ABSTRACT
Plant-parasitic cyst nematodes (Heterodera and Globodera spp.) secrete CLAVATA3/EMBRYO SURROUNDING REGION-RELATED (CLE) effector proteins, which act as ligand mimics of plant CLE peptides to promote successful nematode infection. Previous studies of the Arabidopsis-beet cyst nematode (BCN; H. schachtii) pathosystem showed that Arabidopsis CLE receptors including CLAVATA1 (CLV1), CLV2, and RECEPTOR-LIKE PROTEIN KINASE 2 (RPK2) are required for BCN CLE signaling. Studies further revealed that nematode CLE signaling through GmCLV2 and StCLV2, an Arabidopsis CLV2 orthologue from soybean (Glycines max) and potato (Solanum tuberosum), respectively, is required for the soybean cyst nematode (SCN; H. glycines) and the potato cyst nematode (PCN; G. rostochiensis) to induce disease in their respective host plant. In this study, we identified and characterized two additional potato receptors, StRPK2 and StCLV1, homologues of Arabidopsis RPK2 and CLV1, for a role in PCN parasitism. Using promoter-reporter lines we showed that both StRPK2 and StCLV1 are expressed in the potato root but vary in their spatial expression patterns. Interestingly, StRPK2 but not StCLV1 was found to be expressed and upregulated at PCN infection sites. Nematode infection assays on StRPK2-knockdown lines revealed a decrease in nematode infection. Collectively, our results suggest that parallel CLE signaling pathways involving StCLV2 and StRPK2 are important for PCN parasitism and that manipulation of nematode CLE signaling may represent a viable means to engineer nematode resistance in crop plants including potato.
Introduction
Potato (Solanum tuberosum) is the most important non-cereal food crop in the world. Effective pest and disease management is crucial for the sustainability of this crop. Potato cyst nematodes (PCN), Globodera rostochiensis and G. pallida, are devastating pathogens of potato that cause an estimated yield loss of 9% worldwide.Citation1 PCN are obligate, endoparasitic nematodes that induce the formation of a feeding structure called a syncytium, a multinucleate and metabolically highly active structure of fused cells around the vasculature in potato roots.Citation2,Citation3 The syncytium serves as the sole nutrient source that sustains the growth and development of the nematode. It is now known that effector proteins delivered into the host root through the nematode’s stylet are the major signaling molecules responsible for the formation and maintenance of the syncytium.Citation3,Citation4
Effector genes encoding the CLAVATA3/EMBRYO SURROUNDING REGION-RELATED (CLE) proteins are among the best studied groups of effectors from plant-parasitic nematodes. Nematode CLE genes are widely distributed in various cyst nematode species, including PCN, the soybean cyst nematode (SCN; Heterodera glycines), and the beet cyst nematode (BCN; H. schachtii) that infects sugarbeet and other important vegetable crops.Citation3,Citation5–11 Previous studies have demonstrated that nematode-secreted CLE peptides function as mimics of plant CLE peptides to promote nematode parasitism.Citation6,Citation8–12 Plant CLEs are a large family of extracellular signaling peptides that stimulate receptor-mediated signaling pathways to modulate various developmental and physiological programs.Citation13,Citation14 Plant CLEs are grouped into either A-type or B-type peptides.Citation13,Citation14 The A-type peptides promote cell differentiation, the B-type peptides, on the other hand, inhibit differentiation of tracheary elements and promote procambial cell division. Leucine-rich repeat (LRR) receptor-like kinase (RLK) family members are the major players in perceiving CLE peptides. Of the 27 unique CLE peptides encoded by the Arabidopsis genome, CLAVATA3 (CLV3), an A-type peptide, is the best characterized member. Multiple receptor complexes, including CLV1, the LRR receptor-like protein CLV2 and the coreceptor CORYNE (CRN), BARELY ANY MERISTEM (BAM) receptor kinases, and BAM1 associated RECEPTOR-LIKE PROTEIN KINASE 2 (RPK2), are involved in perceiving the CLV3 peptide to regulate stem cell homeostasis in the shoot apical meristem.Citation14,Citation15 CLE signaling pathways also regulate many aspects of Arabidopsis root development. For example, many CLE peptides including CLE17 and CLE19 can induce premature termination of the root apical meristem (RAM) and a short root phenotype.Citation16 It was recently showed that RPK2 mediates the root growth-arrest phenotype triggered by exogenous application of CLE17 or CLE19 peptide.Citation17 Studies also revealed that CLV1 is involved in CLE40 signaling to regulate stem cell homeostasis in the distal region of the RAM as well as CLE3 signaling in roots under nitrogen-deficient conditions.Citation18,Citation19
