ABSTRACT
During plant growth and development, the YABBY gene plays a crucial role in the morphological structure, hormone signaling, stress resistance, crop breeding, and agricultural production of plant lateral organs, leaves, flowers, and fruits. Astragalus mongholicus is a perennial herbaceous plant in the legume family, widely used worldwide due to its high medicinal and edible value. However, there have been no reports of the YABBY gene family in A. mongholicus. This study used bioinformatics methods, combined with databases and analysis websites, to systematically analyze the AmYABBY gene family in the entire genome of A. mongholicus and verified its expression patterns in different tissues of A. mongholicus through transcriptome data and qRT-PCR experiments. A total of seven AmYABBY genes were identified, which can be divided into five subfamilies and distributed on three chromosomes. Two pairs of AmYABBY genes may be involved in fragment duplication on three chromosomes. All AmYABBY proteins have a zinc finger YABBY domain, and members of the same group have similar motif composition and intron – exon structure. In the promoter region of the genes, light-responsive and MeJa-response cis-elements are dominant. AmYABBY is highly expressed in stems and leaves, especially AmYABBY1, AmYABBY2, and AmYABBY3, which play important roles in the growth and development of stems and leaves. The AmYABBY gene family regulates the growth and development of A. mongholicus. In summary, this study provides a theoretical basis for in-depth research on the function of the AmYABBY gene and new insights into the molecular response mechanism of the growth and development of the traditional Chinese medicine A. mongholicus.
Introduction
Astragalus memeranaceus (Fisch.) Bge. Var. mongholicus (Bge.) Hsiao is a perennial herbaceous plant of the family Leguminosae and is widely used worldwide due to its very high medicinal and food value.Citation1 A. mongholicus is the basal plant of the Chinese medicine astragalus (ASTRAGALI RADIX), and its dried root is the medicinal part, which is used in more than 200 herbal formulas, with a history of application of more than 2,000 years.Citation2 A. mongholicus is a tonic herb that is used to improve vital energy, strengthen the spleen, tranquilize the heart, moisten the intestines and act as a laxative, and astringe the yin to stop sweating.Citation3 The chemical composition of the A. mongholicus complex mainly includes saponins, flavonoids, and polysaccharides. It is also rich in amino acids and trace elements.Citation4,Citation5 Pharmacological activities can improve the body’s immunity and number of scavenging free radicals and promote anti-inflammatory, anti-tumor, anti-diabetic, antioxidant, and other effects.Citation6,Citation7 The growth of A. mongholicus is a delicate process,Citation8 and the study of the molecular mechanism of its growth and development is of great significance in guiding production in the face of an increasingly severe natural environment.
The YABBY gene family belongs to the zinc finger protein superfamily and is a plant-specific transcription factor.Citation9 All YABBY members share two highly conserved structural domains characterized by an N-terminal zinc finger structural domain (C2-C2) and a C-terminal YABBY structural domain (helix – loop – helix).Citation10 There are five subfamilies of YABBY genes, namely, CRC (CRABS CLAW), FIL (FILAMENTOUS FLOWER)/YAB3, INO (INNER NO OUTER), YAB2, and YAB5.Citation11 YABBY genes play a crucial role in morphogenesis, including lateral organs, leaf, flower, and fruit development.Citation12 The YABBY gene family is widely involved in plant life processes. Studies have shown that tomato SlCRCa is highly expressed in petals and stamens and is sensitive to gibberellin (GA) treatment. Overexpression of SlCRCa made tomato petals, stamens and fruits smaller, while knock-out SlCRCa showed the opposite phenotype.Citation13 INO in Arabidopsis thaliana protects reproductive development by reducing iron load in developing seeds by inhibiting NRAMP1 expression.Citation14 Overexpression of the BcYAB3 gene in Chinese cabbage in A. thaliana can lead to downward curling of leaves, delayed bolting and flowering.Citation15 It has been shown that FIL genes are associated with floral organ formation and leaf development,Citation16 YAB2, YAB3 and YAB5 are specifically expressed in plant trophic tissues.Citation17,Citation18 In A.thaliana, the CRC gene, a member of the YABBY gene family, is involved in the development of nectaries and carpels, and the INO gene promotes the development of the external bead cover.Citation19 Overexpression of the soybean GmFILa transcription factor in A. thaliana leads to changes in the dorsal and ventral polarity of the epidermal leaf tissues of the transgenic plants, and the apical meristem development is inhibited.Citation20 Currently, reports on YABBY genes have been carried out in various plants; however, whether YABBY genes are involved in the growth and development of A. mongholicus remains largely unknown. Currently, a high-quality genome of A. mongholicus has been released,Citation21 therefore, it is of great significance to mine the potential function of the AmYABBY gene in the growth and development of A. mongolianus based on whole-genome data and transcriptome data.
