ABSTRACT
BT4 gene was identified to play an important role in Arabidopsis resistance to pst DC3000 in preliminary studies. However, the specific function and molecular mechanism of BT4 gene in regulation of Arabidopsis resistance to Botrytis cinerea had not been described to date. In this study, we found that the expression of BT4 was induced by wounding and B. cinerea inoculation in Arabidopsis. After inoculated with B. cinerea, T-DNA insertion mutants of the BT4 gene, bt4, showed significant susceptibility symptoms, whereas no significant symptoms were found in wild-type (WT), the complemented transgenic plants (CE), and the overexpression transgenic plants (OE). After inoculated with B. cinerea, the expression levels of JAR1 and PDF1.2 genes in bt4 mutant were induced; however, the expression levels of these genes in bt4 mutant were significantly lower than those in the WT, CE, and OE. These results indicated that the BT4 positively regulate the expression of genes in JA/ET signaling pathways. Therefore, the BT4 may be involved in the regulation of JA/ET signaling pathways to affect Arabidopsis resistance to B. cinerea.
Introduction
Gray mold caused by Botrytis cinerea is a world-wide plant disease, which seriously affects the yield and quality of crops. The isolation of resistant-related genes against B. cinerea and the molecular mechanism of its disease-resistant provide the theoretical basis for the guidance of disease control and research of the molecular mechanism of interaction between plants and pathogens. At present, a lot of resistance-related genes of Arabidopsis against B. cinerea were identified. These genes regulate the resistance of Arabidopsis to B. cinerea by regulating SA and JA/ET signaling pathways. ERF1 is the activation factor of JA/ET signaling pathway in Arabidopsis. Overexpression of ERF1 could activate the expression of resistance-related genes PDF1.2 and ChiB and increase resistance to B. cinerea.Citation1 In Arabidopsis MYB30 is a positive regulator of hypersensitive response (HR) in Arabidopsis. Overexpression of MYB30 in tobacco and Arabidopsis were more resistant to Pseudomonas syringae pv. tomato DC3000 (Pst DC3000).Citation2 MYB46 could bound to the cis site of the promoter of cell wall peroxidase encoding gene Ep5C in Arabidopsis to regulate the synthesis of secondary cell wall in vascular bundles, which affected the resistance of Arabidopsis to B. cinerea.Citation3 The BOS1 (MYB108) gene is mainly involved in JA signaling pathway of Arabidopsis. Loss of BOS1 displayed more sensitive to B. cinerea and increased the resistance to Pst DC3000.Citation4 ERF19 as a novel player in the mitigation of PTI, and highlights a potential role for NINJA in fine-tuning ERF19-mediated regulation of Arabidopsis against B. cinerea.Citation5 WRKY33 is a key transcriptional regulator of hormonal and metabolic processes toward B. cinerea infection and is essential for resistance.Citation6,Citation7
Approximately 80 BTB proteins were identified in Arabidopsis, and several reports showed that BTB protein play a variety of functions in organisms, including transcriptional activation and inhibition,Citation8 regulation of cytoskeleton,Citation9 regulation of ion channels,Citation10 and ubiquitination of protein.Citation11 Five BT proteins (BT1–BT5) with BTB and TAZ domains were grouped into a small subfamily that interacts with calmodulin. Among them, BT1, BT2, and BT4 were found to bind to bromo-domain transcriptional regulators and the BT2 promoter binds to transcription factor TELOMERASE ACTIVATOR1 (TAC1) to regulate telomerase activity in mature vegetative organs.Citation12,Citation13 Some genes containing BTB domains were involved in plant disease resistance, such as NPR1 of Arabidopsis.Citation14
Previously, the BT4 gene, encoding a transcriptional regulator with BTB (broad-complex, tramtrack, and bric-a-brac) and TAZ (a transcriptional adapter zinc finger) domains, was isolated and identified as a resistant-related gene in response to Pst DC3000; meanwhile, BT4 also could be induced by SA and JACitation15 . In this study, we investigated the function of BT4 in Arabidopsis resistance to B. cinerea and further analyzed the probable mechanism of BT4 gene regulation in Arabidopsis resistance to B. cinerea.