The Arabidopsis-BCN pathosystem was initially used to investigate the mechanistic details of nematode CLE signaling during plant parasitism. Studies revealed that Arabidopsis receptors CLV2/CRN, CLV1, and RPK2 are involved in perceiving BCN CLEs and that this perception is required for successful nematode infection and syncytium development.Citation20,Citation21 Orthologues of these Arabidopsis receptors were identified from soybean (Glycine max), a host for SCN.Citation11 All these CLE receptors were found to be expressed in SCN-induced syncytia in soybean roots, and nematode development was compromised in transgenic soybean hairy roots with a reduced expression of GmCLV2, GmCRN, or GmCLV1, indicating that soybean receptors GmCLV2/GmCRN and GmCLV1 play a critical important role in SCN parasitism.Citation11 We previously characterized StCLV2 from potato, an Arabidopsis CLV2 orthologue. StCLV2 was found to be highly upregulated at PCN infection sites and transgenic potato lines with reduced StCLV2 expression showed increased resistance to PCN infection.Citation12 StCLV2 was the only CLE receptor from potato that has been characterized and demonstrated to be directly involved in CLE signaling that is crucial for PCN to induce disease in potato.Citation12 Parallel signaling pathways involving multiple CLE receptors are important for BCN and SCN to successfully infect their respective host plants.Citation20,Citation21 We thus hypothesized that additional CLE receptors from potato are likely involved in perceiving PCN CLE signals to promote successful nematode infection. In this study, we cloned and characterized StRPK2 and StCLV1 from potato, homologues of Arabidopsis RPK2 and CLV1 receptor genes, and investigated their role in PCN parasitism.
Results and discussion
Cloning and characterization of StRPK2 and StCLV1 genes from potato
Searching the potato genomeCitation22 through BLAST analysis identified two potato genes Soltu.DM.03G007830.1 (named StRPK2; Genbank accession number OP171928) and Soltu.DM.04G036750.1 (named StCLV1; Genbank accession number OP171929), homologous to Arabidopsis RPK2 and CLV1, respectively. StRPK2 encodes an 1126-amino acid (aa) protein consisting of a predicted signal peptide, an extracellular domain of 16 LRRs, a transmembrane region, and a putative kinase domain located at the C-terminus (). StRPK2 is 76% similar and 64% identical to RPK2 from Arabidopsis. StCLV1 encodes a 982-aa protein that contains a predicted signal peptide, an extracellular domain of 19 LRRs, a transmembrane region, and a C-terminal putative kinase domain (). The StCLV1 protein sequence is 76% similar and 63% identical to CLV1 from Arabidopsis. We further conducted phylogenetic analysis of these two receptors from Arabidopsis, potato, and soybean. The analysis revealed that StRPK2 and StCLV1 are more similar to their respective homologues from Arabidopsis than to those from soybean (). A recent study of LRR-RLK family proteins encoded by potato showed that StCLV1 we identified was grouped with CLV1 from Arabidopsis, rice, and tomato,Citation25 further supporting our conclusion that StCLV1 is the orthologue of Arabidopsis CLV1.
Figure 1. Characterization of StRPK2 and StCLV1. (a) Schematic representation of predicted domains in StRPK2 and StCLV1 in comparison with Arabidopsis RPK2 and CLV1, respectively. All these plant CLE receptors contain an N-terminal signal peptide, a LRR domain, a transmembrane domain, and a C-terminal kinase domain. (b) Phylogenetic relationship of StRPK2 and StCLV1 with their homologues from Arabidopsis and soybean (Glycines max). Multiple sequence alignment was obtained using the ClustalX program,Citation23 and the unrooted consensus tree from 1,000 bootstrap replicates was generated using the PHYLIP 3.61 program.Citation24 Sequences included are CLV1 (AT1G75820), RPK2 (AT3G02130), CRN (AT5G13290), Glycine max GmRPK2A, GmRPK2B, GmCLV1A, GmCLV1B, and GmCLV1C.Citation11 (c) Subcellular localization of StRPK2 and StCLV1 in the plant cell. The StRPK2-GFP:HA or StCLV1-GFP:HA fusion construct was transiently expressed in Nicotiana benthamiana leaves, and GFP signals that colocalized with FM4-64 was observed in the plasma membrane. (d) RT-PCR analysis of StRPK2 and StCLV1 expression in various potato tissues. The potato StUBI gene (XM_006360024) was used as an internal control.