In this study, we performed genome-wide identification and characterization of the A. mongholicus YABBY gene family members of A. mongholicus. We also comprehensively analyzed the physicochemical properties, collinearity, phylogenetic evolution, gene structure, and cis-regulatory elements of the AmYABBY genes. In addition, the expression patterns of AmYABBY genes during the growth of A. mongholicus were comprehensively analyzed by transcriptome data combined with RT-qPCR experiments, which provided valuable information for screening candidate YABBY genes involved in the regulation of the growth and development of A. mongholicus.
Materials and methods
Plant material
The plant material, A. mongholicus seeds, was identified by the Wei Ma Research Institute of Heilongjiang University of Chinese Medicine. Soil-less cultivation was carried out in the light incubator of the Medicinal Molecular Laboratory of Heilongjiang University of Chinese Medicine. The nutrient solution used was an improved Hoagland nutrient solution (Coolaber Technology Co., Ltd., Beijing, China), with a light intensity of 2000–2500 Lux, a temperature of 25°C, and a light cycle of 16 hours (light)/8 hours (dark). When the seedlings of A. mongholicus were 40 days old, healthy plants with similar growth conditions were selected, and roots, stems, and leaves of the same plant were sampled. After collection, they were frozen with liquid nitrogen and stored in a −80°C ultra-low temperature refrigerator for further analysis.
Data sources
The whole-genome sequence and annotation files of A. mongholicus were downloaded from the Glo-bal Pharmacopoeia Genome Database (GPGD)Citation22 website (http://www.gpgenome.com/species/109). A. thaliana (GCA_000005425. 2), Malus domestica (GCF_002114115.1), Codonopsis lanceolata (GCA_013146195.2), and O.sativa (GCA_001433935.1) genome files and annotation files were downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/).
Identification of the AmYABBY gene family
In order to identify all members of the YABBY gene family in the genome of A. mongholicus, all protein sequences in the genome of A. mongholicus were extracted using TBtools softwareCitation23 and submitted to the Plant Transcription Factor Database (http://planttfdb.gao-lab.org/index.php) to obtain the YABBY gene candidate sequences, and the structural domains of the candidate sequences were confirmed using the CD-search (https://www.ncbi.nlm.nih.gov/cdd/) online tool. After removing redundant and structurally domain incomplete sequences, the final AmYABBY gene was obtained. Physicochemical properties of the AmYABBY protein, including molecular mass, isoelectric point, and hydrophobicity, were analyzed using the online analysis software ExPASy ProtParam (http://web.expasy.org/ProtParam). On another online platform (http://www.csbio.sjtu.edu.cn/bioinf/euk-multi-2/), AmYABBY protein subcellular localization results were used to obtain the prediction.
Secondary and tertiary structure prediction of gene-encoded proteins
We used TBtools software to extract and obtain protein sequences of A. mongholicus YABBY gene family members and used the SOPMA (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl.page=npsa_sopma.html) online website to predict the A. mongholicus YABBY protein secondary structure. The tertiary structure of the A. mongholicus YABBY protein (https://swissmodel.expasy.org/) was constructed using the SWISS-MODEL online site.