Results
BT4 positively regulates the resistance to B. cinerea in Arabidopsis
Wounding provides nutrients to pathogens and facilitates their entry into the tissue and subsequent infection. Plants have evolved constitutive and induced defense mechanisms to properly respond to wounding and prevent infection. To explore the expression level of BT4 under wounding stress, we analyzed the datasets related to wounding stress from the GEO database (GSE101422). It was found that BT4 was significantly induced by wounding across different timepoint (). To study the function of the BT4 gene in Arabidopsis resistance to B. cinerea, the Col-0, bt4, CE, and OE plants were inoculated with B. cinerea to investigate the resistance of mutants to B. cinerea. The results have shown that bt4 mutant exhibited enhanced susceptibility to B. cinerea, whereas the Col-0, CE and OE plants showed obvious resistance to B. cinerea (). Trypan blue and DAB staining revealed that there were numerous dead cells and ROS in the leaves of the bt4 mutant inoculated with B. cinerea, with fewer dead cells and ROS were observed in leaves of Col-0, CE and OE plants (). Consistent with this phenotype, the expression levels of BcACTIN were significantly up-regulated in bt4 mutant (), and the content of chlorophyll were decreased obviously in bt4 mutant (). In addition, the plant hormone JA in bt4 mutants were also detected, and it was found that the contents of JA were also affected by the expression of BT4 gene. The contents of the plant hormone JA were significantly reduced in bt4, partially recovered in CE, and the highest in OE, exceeding Col-0 (). The results showed that bt4 mutant had strong susceptibility to B. cinerea, and indicated that the BT4 gene played a positive role in Arabidopsis resistance to B. cinerea.
Figure 1. The BT4 gene positively regulate the resistance of Arabidopsis to Botrytis cinerea. (a) The expression level of BT4 gene under wounding treatment. (b) Trypan blue and DAB observation of Col-0, bt4 mutant, CE and OE. (c) Expression level BcACTIN in Col-0, bt4 mutant, CE and OE inoculated by B. cinerea. (d) Evaluation of Col-0, bt4 mutant, CE and OE chlorophyll content after inoculation of B. cinerea.
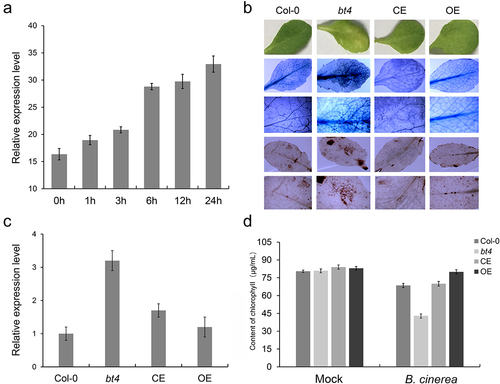
Figure 2. The expression of resistance-related genes inoculated with Botrytis cinerea. (a) Contents of JA in Col-0, bt4 mutant, CE and OE. (b) The expression level of JAR1 inoculated by B. cinerea in different timepoint. (c) The expression level of PDF1.2 inoculated by B. cinerea in different timepoint. (d) The expression level of PR3 inoculated by B. cinerea in different timepoint. (e) The expression level of JAL35 inoculated by B. cinerea in different timepoint. (f) The expression level of LOX2 inoculated by B. cinerea in different timepoint.
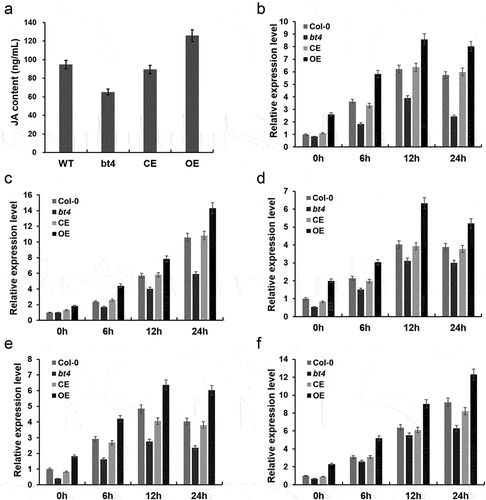
BT4 deficiency alters the expression of key JA signaling pathway genes in response to B. cinerea
To investigate whether the mutation of the BT4 gene affects the expression of key genes in JA signaling pathways, we further detected the expression level of JAR1 (Jasmonic acid-amido synthetase), PDF1.2 (Plant defensin 1.2), PR3 (Pathogenesis-related 3), JAL35 (Jacalin-related Lectin 35) and LOX2 (lipoxygenase 2) in the Col-0, bt4, CE, and OE plants inoculated with B. cinerea. The expression levels of JAR1, PDF1.2, PR3, JAL35 and LOX2 were obviously reduced by B. cinerea in the Col-0, bt4, CE, and OE plants ()). However, the expression levels of these genes were significantly down-regulated in bt4 mutants compared to those in the Col-0, CE, and OE plants. These results suggested that the BT4 gene might affect the expression of JA/ET signaling pathway genes in response to B. cinerea infection.