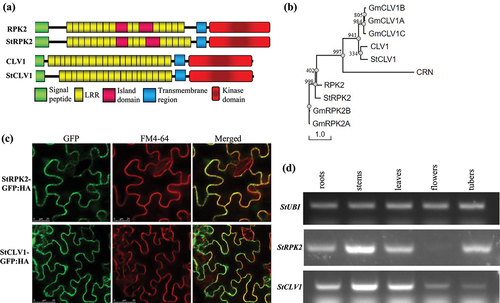
To test the predicted membrane localization of StRPK2 and StCLV1, we transiently expressed an StRPK2-GFP:HA (hemagglutinin-tagged GFP) or StCLV1-GFP:HA fusion protein in Nicotiana benthamiana leaves. Both StRPK2-GFP:HA and StCLV1-GFP:HA were found to be accumulated predominantly at the cell periphery and colocalized with the styryl dye FM4-64 plasma membrane counterstain ().
The Arabidopsis RPK2 and CLV1 genes are expressed in multiple tissues including roots.Citation21,Citation26,Citation27 We also examined the expression of StRPK2 and StCLV1 in various potato tissues using RT-PCR. Similar to RPK2 and CLV1, StRPK2 and StCLV1 were found to be expressed in multiple potato tissues including roots although both genes showed the highest expression in the stem tissue that contained emerging branches in comparison with other tested tissues ( and S1). Collectively, our data indicated that StCLV1 and StRPK2 may serve as functional receptors capable of perceiving PCN CLE peptides during nematode parasitism. Further studies, including confirming the binding of PCN CLE peptides to these receptors, are needed to verify this hypothesis.
StRPK2 and StCLV1 have different spatial expression patterns in potato roots
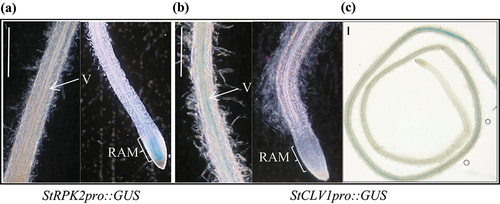
Prior to this study, knowledge on the spatial expression of StRPK2 and StCLV1 in the potato root tissue was lacking. For StCLV1 and StRPK2 to function as receptors that perceive PCN-secreted CLE signals, they should be expressed in the appropriate root tissue during nematode infection. We utilized transgenic potato lines expressing a promoter::β-glucuronidase (GUS) construct, StCLV1pro::GUS or StRPK2pro::GUS, to investigate the expression of these two receptor genes in potato roots under normal growing conditions. In Arabidopsis roots, CLV1 is expressed in the RAM and throughout the root vasculature starting at the maturation zone whereas RPK2 is specifically expressed in the RAM.Citation21,Citation26 Through analyzing the promoter::GUS lines we found that StCLV1 and StRPK2 have different spatial expression patterns in potato roots. StRPK2 appeared to be specifically expressed in the RAM whereas StCLV1 only had expression in the vascular tissue at the maturation region of potato roots (). The difference in root expression patterns between Arabidopsis CLV1 and StCLV1 might be due to the presence of different cis-acting elements identified in the promoter regions of CLV1 and StCLV1 (Table S1). StRPK2 and StCLV1 do not have overlapping expression regions. Thus, it would be interesting to determine whether these two receptors have similar or different functions in potato roots.
Figure 2. StRPK2 and StCLV1 show different spatial expression patterns in potato roots. Transgenic potato lines expressing StRPK2pro::GUS or StCLV1pro::GUS were generated and used to evaluate StRPK2 and StCLV1 expression in potato roots. StRPK2pro::GUS expression was observed primarily in the root apical meristem (a) whereas StCLV1pro::GUS expression was observed only in the root vascular tissue at the maturation region of potato roots under normal growth conditions (b,c). V, vasculature; RAM, root apical meristem. Bar = 1 mm.