Chromosome location and evolution analysis
The chromosome location information of the A. mongholicus YABBY gene was obtained from the genome annotation file of A. mongholicus. Its corresponding chromosome physical location was mapped using TBtools software, and the genes were named according to the order of their chromosome locations. The evolution relationships within the species of A. mongholicus and with the other five species were analyzed using MCScanX.Citation24 The Advanced Circos function of TBtools software was used to visualize AmYABBY gene-replication events, and the Dual Systeny Plot function was used to visualize the covariate relationships between A. mongholicus and other species.
Phylogenetic analysis of AmYABBY and AtYABBY proteins
The A. thaliana YABBY protein sequences were downloaded from the TAIR database (https://www.arabidopsis.org/). The O. sativa and Glycine max YABBY protein sequences were downloaded from the Phytozome (https://phytozome-next.jgi.doe.gov/). The full-length protein sequences of AmYABBY and AtYABBY were aligned by multiple sequence comparison using MEGA-Ⅹ11 software,Citation25 and the phylogenetic tree was constructed using the neighbor-joining (NJ) method with the bootstrap replicated 1000 times. The AmYABBY proteins were divided into different subfamilies based on the classification of A. thaliana YABBY proteins. AmYABBY proteins were categorized into subfamilies based on the A. thaliana YABBY protein classification and beautified through the Evolview website (www://evolgenius.info/evolview-v3/).
AmYABBY gene motif, structure, and cis-element analysis
DNAMAN software was used to analyze the conserved domain of the AmYABBY protein. Motif analysis of AmYABBY gene family members was performed using the MEME website (http://meme-suite.org/tools/meme)Citation26 with parameters set to the Classic mode, ZOOPS, and number of motifs set to ten. All AmYABBY gene upstream 2000 bp sequences were used as the promoter region and submitted to the PlantCRAE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/)Citation27 to analyze their cis-element composition. TBtools software was applied to visualize the phylogeny, motif, conserved structural domains, gene structure, and cis-element composition of the AmYABBY gene.
Analysis of RNA-Seq data and real-time quantitative PCR (qRT-PCR) validation
The transcriptome data of roots, stems, and leaves of A. mongholicus used in this study were measured by our group. The expression of each AmYABBY gene was quantified in terms of reads per kilobase of transcription per million mapped reads (FPKM) values. All FPKM values were processed using log2(FPKM +1) log transformation, and heatmaps were generated using Tbtools software.Total RNA was extracted from the roots, stems, and leaves of A. mongholicus using the Plant Total RNA Extraction Kit Plant Total RNA Extraction Kit (Tiangen Biochemical Technology Co., Ltd., Beijing, China). Then, the first strand of cDNA was synthesized by reverse transcription using a cDNA First Strand Synthesis Kit (Msunflowers Biotech Co., Ltd., Beijing, China) and then synthesized using a Real-Time Fluorescence PCR Kit (Vazyme Biotech Co. Ltd., Nanjing, China) to perform qRT-PCR experiments. The AriaMx real-time PCR system (Agilent Technologies) was used for qRT-PCR experiments. The 18s gene was used as the internal reference gene, and three technical replicates were set up for each experiment. The 2−ΔΔCt methodCitation28 was used to calculate the relative gene expression in each tissue. Primer Premier (version 5.0) software was used to design qRT-PCR specific primers. Then, GraphPad prism (v8.0.2) software was used to plot relative gene expression histograms and analyze them via t-test. The primer sequences used for qRT-PCR can be found in Supplementary Table S1 (Table S1).
Results
Identification and physicochemical property analysis of AmYABBY genes
After removing redundant and incomplete sequences with structural domains, seven AmYABBY genes were finally identified (, File S1). In subsequent analyses, we named these genes AmYABBY1 to AmYABBY7 based on the gene’s position in the chromosome or genome. Detailed physicochemical property analyses were subsequently performed, which indicated that the amino acid lengths of the AmYABBY proteins ranged from 750 (AmYABBY5) to 495 (AmYABBY1). The predicted molecular weight of AmYABBY proteins ranges from 62,531.52 Da −41810.41 Da, and the isoelectric point is 5.10–5.18. The mean hydrophilicity of all AmYABBY proteins is less than 0, and probably all of them are hydrophilic.
Table 1. Physicochemical properties and secondary structure analysis of AmYABBY gene family.
