and CS Byrt. Adaptable and multifunctional ion-conducting aquaporins. Annual Review of Plant Biology. 2021;72:703-736. doi:https://doi.org/10.1146/annurev-arplant-081720-013608.
- Hove RM, Bhave M. Plant aquaporins with non-aqua functions: deciphering the signature sequences. Plant Molecular Biology. 2011;75(4–5):413–430. doi:https://doi.org/10.1007/s11103-011-9737-5.
- Verbavatz J-M, Brown D, Sabolić I, Valenti G, Ausiello D, Van Hoek AN, Ma T, Verkman AS. Tetrameric assembly of CHIP28 water channels in liposomes and cell membranes: a freeze-fracture study. Journal of Cell Biology. 1993;123(3):605–618. doi:https://doi.org/10.1083/jcb.123.3.605.
- Jung JS, Preston GM, Smith BL, Guggino WB, Agre P. WB Guggino and P Agre. Molecular structure of the water channel through aquaporin CHIP. The Hourglass Model. Journal of Biological Chemistry. 1994;269(20):14648–14654. doi:https://doi.org/10.1016/S0021-9258(17)36674-7.
- Forrest KL, Bhave M. Major intrinsic proteins (MIPs) in plants: a complex gene family with major impacts on plant phenotype. Functional & Integrative Genomics. 2007;7(4):263–289. doi:https://doi.org/10.1007/s10142-007-0049-4.
- de Groot BL, Grubmüller H. Water permeation across biological membranes: mechanism and dynamics of aquaporin-1 and GlpF. Science. 2001;294(5550):2353–2357. doi:https://doi.org/10.1126/science.1066115.
- Wallace IS, Roberts DM. Homology modeling of representative subfamilies of arabidopsis major intrinsic proteins. Classification Based on the Aromatic/arginine Selectivity Filter. Plant Physiology. 2004;135:1059–1068.
- Wallace IS, Choi W-G, Roberts DM. The structure, function and regulation of the nodulin 26-like intrinsic protein family of plant aquaglyceroporins. Biochimica Et Biophysica Acta (BBA) - Biomembranes. 2006;1758(8):1165–1175. doi:https://doi.org/10.1016/j.bbamem.2006.03.024.
- Fu D, Libson A, Miercke LJW, Weitzman C, Nollert P, Krucinski J, Stroud RM. Structure of a glycerol-conducting channel and the basis for its selectivity. Science. 2000;290(5491):481–486. doi:https://doi.org/10.1126/science.290.5491.481.
- Sui H, Han B-G, Lee JK, Walian P, Jap. BK. Structural basis of water-specific transport through the AQP1 water channel. Nature. 2001;414(6866):872–878. doi:https://doi.org/10.1038/414872a.
- Ilan B, Tajkhorshid E, Schulten K, Voth GA. The mechanism of proton exclusion in aquaporin channels. Proteins: Structure, Function, and Bioinformatics. 2004;55(2):223–228. doi:https://doi.org/10.1002/prot.20038.
- Tajkhorshid E, Nollert P, Jensen MØ, Miercke LJ, O’Connell J, Stroud RM, Schulten K. Control of the selectivity of the aquaporin water channel family by global orientational tuning. Science. 2002;296(5567):525–530. doi:https://doi.org/10.1126/science.1067778.
- Kong Y, Ma J. Dynamic mechanisms of the membrane water channel aquaporin-1 (AQP1). Proceedings of the National Academy of Sciences. 2001;98:14345–14349. doi:https://doi.org/10.1073/pnas.251507998.
- Thomas D, Bron P, Ranchy G, Duchesne L, Cavalier A, Rolland J-P, Raguénès-Nicol C, Hubert J-F, Haase W, Delamarche C, et al. Aquaglyceroporins, one channel for two molecules. Biochimica et Biophysica Acta (BBA)-Bioenergetics. 2002;1555(1–3):181–186. doi:https://doi.org/10.1016/S0005-2728(02)00275-X.
- Stroud RM, Savage D, Miercke LJ, Lee JK, Khademi S, Harries W. Selectivity and conductance among the glycerol and water conducting aquaporin family of channels. FEBS Letters. 2003;555(1):79–84. doi:https://doi.org/10.1016/S0014-5793(03)01195-5.
- Park J, Saier M Jr. Phylogenetic characterization of the MIP family of transmembrane channel proteins. The Journal of Membrane Biology. 1996;153(3):171–180. doi:https://doi.org/10.1007/s002329900120.