StRPK2 but not StCLV1 is responsive to PCN infection
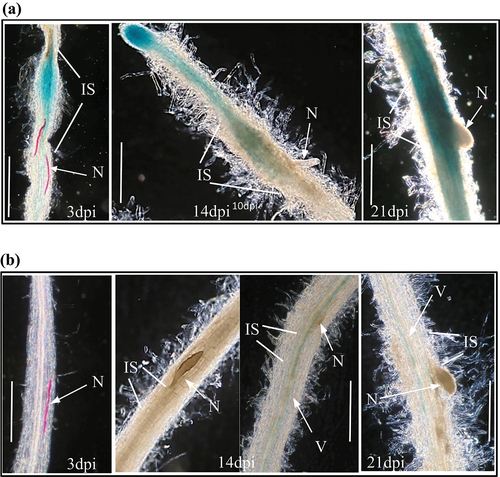
To determine whether StRPK2 and StCLV1 are responsive to nematode infection, transgenic StRPK2pro::GUS and StCLV1pro::GUS lines were infected with PCN and monitored during nematode development. For the StRPK2pro::GUS lines, strong GUS expression was detected at and around nematode infection sites as soon as the second-stage juveniles began to feed, and the expression level remained high at the infection sites associated with parasitic nematodes at later developmental stages (). GUS expression was also observed in the vasculature adjacent to nematode infection sites (). The induced expression of StRPK2 at nematode infection sites indicated that StRPK2 may be involved in perceiving PCN CLE peptides during nematode infection. We previously showed that StCLV2 is also upregulated at nematode infection sites.Citation12 PCN CLE genes are expressed throughout nematode parasitic stages.Citation8 The continuous secretion of CLE effectors might allow the nematode to activate and interact with host endogenous CLE receptors including StCLV2 and StRPK2 to promote syncytium formation and successful nematode parasitism. For the StCLV1pro::GUS lines, however, almost no GUS expression was observed at nematode infection sites over the course of nematode development (), indicating that StCLV1 may not have role in PCN parasitism.
Figure 3. StRPK2 but not StCLV1 is responsive to nematode infection. Transgenic potato lines expressing StRPK2pro::GUS or StCLV1pro::GUS were generated and used to determine StRPK2 or StCLV1 expression during nematode infection. (a) Roots of transgenic potato lines expressing StRPK2pro::GUS were infected with the potato cyst nematode (Globodera rostochiensis), and GUS activity was observed at nematode infection sites associated with parasitic second-stage juveniles (J2) (at 3 days post-inoculation, dpi) (left), third-stage juveniles (at 14 dpi) (middle), and fourth-stage juveniles (at 21 dpi) (right). (b) Roots of transgenic potato lines expressing StCLV1pro::GUS were infected with the potato cyst nematode (Globodera rostochiensis), but no obvious GUS activity was detected at nematode infection sites associated with parasitic J2 (at 3 dpi) (left), parasitic J3 (at 14 dpi) (middle), and parasitic J4 (at 21 dpi) (right). N, nematode; IS, infection site; V, vasculature. Bar = 1 mm.
Silencing StRPK2 reduces PCN infection
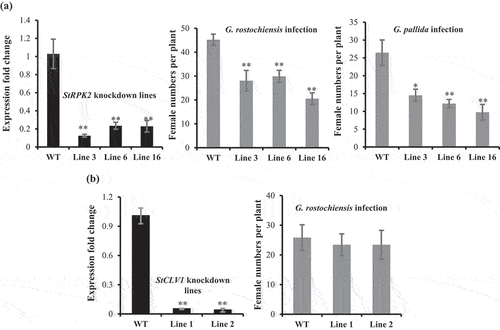
To further investigate a role of StRPK2 and StCLV1 in nematode parasitism, we made artificial microRNA constructs specifically targeting StRPK2 and StCLV1 and transformed them individually into potato. Screening the obtained independent transgenic lines identified two to three lines where the expression of the targeted gene was dramatically reduced in roots compared with wild-type potato lines (, a and b). StRPK2 and StCLV1 knockdown lines did not show obvious phenotypic difference in roots and aboveground tissues in comparison with the control plants. We then tested nematode infection on StRPK2 and StCLV1 knockdown lines along with wild-type plants. At 35 days after nematode infection, nematode females formed on the roots were counted. Our results showed that the three StRPK2 knockdown lines were more resistant to infection by both PCN species as the number of nematode females recovered from the three lines was approximately 36% to 62% less than those from the control lines (). The decreased susceptibility to PCN supports a role of StRPK2 in nematode parasitism. As for the StCLV1 knockdown lines, although a reduced number of nematode females was observed, our statistical analysis revealed that the female number observed on the StCLV1 knockdown lines was not significantly different from that observed on the control lines (). Meanwhile, StCLV1 was found not to be responsive to nematode infection. Collectively, our results indicated that StCLV1 may not have a role in PCN parasitism.