According to the predicted data of secondary structure, the values of α-helices of the proteins encoded by the YABBY gene were all greater than 0.1512. Among them, the α-helices of AmYABBY1, AmYABBY4, and AmYABBY7 were very close to each other, and the α-helices of AmYABBY2 and AmYABBY3 showed similarity. The results in terms of β-turning angle showed that AmYABBY1 to AmYABBY7 proteins all had a get-turn angle degree of 0. The elongated chains were 0.4735 to 0.6223, of which AmYABBY3 and AmYABBY4, AmYABBY5 and AmYABBY6 had similar elongation lengths, respectively. The Irregular curls were 0.5743 ~ 0.7209, with similar degrees of irregular curls for AmYABBY1, AmYABBY2, AmYABBY4, and AmYABBY7 ().
The tertiary structures of AmYABBY4, AmYABBY7, amyabby1, and AmYABBY6 proteins were similar, and the tertiary structures of AmYABBY2, AmYABBY3, AmYABBY4, and AmYABBY5 proteins were consistent. The results showed that some gene family members had the same tertiary protein structure (Supplementary Figure S1).
Chromosomal localization and fragment-duplication events of AmYABBY genes
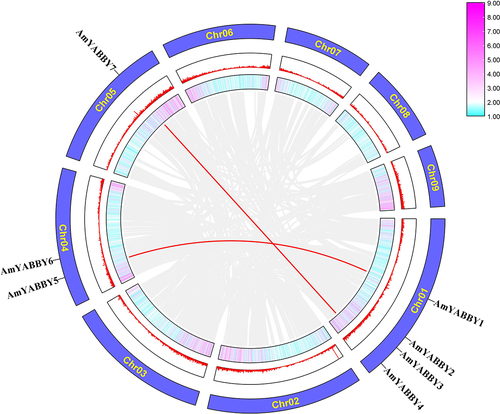
The distribution of AmYABBY genes on the chromosomes of A. mongholicus did not have a clear pattern. Seven AmYABBY genes were unevenly distributed on the three chromosomes of A. mongholicus. Chromosome one contained the largest number of AmYABBY gene family members (AmYABBY1, AmYABBY2, AmYABBY3, and AmYABBY4), and chromosome five contained the smallest number of AmYABBY genes (). To understand the expansion of the AmYABBY gene family, an investigation of the gene-duplication event in A. mongholicus was carried out. Two pairs of AmYABBY genes in the AmYABBY gene family were generated by segmental duplication: AmYABBY1 AmYABBY5 and AmYABBY4, AmYABBY7.
Figure 1. Chromosome distribution and fragment-replication event analysis of the AmYABBY gene family. Red-line generations indicate tandem duplication relationships between AmYABBY genes. The two outer circles represent gene density information: pink indicates high gene density, while blue indicates low gene density.
Phylogenetic analysis of the YABBY gene family in A. mongholicus
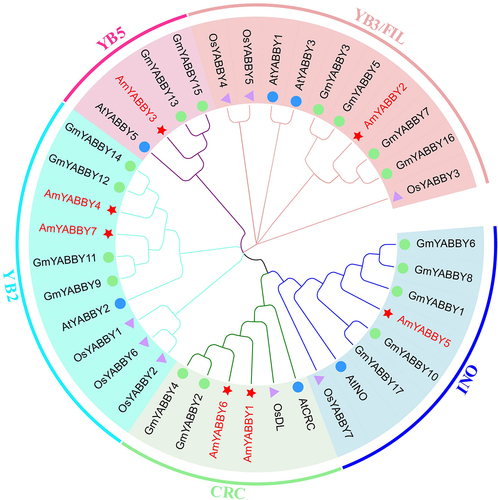
The amino acid sequences of A. thaliana, O. sativa and G. max YABBY family members were downloaded to explore the evolutionary relationship and classification of the AmYABBY gene family. We constructed a neighbor-joining phylogenetic tree using the YABBY protein sequences of A. mongholicus, O. sativa, G. max and A. thaliana. Phylogenetic analysis showed that seven AmYABBY proteins were divided into five subfamilies, including CRC containing AmYABBY1 and AmYABBY6, YB5 containing AmYABBY3, INO containing AmYABBY5, YB2 containing AmYABBY4 and YABBY7, and YB3/FIL containing AmYABBY1. The results of phylogenetic tree analysis showed that the AmYABBY Proteins of A. mongholicus and A. thaliana had high homology in each cluster ().