- Froger A, Thomas D, Delamarche C, Tallur B. Prediction of functional residues in water channels and related proteins. Protein Science. 1998;7(6):1458–1468. doi:https://doi.org/10.1002/pro.5560070623.
- Azad AK, Yoshikawa N, Ishikawa T, Sawa Y, Shibata H. Substitution of a single amino acid residue in the aromatic/arginine selectivity filter alters the transport profiles of tonoplast aquaporin homologs. Biochimica Et Biophysica Acta (Bba)-biomembranes. 2012;1818(5491):1–11. doi:https://doi.org/10.1016/j.bbamem.2011.09.014.
- Johanson U, Karlsson M, Johansson I, Gustavsson S, Sjövall S, Fraysse L, Weig AR, Kjellbom P. The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiology. 2001;126(4):1358–1369. doi:https://doi.org/10.1104/pp.126.4.1358.
- Dean RM, Rivers RL, Zeidel ML, Roberts DM. Purification and functional reconstitution of soybean nodulin 26. An Aquaporin with Water and Glycerol Transport Properties. Biochemistry. 1999;38:347–353.
- Weig AR, Jakob C. Functional identification of the glycerol permease activity of Arabidopsis thaliana NLM1 and NLM2 proteins by heterologous expression in Saccharomyces cerevisiae. FEBS Letters. 2000;481(3):293–298. doi:https://doi.org/10.1016/S0014-5793(00)02027-5.
- Choi W-G, Roberts DM. Arabidopsis NIP2;1, a major intrinsic protein transporter of lactic acid induced by anoxic stress. Journal of Biological Chemistry. 2007;282(33):24209–24218. doi:https://doi.org/10.1074/jbc.M700982200.
- Wallace IS, Roberts DM. Distinct transport selectivity of two structural subclasses of the nodulin-like intrinsic protein family of plant aquaglyceroporin channels. Biochemistry. 2005;44(51):16826–16834. doi:https://doi.org/10.1021/bi0511888.
- Ma JF, Tamai K, Yamaji N, Mitani N, Konishi S, Katsuhara M, Ishiguro M, Murata Y, Yano M. A silicon transporter in rice. Nature. 2006;440(7084):688–691. doi:https://doi.org/10.1038/nature04590.
- Zhao XQ, Mitani N, Yamaji N, Shen RF, Ma. JF. Involvement of silicon influx transporter OsNIP2;1 in selenite uptake in rice. Plant Physiology. 2010;153(4):1871–1877. doi:https://doi.org/10.1104/pp.110.157867.
- Wang Y, Li R, Li D, Jia X, Zhou D, Li J, Lyi SM, Hou S, Huang Y, Kochian LV, Liu J. NIP1;2 is a plasma membrane-localized transporter mediating aluminum uptake, translocation, and tolerance in Arabidopsis. Proceedings of the National Academy of Sciences. 2017; 114:5047–5052. doi:https://doi.org/10.1073/pnas.1618557114.
- Bienert GP, Thorsen M, Schüssler MD, Nilsson HR, Wagner A, Tamás MJ, Jahn TP. A subgroup of plant aquaporins facilitate the bi-directional diffusion of As(OH)3 and Sb(OH)3 across membranes. BMC Biology. 2008;6(1):1–15. doi:https://doi.org/10.1186/1741-7007-6-26.
- Isayenkov SV, Maathuis FJ. The Arabidopsis thaliana aquaglyceroporin AtNIP7;1 is a pathway for arsenite uptake. FEBS Letters. 2008;582(11):1625–1628. doi:https://doi.org/10.1016/j.febslet.2008.04.022.
- Kamiya T, Fujiwara T. Arabidopsis NIP1;1 transports antimonite and determines antimonite sensitivity. Plant and Cell Physiology. 2009;50(11):1977–1981. doi:https://doi.org/10.1093/pcp/pcp130.
- Kamiya T, Tanaka M, Mitani N, Ma JF, Maeshima M, Fujiwara T. NIP1;1, an aquaporin homolog, determines the arsenite sensitivity of Arabidopsis thaliana. Journal of Biological Chemistry. 2009;284(4):2114–2120. doi:https://doi.org/10.1074/jbc.M806881200.
- Kato Y, Miwa K, Takano J, Wada M, Fujiwara T. Highly boron deficiency-tolerant plants generated by enhanced expression of NIP5;1, a boric acid channel. Plant and Cell Physiology. 2009;50(1):58–66. doi:https://doi.org/10.1093/pcp/pcn168.