Figure 4. StRPK2 but not StCLV1 plays an important role in nematode parasitism. Transgenic potato lines expressing an artificial microRNA construct targeting StRPK2 or StCLV1 were generated. Quantitative RT-PCR was used to evaluate StRPK2 or StCLV1 expression in the obtained transgenic lines. Three independent StRPK2 knockdown lines (lines 3, 6, and 16) and two independent StCLV1 knockdown lines (lines 1 and 2) were found to have dramatically reduced expression of the target gene compared with the wild-type (WT) plant. Data represents means ± SD of three biological replicates, normalized to the potato StUBI gene (XM_006360024) and relative to expression in the WT plant (**P < .001, Student’s t test) (a: left; b: left). (a) StRPK2 knockdown lines showed reduced susceptibility to nematode infection. Wild-type and transgenic lines (20 vegetatively propagated plantlets for each line) were inoculated with Globodera rostochiensis or G. pallida juveniles, and nematode females were counted at 5 weeks after nematode inoculation. Values are means ± SD of two independent experiments (*0.01 < P < .05, **P < .01, Student’s t test). (b) StCLV1 knockdown lines showed a nematode susceptibility level similar to that of the WT plants. WT and StCLV1 knockdown lines (7-9 vegetatively propagated plantlets for each line) were inoculated with G. rostochiensis juveniles, and nematode females were counted at 5 weeks after nematode inoculation. Values are means ± SD (n = 7–9). Similar results were obtained from two independent experiments and one representative experiment is shown. No statistical difference was observed between the StCLV1 knockdown lines and the WT plants.
Conclusion
Previously, we reported that in the potato-PCN pathosystem, nematode CLE signaling through StCLV2 is required for successful nematode parasitism.Citation12 In this study, we characterized two candidate CLE receptors from potato, StCLV1 and StRPK2, and determined that StRPK2 but not StCLV1 is important for PCN parasitism. StRPK2 knockdown lines showed enhanced resistance to both PCN species, thus targeting StRPK2 may represent a viable means for generating potatoes with broad-spectrum nematode resistance. PCN encodes multidomain CLE effectors, which likely give rise to multiple different bioactive CLE peptides when delivered into potato roots during nematode infection.Citation12 Therefore, it is highly likely that more members of the LRR-RLK family in potato are involved in perceiving and relaying PCN-secreted CLE signals. The GrCLE1 peptide from PCN was previously shown to be able to bind Arabidopsis BAM1 and BAM2 receptors.Citation28 Guo et al. (2017) recently identified the expression of B-type CLE genes from SCN and BCN.Citation29 The study further revealed that the TDIF (tracheary element differentiation inhibitory factor)-TDR (TDIF receptor)-WOX4 pathway, which promotes procambial meristem cell proliferation, is involved in BCN parasitism on Arabidopsis.Citation29 B-type CLEs have not been identified in PCN. Further studies are needed to determine if any of the BAM and TDR homologues from potato and soybean have a role in PCN and SCN parasitism, respectively. Meanwhile, it would be interesting to investigate if there is any connection among CLE signaling pathways mediated by different CLE receptors during nematode infection. Cyst nematode secreted-CLE effectors are the key signaling molecules crucial to nematode parasitism. Identification of host receptors targeted by nematode CLE peptides as well as the associated downstream signaling components regulated by host receptors may provide novel means to engineer nematode resistance in economically important crops including potato.
Materials and methods
Nematode culture and plant materials
Maintenance of the potato cyst nematode (Globodera rostochiensis and G. pallida) culture and nematode inoculation on cultured potato plantlets were conducted as described previously.Citation30,Citation31 Potato (Solanum tuberosum) cv. Désirée was used for generating transgenic plants and Nicotiana benthamiana was used for transient expression assays.
Sequence and phylogenetic analysis of StRPK2 and StCLV1
Arabidopsis RPK2 and CLV1 gene sequences were used to search the potato genome via BLAST analysis to identify the respective homologous sequences from potato and then named them as StRPK2 and StCLV1, respectively. The SMART (simple modular architecture research tool) database (http://smart.embl-heidelberg.de/) and NCBI-CDD (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) were used to identify the signal peptide, the transmembrane region, and LRR and kinase domains in Soltu.DM.03G007830.1 (named StRPK2) and Soltu.DM.04G036750.1 (named StCLV1) encoded protein sequences. Phylogenetic analysis of Arabidopsis RPK2 and CLV1 and their homologues from potato and soybean was conducted as described previously.Citation30 Full-length protein sequences were used for the analysis.
Characterization of the CLV1 promoter region
The PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/)Citation32 was used to identify cis-acting elements in the Arabidopsis CLV1 (−3450 bp to −1 bp) and StCLV1 (−3465 bp to −1 bp) promoter regions (Table S1).