AmYABBY gene structure and cis-element analysis
Sequence alignment of AmYABBY family members was performed. The alignment results showed that both AmYABBY1-AmYABBY7 proteins contained a YABBY domain and a zinc finger domain (). There are conserved amino acids such as cysteine, leucine, proline, and glycine.
Figure 3. Alignment of conserved sequences of AmYABBY proteins, which are the YABBY conserved domain and zinc finger domain, respectively.

To explore the similarity of AmYABBY protein structures, 10 conserved motifs were identified using the MEME tool. These conserved motifs ranged from 7 to 50 amino acids in length, with at least 4 to 7 conserved motifs distributed among 7 AmYABBY proteins (). All sequences had motif1 (YABBY domain) and motif2 (zinc finger domain) (Supplementary Figure S2), which were further analyzed as the zinc finger structural domain and the YABBY structural domain, respectively. Evolutionarily related members have similar motif composition. For example, all members of the YB2 subfamily have seven identical motifs.
Figure 4. Analysis of gene motif, gene structure, and cis-element of AmYABBY gene family. (a) AmYABBY protein motif composition; different colors represent different motifs. (b) AmYABBY gene structure. CDS: coding sequence, UTR: in the untranslated region, lines denote introns. (c) The cis-acting element contained in the AmYABBY gene family.
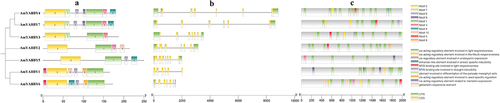
To further understand the evolution of AmYABBY genes, we compared the sequences of AmYABBY genes in A. mongholicus and analyzed their coding regions and introns (). All members contained introns, and the number of introns was distributed in the range of five to six. As expected, the number of introns was relatively conserved among members of the same subfamily, the YB2 subfamily and the CRC subfamily, which both featured five to six introns.
To investigate the potential transcriptional regulatory role of AmYABBY, cis-elements were extracted from the PlantCARE database in this study using a 2000 bp upstream element of the AmYABBY gene as the promoter region. A total of 95 valuable cis-elements were identified in the promoter regions of 7 AmYABBY genes, and these cis-elements can be broadly categorized into 3 groups (). Elements related to plant growth and development included light response, meristematic tissue expression, endosperm expression, seed-specific regulatory response elements, and fenestrated chloroplast differentiation. Cis-elements associated with hormones include gibberellin and methyl jasmonate response elements. Those with involvement in the abiotic stress-related response include cis-elements that bind sites with MYB genes to participate in drought response. This suggests that AmMYB genes may regulate AmYABBY genes to form a regulatory network for different biological functions.
Synteny analysis of AmYABBY genes
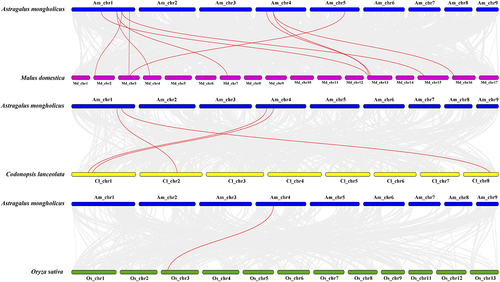
In order to further understand the evolutionary relationship of the YABBY gene family among different species, we constructed a covariance between A. mongholicus and two dicotyledons (M. domestica and C. lanceolata) and one monocotyledon (O.sativa). The results showed that AmYABBY and MdYABBY were the most closely co-related with ten gene pairs, AmYABBY and ClYABBY were the most closely co-related with four pairs, and AmYABBY and OsYABBY were poorly co-related with only one pair. These results indicate that most of these homologous YABBY gene pairs occurred after the differentiation of dicotyledonous and monocotyledonous plants ().