Conclusion
The JA signaling pathways play critical roles in protecting plant against pathogens.Citation16 JA and ET are also involved in induced systemic resistance of plants.Citation17 In this study, we found that wounding could induce the expression level of BT4 gene. After inoculation with B. cinerea, the expression levels of key genes of the JA signaling pathways in the bt4 mutant were significantly lower than those in the wild-type, CE, and OE mutant. These results indicated that the BT4 gene might be involved in the regulation of JA signaling pathways and further regulated the resistance to B. cinerea in Arabidopsis.
Materials and methods
Plant materials and growth conditions
Arabidopsis wild-type Col-0, BT4 gene T-DNA insertion mutant bt4 (SALK-015577), BT4 gene complemented transgenic plants CE, and the BT4 gene overexpression transgenic plants OE were provided by Mycotoxin and Molecular Plant Pathology Laboratory (Hebei Agricultural University, Baoding, China). Arabidopsis were grown on mixture of vermiculite: plant ash: perlite (6:2:1) in a greenhouse with a rhythm of 16 h light/8 h dark.
B. cinerea inoculation
B. cinerea (B05.10), provided by Hebei Key Laboratory of Plant Physiology and Molecular Pathology (Hebei Agricultural University, Baoding, China), was grown on PDA media and incubated at 22°C for 2 weeks. The conidia suspension of B. cinerea was prepared by brushing the B. cinerea medium with distilled water. The conidia suspension of B. cinerea was filtered and adjusted the final concentration of 4 ~ 8 × 106 conidia/ml. Four-week-old of Arabidopsis plants were used to inoculate with 5 μl conidia suspension of B. cinerea placed onto individual leaves.
Inoculated Arabidopsis plants were moisturized at 22°C for 24 h in darkness and then incubated under normal conditions. At 0, 3, 6, 12, and 24 h post-inoculation (hpi), Arabidopsis plants were collected and used for Real-time PCR analysis. At 48 hpi, lesion formation was observed and the inoculated leaves were stained with trypan blue and DAB, and used for and relative expression level of B. cinerea BcACTIN and determination of chlorophyll content.
RNA extraction and real-time PCR analysis of gene expression
Total RNA of Arabidopsis plants were extracted with Trizol regent (TaKaRa, Dalian, China) according to the manufacturer’s instructions and treated with RNase-free DNase I (Omega, USA) to remove genomic DNAs. The first strand cDNA was obtained using PrimeScript RT regent kit (TaKaRa, Dalian, China). The Real-time PCR reaction was done on a CFX96 real-time PCR system (BioRad, Hercules, CA, USA) in 10 μL reactions containing 0.2 μL SYBR Premix Ex TaqTM (TaKaRa, Dalian, China) and each gene-specific primer (Table S1). Relative gene expression levels were calculated using 2−CT method with three independent biological replicates.
Supplemental Material
Download MS Word (16.1 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15592324.2022.2104003
Additional information
Funding
References
- Lorenzo O, Piqueras R, Sanchez-Serrano JJ, Solano R. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell. 2003;15(1):1–4. doi:10.1105/tpc.007468.
- Froidure S, Canonne J, Daniel X, Jauneau A, Briere C, Roby D, Rivas S. At sPLA 2 -α nuclear relocalization by the Arabidopsis transcription factor AtMYB30 leads to repression of the plant defense response. Proc Natl Acad Sci. 2010;107(34):15281–15286. doi:10.1073/pnas.1009056107.