Subcellular localization
The StRPK2 (OP171928) and StCLV1 (OP171929) genes were identified in the annotated potato genome sequence and amplified from complementary DNA derived from potato roots using primer pairs StRPK2_ATGF and StRPK2_TGAR, and StCLV1_ATGF and StCLV1_TGAR (Table S2). The amplified PCR products were cloned into the pBIN61-GFP:HA binary vectorCitation33 at the XbaI and BamHI sites to generate StRPK2-GFP:HA and StCLV1-GFP:HA fusion protein constructs. Agrobacterium tumefaciens strain C58C1 transformed with StRPK2-GFP:HA or StCLV1-GFP:HA was infiltrated into N. benthamiana leaves.Citation31 Two days after infiltration, leaves were collected and stained in FM4-64 solution (Invitrogen). Leaf sections were then visualized with an SP5 Leica confocal microscope.
Histochemical GUS assay and nematode staining
A 2484-bp DNA sequence upstream of the start codon of StRPK2 that likely covers all the cis-regulatory elements of the gene and a 3465-bp DNA sequence upstream of the start codon of StCLV1 containing all the possible cis-regulatory elements were amplified from potato genomic DNA by PCR using primer pairs StRPK2pr_SalI-2484 F and StRPK2pr_BamHI-2 R for StRPK2 and StCLV1_SalI_-f3465F and StCLV1_BamHI_R for StCLV1 (Table S2). The amplified DNA fragments were cloned into the binary vector pBI101.2Citation34 at the SalI and BamHI sites to generate the StRPK2pro::GUS and StCLV1pro::GUS constructs. Transgenic potato lines expressing the individual promoter-GUS construct were generated as described.Citation31 Nematode infection on transgenic potato plantlets as well as nematode staining and histochemical GUS assays were performed as described previously.Citation12
Generation of gene knockdown lines and nematode infection assays
To make the artificial microRNA (amiRNA) construct, a 21-mer sequence (5’-TAATACATGGCACGATTGCCG-3’) targeting StRPK2 and a 21-mer sequence (5’- TATCTGACTATATCTGCGCCG-3’) targeting StCLV1, designed according to the Web artificial microRNA designer interface WMD3 (wmd3.weigelworld.org), was engineered into the miR319a precursor by replacing the endogenous MIR319a sequence in pRS300Citation35 using overlapping PCRCitation36 with primers as listed in Table S2. The new amiR319a precursor was then cloned into the modified binary vector pSMD1 at the XhoI and SacI sites to make the pSMD1-StRPK2ami and the pSMD1-StCLV1ami artificial microRNA constructs.Citation12 These constructs were transformed individually into A. tumefaciens strain LBA4404 and used for potato transformation. Transgenic potato lines expressing the amiRNA constructs were generated as described.Citation31 Nematode infection assays on transgenic potato plantlets were conducted and the number of nematode females was counted on each plantlet at 5 weeks after nematode inoculation.Citation30,Citation31 The experiments were conducted at least two times with a minimum of 15 replicates for each line, and similar results were obtained. The results were analyzed by the student t-test.
Reverse transcription-PCR and quantitative reverse transcription-PCR
mRNA from roots, leaves, and stems of cultured potato plantlets and flowers and tubers from a potted potato plant was extracted and used to determine StRPK2 and StCLV1 expression in various potato tissues by RT-PCR. mRNA from roots of cultured plantlets of transgenic potato lines expressing pSMD1-StRPK2ami or pSMD1-StCLV1ami as well as a wild-type control plant was extracted and used to quantify StRPK2 and StCLV1 expression by quantitative RT-PCR as described previously.Citation30 mRNA was treated with DNase I and confirmed with no genomic DNA contamination by PCR before cDNA synthesis. The potato StUBI gene (XM_006360024) was used as an endogenous reference. Primers used are listed in Table S2. The RT-qPCR data were obtained from three independent experiments with three technical replicates for each cDNA sample.
Acknowledgments
This work was supported by a grant from the National Science Foundation (grant no. IOS-1456047 to MGM and XW), a grant from the USDA-NIFA-AFRI (grant no. 2009-35302-05304 to MGM and XW), and funding from the US Department of Agriculture, Agricultural Research Service.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Turner SJ, Subbotin SA. Cyst nematodes. In: Perry RN, Moens M, editors. Plant nematology. 2nd. Wallingford (U.K): CAB International; 2013:109–8.
- Hussey RS, Grundler FM. Nematode parasitism of plants. In: Perry RN, Wright J, editors. Physiology and biochemistry of free-living and plant parasitic nematodes. Oxford (UK): CAB International Press; 1998. p. 213–243.