Figure 5. Genomic covariance of A. mongholicus with M. domestica and C. lanceolata in O. sativa. The red line indicates the co-linearity relationship of the YABBY gene from top to bottom. Blue, purple, yellow, and green colors represent A. mongholicus, M. domestica, and C. lanceolata in O. sativa, respectively.
Expression pattern of AmYABBY genes in different tissues of A. mongholicus qRT-PCR verification
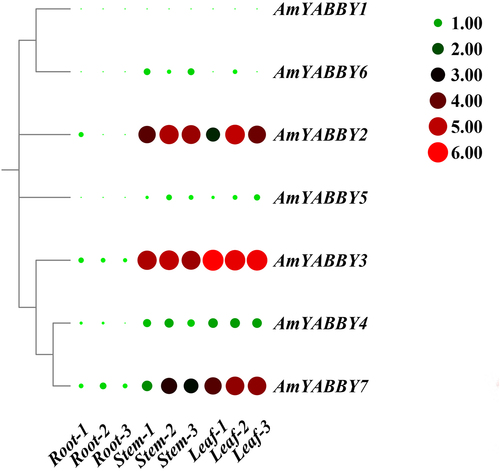
In order to investigate the expression pattern of AmYABBY genes in different tissues, the gene expression levels of seven AmYABBY genes were analyzed based on the stem transcriptome data of leaves and roots of A. mongholicus. The results showed that five genes were expressed in stems and leaves (RPKM >0.5), and all the genes were expressed at low levels in roots. Two genes were expressed at very high levels in leaves (AmYABBY2 and AmYABBY3), and four genes were expressed at very high levels in leaves (AmYABBY1, AmYABBY2, AmYABBY3, and AmYABBY7). AmYABBY1, AmYABBY3, and AmYABBY7 were expressed in all tissue parts, and these genes may be involved in the entire growth and development cycle of A. mongholicus. Two genes (AmYABBY5 and AmYABBY6) were not expressed in all tissue sites (RPKM <0.5) and may be pseudogenes or require specific conditions to activate expression (, Table S2).
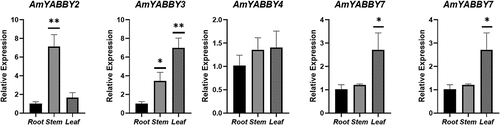
Based on the transcriptome data, these five AmYABBY genes were selected as candidate genes for qRT-PCR analysis in this study. The results showed that the expression patterns of the five candidate genes were basically consistent with the expression trends obtained from the RNA-seq data. Notably, AmYABBY1 and AmYABBY2 had higher expression in stems, and AmYABBY3 and AmYABBY7 had higher expression levels in leaves. All of the above indicated that these five AmYABBY genes might be closely related to the growth and development of A. mongholicus ().
Discussion
YABBY genes are a unique gene family in plants, which are more thoroughly studied in the field of plants.Citation29 The YABBY gene family is not only important for the regulation of plant growth and development but also involved in the regulation of plant secondary metabolism and abiotic stressesCitation30; for example, overexpression of AaYABBY5 in Artemisia annua significantly enhanced the content of artemisinin, and MsYABBY5 expression also affects the synthesis of monoterpenes and other terpene compounds in Mentha spicata.Citation31 The polar expression of the YABBY family is more conserved in dicotyledonous plants, but there is a greater divergence in monocotyledonous plants.Citation32 The YABBY gene family has been heavily studied in various plants; however, it has not yet been studied in A. mongholicus. Therefore, in order to obtain a comprehensive understanding of the basic characteristics of the A. mongholicus gene family, we analyzed seven YABBY gene family members from the A. mongholicus genome. In this study, based on genome-wide data, seven AmYABBY genes were identified, and their chromosomal distribution, phylogenetic relationships, motif prediction, gene structure, anterograde element prediction, and expression patterns were comprehensively analyzed. This study provides new insight into further study on the growth and development of the YABBY gene in A. mongholicus and its process of molecular mechanism regulation.