- Ramirez V, Agorio A, Coego A, Garcia-Andrade J, Hernandez MJ, Balaguer B, Ouwerkerk PBF, Zarra I, Vera P. MYB46 modulates disease susceptibility to Botrytis cinerea in Arabidopsis. Plant Physiol. 2011;155(4):1920–1935. doi:10.1104/pp.110.171843.
- Mengiste T, Chen X, Salmeron J, Dietrich R. The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell. 2003;15(11):2551–2565. doi:10.1105/tpc.014167.
- Huang PY, Zhang J, Jiang B, Chan C, Yu JH, Lu YP, Chung K, Zimmerli L. NINJA-associated ERF19 negatively regulates Arabidopsis pattern-triggered immunity. J Exp Bot. 2019;70(3):1033–1047. doi:10.1093/jxb/ery414.
- Sham A, Moustafa K, Al-Shamisi S, Alyan S, Iratni R, AbuQamar S, Balestrini R. Microarray analysis of Arabidopsis WRKY33 mutants in response to the necrotrophic fungus Botrytis cinerea. PloS One. 2017;12(2):e0172343. doi:10.1371/journal.pone.0172343.
- Liu S, Ziegler J, Zeier J, Birkenbihl RP, Somssich IE. Botrytis cinerea B05.10 promotes disease development in Arabidopsis by suppressing WRKY33-mediated host immunity. Plant Cell Environ. 2017;40(10):2189–2206. doi:10.1111/pce.13022.
- Melnick A, Ahmad KF, Arai S, Polinger A, Ball H, Borden KL, Carlile GW, Prive GG, Licht JD. In-depth mutational analysis of the promyelocytic leukemia zinc finger BTB/POZ domain reveals motifs and residues required for biological and transcriptional functions. Mol Cell Biol. 2000;20(17):6550–6567. doi:10.1128/MCB.20.17.6550-6567.2000.
- Bomont P, Cavalier L, Blondeau F, Ben Hamida C, Belal S, Tazir M, Demir E, Topaloglu H, Korinthenberg R, Tüysüz B, et al. The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat family, is mutated in giant axonal neuropathy. Nat Genet. 2000;26(3):370–374. doi:10.1038/81701.
- Kobayashi A, Kang M-I, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24(16):7130–7139. doi:10.1128/MCB.24.16.7130-7139.2004.
- Wilkins A, Ping Q, Carpenter CL. RhoBTB2 is a substrate of the mammalian Cul3 ubiquitin ligase complex. Gene Dev. 2004;18(8):856–861. doi:10.1101/gad.1177904.
- Du L, Poovaiah BW. A novel family of Ca 2+ /Calmodulin-binding proteins involved in transcriptional regulation: interaction with fsh/Ring3 class transcription activators. Plant Mol Biol. 2004;54(4):549–569. doi:10.1023/B:PLAN.0000038269.98972.bb.
- Ren SX, Mandadi KK, Boedeker AL, Rathore KS, McKnight TD. Regulation of Telomerase in Arabidopsis by BT2, an apparent target of TELOMERASE ACTIVATOR1. Plant Cell. 2007;19(1):23–31. doi:10.1105/tpc.106.044321.
- Boyle P, Le Su E, Rochon A, Shearer HL, Murmu J, Chu JY, Fobert PR, Despres C. The BTB/POZ Domain of the Arabidopsis Disease Resistance Protein NPR1 interacts with the repression domain of TGA2 to Negate Its Function. Plant Cell. 2009;21(11):3700–3713. doi:10.1105/tpc.109.069971.
- Zheng X, Xing J, Zhang K, Pang X, Zhao Y, Wang G, Zang J, Huang R, Dong J. Ethylene Response Factor ERF11 activates BT4 transcription to regulate immunity to Pseudomonas syringae. Plant Physiol. 2019;180(2):1132–1151. doi:10.1104/pp.18.01209.
- Bari R, Jones JD. Role of plant hormones in plant defence responses. Plant Mol Biol. 2009;69(4):473–488. doi:10.1007/s11103-008-9435-0.
- Pieterse CM, Leon-Reyes A, Van der Ent S, Van Wees SC. Networking by small-molecule hormones in plant immunity. Nat Chem Biol. 2009;5(5):308–316. doi:10.1038/nchembio.164.