- Mitchum MG, Hussey RS, Baum TJ, Wang X, Elling AA, Wubben M, Davis EL. Nematode effector proteins: an emerging paradigm of parasitism. New Phytol. 2013;199(4):879–894. doi:10.1111/nph.12323.
- Hewezi T. Cellular signaling pathways and posttranslational modifications mediated by nematode effector proteins. Plant Physiol. 2015;169(2):1018–1026. doi:10.1104/pp.15.00923.
- Wang X, Allen R, Ding X, Goellner M, Maier T, de Boer JM, Baum TJ, Hussey RS, Davis EL. Signal peptide-selection of cDNA cloned directly from the esophageal gland cells of the soybean cyst nematode Heterodera glycines. Mol Plant Microbe Interact. 2001;14(4):536–544. doi:10.1094/MPMI.2001.14.4.536.
- Wang X, Mitchum MG, Gao B, Li C, Diab H, Baum TJ, Hussey RS, Davis EL. A parasitism gene from a plant-parasitic nematode with function similar to CLAVATA3/ESR (CLE) of Arabidopsis thaliana. Mol Plant Pathol. 2005;6(2):187–191. doi:10.1111/j.1364-3703.
- Mitchum MG, Wang X, Davis EL. Diverse and conserved roles of CLE peptides. Curr Opin Plant Biol. 2008;11(1):75–81. doi:10.1016/j.pbi.2007.10.010.
- Lu SW, Chen S, Wang J, Yu H, Chronis D, Mitchum MG, Wang X. Structural and functional diversity of CLAVATA3/ESR (CLE)-like genes from the potato cyst nematode Globodera rostochiensis. Mol Plant Microbe Interact. 2009;22(9):1128–1142. doi:10.1094/MPMI-22-9-1128.
- Wang J, Lee C, Replogle A, Joshi S, Korkin D, Hussey R, Baum TJ, Davis EL, Wang X, Mitchum MG. Dual roles for the variable domain in protein trafficking and host-specific recognition of Heterodera glycines CLE effector proteins. New Phytol. 2010;187(4):1003–1017. doi:10.1111/j.1469-8137.
- Wang J, Replogle A, Hussey R, Baum T, Wang X, Davis EL, Mitchum MG. Identification of potential host plant mimics of CLAVATA3/ESR (CLE)-like peptides from the plant-parasitic nematode Heterodera schachtii. Mol Plant Pathol. 2011;12(2):177–186. doi:10.1111/j.1364-3703.
- Guo X, Chronis D, De La Torre CM, Smeda J, Wang X, Mitchum MG. Enhanced resistance to soybean cyst nematode Heterodera glycines in transgenic soybean by silencing putative CLE receptors. Plant Biotechnol J. 2015;13(6):801–810. doi:10.1111/pbi.12313.
- Chen S, Lang P, Chronis D, Zhang S, De Jong WS, Mitchum MG, Wang X. In planta processing and glycosylation of a nematode CLAVATA3/ENDOSPERM SURROUNDING REGION-like effector and its interaction with a host CLAVATA2-like receptor to promote parasitism. Plant Physiol. 2015;167(1):262–272. doi:10.1104/pp.114.251637.
- Yamada M, Sawa S. The roles of peptide hormones during plant root development. Curr Opin Plant Biol. 2013;16(1):56–61. doi:10.1016/j.pbi.2012.11.004.
- Fletcher JC. Recent advances in Arabidopsis CLE peptide signaling. Trends Plant Sci. 2020;25(10):1005–1016. doi:10.1016/j.tplants.2020.04.014.
- Song XF, Hou XL, Liu CM. CLE peptides: critical regulators for stem cell maintenance in plants. Planta. 2021;255(1):5. doi:10.1007/s00425-021-03791-1.
- Whitford R, Fernandez A, De Groodt R, Ortega E, Hilson P. Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proc Natl Acad Sci U S A. 2008;105(47):18625–18630. doi:10.1073/pnas.0809395105.
- Racolta A, Nodine MD, Davies K, Lee C, Rowe S, Velazco Y, Wellington R, Tax FE. A common pathway of root growth control and response to CLE peptides through two receptor kinases in Arabidopsis. Genetics. 2018;208(2):687–704. doi:10.1534/genetics.117.300148.
- Stahl Y, Grabowski S, Bleckmann A, Kühnemuth R, Weidtkamp-Peters S, Pinto KG, Kirschner GK, Schmid JB, Wink RH, Hülsewede A, et al. Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr Biol. 2013;23(5):362–371. doi:10.1016/j.cub.2013.01.045.