- Takano J, Wada M, Ludewig U, Schaaf G, Wirén NV, Fujiwara. T. Arabidopsis major intrinsic protein NIP5;1 Is essential for efficient boron uptake and plant development under boron limitation. Plant Cell. 2006;18(6):1498–1509. doi:https://doi.org/10.1105/tpc.106.041640.
- Mitani N, Yamaji N, Ma. JF. Characterization of substrate specificity of a rice silicon transporter, Lsi1. Pflügers Archiv-European Journal of Physiology. 2008;456(4):679–686. doi:https://doi.org/10.1007/s00424-007-0408-y.
- Wallace IS, Wills DM, Guenther JF, Roberts DM. Functional selectivity for glycerol of the nodulin 26 subfamily of plant membrane intrinsic proteins. FEBS Letters. 2002;523(1–3):109–112. doi:https://doi.org/10.1016/S0014-5793(02)02955-1.
- Kochian LV, Piñeros MA, Liu J, Magalhaes JV. Plant adaptation to acid soils: the molecular basis for crop aluminum resistance. Annual Review of Plant Biology. 2015;66(1):571–598. doi:https://doi.org/10.1146/annurev-arplant-043014-114822.
- Wang Y, Cai Y, Cao Y, Liu. J. Aluminum-activated root malate and citrate exudation is independent of NIP1;2-facilitated root-cell-wall aluminum removal in Arabidopsis. Plant Signaling & Behavior. 2018;13(1):e1422469. doi:https://doi.org/10.1080/15592324.2017.1422469.
- Liu J, Magalhaes JV, Shaff J, Kochian LV. Aluminum‐activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant Journal. 2009;57(3):389–399. doi:https://doi.org/10.1111/j.1365-313X.2008.03696.x.
- Liu J, Luo X, Shaff J, Liang C, Jia X, Li Z, Magalhaes J, Kochian LV. A promoter‐swap strategy between the AtALMT and AtMATE genes increased Arabidopsis aluminum resistance and improved carbon‐use efficiency for aluminum resistance. Plant Journal. 2012;71(2):327–337. doi:https://doi.org/10.1111/j.1365-313X.2012.04994.x.
- Hoekenga OA, Maron LG, Piñeros MA, Cançado GM, Shaff J, Kobayashi Y, Ryan PR, Dong B, Delhaize E, Sasaki T, Matsumoto, H, Yamamoto Y, Koyama, H, Kochian LV. AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proceedings of the National Academy of Sciences 2006;103(25):9738–9743. doi:https://doi.org/10.1073/pnas.0602868103.
- Wang Y, Yu W, Cao Y, Cai Y, Lyi SM, Wu W, Kang Y, Liang C, Liu J. An exclusion mechanism is epistatic to an internal detoxification mechanism in aluminum resistance in Arabidopsis. BMC Plant Biology. 2020;20:122. doi:https://doi.org/10.1186/s12870-020-02338-y.
- Miwa K, Tanaka M, Kamiya T, Fujiwara T. Molecular Mechanisms of Boron Transport in Plants: Involvement of Arabidopsis NIP5;1 and NIP6;1. In: Jahn T.P., Bienert G.P. (eds) MIPs and Their Role in the Exchange of Metalloids. Advances in Experimental Medicine and Biology, 2010;679:83-96. Springer, New York, NY. doi:https://doi.org/10.1007/978-1-4419-6315-4_7
- Johnson KD, Chrispeels MJ. Tonoplast-bound protein kinase phosphorylates tonoplast intrinsic protein. Plant Physiology. 1992;100(4):1787–1795. doi:https://doi.org/10.1104/pp.100.4.1787.
- Törnroth-Horsefield S, Wang Y, Hedfalk K, Johanson U, Karlsson M, Tajkhorshid E, Neutze R, Kjellbom P. Structural mechanism of plant aquaporin gating. Nature. 2006;439(7077):688–694. doi:https://doi.org/10.1038/nature04316.
- Santoni V, Verdoucq L, Sommerer N, Vinh J, Pflieger D, Maurel. C. Methylation of aquaporins in plant plasma membrane. Biochemical Journal. 2006;400(1):189–197. doi:https://doi.org/10.1042/BJ20060569.
- Vera-Estrella R, Barkla BJ, Bohnert HJ, Pantoja O. HJ Bohnert and O Pantoja. Novel regulation of aquaporins during osmotic stress. Plant Physiology. 2004;135(4):2318–2329. doi:https://doi.org/10.1104/pp.104.044891.