- Araya T, Miyamoto M, Wibowo J, Suzuki A, Kojima S, Tsuchiya YN, Sawa S, Fukuda H, von Wirén N, Takahashi H. CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc Natl Acad Sci U S A. 2014;111(5):2029–2034. doi:10.1073/pnas.1319953111.
- Replogle A, Wang J, Bleckmann A, Hussey RS, Baum TJ, Sawa S, Davis EL, Wang X, Simon R, Mitchum MG. Nematode CLE signaling in Arabidopsis requires CLAVATA2 and CORYNE. Plant J. 2011;65(3):430–440. doi:10.1111/j.1365-313X.
- Replogle A, Wang J, Paolillo V, Smeda J, Kinoshita A, Durbak A, Tax FE, Wang X, Sawa S, Mitchum MG. Synergistic interaction of CLAVATA1, CLAVATA2, and RECEPTOR-LIKE PROTEIN KINASE 2 in cyst nematode parasitism of Arabidopsis. Mol Plant Microbe Interact. 2013;26(1):87–96. doi:10.1094/MPMI-05-12-0118-FI.
- Potato Genome Sequencing Consortium; Xu X, Pan S, Cheng S, Zhang B, Mu D, Ni P, Zhang G, Yang S, Li R, Wang, J, et al. Genome sequence and analysis of the tuber crop potato. Nature. 2011;475(7355):189–195. doi:10.1038/nature10158.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi:10.1093/nar/25.24.4876.
- Felsenstein J. PHYLIP: phylogeny inference package (version 32). Cladistics. 1989;5:164–166.
- Li X, Salman A, Guo C, Yu J, Cao S, Gao X, Li W, Li H, Guo Y. Identification and characterization of LRR-RLK family genes in potato reveal their involvement in peptide signaling of cell fate decisions and biotic/abiotic stress responses. Cells. 2018;7(9):120. doi:10.3390/cells7090120.
- Kinoshita A, Betsuyaku S, Osakabe Y, Mizuno S, Nagawa S, Stahl Y, Simon R, Yamaguchi-Shinozaki K, Fukuda H, Sawa S. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development. 2010;137(22):3911–3920. doi:10.1242/dev.048199.
- Mizuno S, Osakabe Y, Maruyama K, Ito T, Osakabe K, Sato T, Shinozaki K, Yamaguchi-Shinozaki K. Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. Plant J. 2007;50(5):751–766. doi:10.1111/j.1365-313X.
- Guo Y, Ni J, Denver R, Wang X, Clark SE. Mechanisms of molecular mimicry of plant CLE peptide ligands by the parasitic nematode Globodera rostochiensis. Plant Physiol. 2011;157(1):476–484. doi:10.1104/pp.111.180554.
- Guo X, Wang J, Gardner M, Fukuda H, Kondo Y, Etchells JP, Wang X, Mitchum MG. Identification of cyst nematode B-type CLE peptides and modulation of the vascular stem cell pathway for feeding cell formation. PLoS Pathog. 2017;13(2):e1006142. doi:10.1371/journal.ppat.1006142.
- Lu SW, Tian D, Borchardt-Wier HB, Wang X. Alternative splicing: a novel mechanism of regulation identified in the chorismate mutase gene of the potato cyst nematode Globodera rostochiensis. Mol Biochem Parasitol. 2008;162(1):1–15. doi:10.1016/j.molbiopara.2008.06.002.
- Chronis D, Chen S, Lu S, Hewezi T, Carpenter SC, Loria R, Baum TJ, Wang X. A ubiquitin carboxyl extension protein secreted from a plant-parasitic nematode Globodera rostochiensis is cleaved in planta to promote plant parasitism. Plant J. 2013;74(2):185–196. doi:10.1111/tpj.12125.
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–327. doi:10.1093/nar/30.1.325.
- Sacco MA, Koropacka K, Grenier E, Jaubert MJ, Blanchard A, Goverse A, Smant G, Moffett P. The cyst nematode SPRYSEC protein RBP-1 elicits Gpa2- and RanGAP2-dependent plant cell death. PLoS Pathog. 2009;5(8):e1000564. doi:10.1371/journal.ppat.1000564.
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6(13):3901–3907. doi:10.1002/j.1460-2075.1987.tb02730.x.
- Ossowski S, Schwab R, Weigel D. Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 2008;53(4):674–690. doi:10.1111/j.1365-313X.
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18(5):1121–1133. doi:10.1105/tpc.105.039834.