In this study, a total of seven AmYABBY genes were identified through bioinformatics methods, which is in line with the majority of the previous reports, such as six on A.thaliana, six on Platycodon grandiflorus, six on Vitis vinifera,Citation33 and six on Punica granatum.Citation34 Further analysis revealed that all AmYABBY genes contained zinc finger structural domains and YABBY structural domains, indicating that the identification results were accurate, reliable, and consistent with those of other plant species.Citation35 Physicochemical characterization showed that the average value of hydrophobicity of all AmYABBY proteins was greater than 0, suggesting that AmYABBY may be hydrophilic proteins which is the same as the identification results of Nelumbo nucifera.Citation36 Hydrophilic proteins can favor plant resistance to abiotic stresses, which may also be the reason why YABBY genes can respond to abiotic stresses. The secondary structures of proteins are mainly composed of four types of spatial structures: the α-helix, β-fold, irregular coil, and extended chain. The secondary structures of proteins of seven genes in the AmYABBY gene family are mainly formed by a random coil. The tertiary structures of proteins of AmYABBY4 and AmYABBY7 genes are similar.
To investigate the evolutionary relationship of the AmYABBY gene family, we constructed a phylogenetic tree using A. mongholicus and rice and identified five subfamilies. This is consistent with the results of the developmental tree constructed using Platycodon grandiflorus. YABBY1 and YABBY6 belong to the CRC subfamily, and various transcription factors in the CRC subfamily can regulate the growth and development process of plants, including the longitudinal and radial growth of the pistil, the influence on the meristem of flowers, the growth of fruit mediastinum, and the formation of carpel wall polarity.Citation37 YABBY3 belongs to the YB5 subfamily, indicating that it can regulate the development of lateral organ polarity, edge formation, leaf maturation, and the stem apical meristem and leaf sequence.Citation38 YABBY1 and YABBY6 are part of the CRC class, which plays a regulatory role in nectary development in the carpel and core dicots of angiosperms.Citation39 AmYABBY5 is part of INO, which may be involved in the development of the outer integument of ovules.Citation40 AmYABBY4 and AmYABBY7 are part of YB2, which may be involved in the direction of cell differentiation at the distal end of the outer organ. AmYABBY2 is part of YB3/FIA, so this gene may has the ability to regulate the formation of plant floral organs and maintain and manage the meristem of floral order. In addition, genes in the same subfamily share the same gene structure and conserved motifs; for example, AmYABBY4, AmYABBY7, AmYABBY1, and AmYABBY6 are evolutionarily close, indicating that they may have the same function. The consistent number of conserved motifs and gene structures suggests that the two genes may have been generated through gene replication during species evolution.
Cis-elements in the promoter region play a key role in regulating gene expression.Citation41 For example, 2000 bp upstream of the promoter, several responding elements of AmYABBY genes were identified. These can be divided into three categories: light response elements, hormone response elements, and abiotic stress response elements. The AmYABBY gene may be closely related to plant hormone signal transduction and plant stress response, especially hormone-induced elements such as gibberellin, abscisic acid, and methyl jasmonate. Expression analysis showed that AmYABBY3 (YB5 class) genes were highly expressed in leaves, proving that YB5 is crucial for leaf growth and development. The RT-PCR results indicate that AmYABBY is highly expressed in stems and leaves. Generally speaking, homologous genes have similar expression patterns, but the expression patterns of AmYABBY1 and AmYABBY6 are completely different. It is speculated that the two have undergone functional differentiation due to different expression patterns during evolution, resulting in dissimilar functions produced by homologous genes. Genes with high expression and specific expression may play a role in corresponding tissue parts, and some genes may also undergo functional redundancy.
Conclusions
Seven AmYABBY genes were successfully identified in this study, which were divided into five subfamilies with six or seven introns. In the upstream 2000 bp position of the promoter, we found multiple light-responsive and hormone-responsive elements, suggesting that the YABBY gene is useful for expression in various plants. The high expression of AmYABBY genes in leaves and stems proves that they play an integral role in the growth and development of A. mongholicus. Therefore, the results of this study help to further understand the role of AmYABBY genes, lay a solid foundation for the taxonomic and functional studies of YABBY in other plants, and provide an important data basis for future studies on the growth, developmental regulation, and quality improvement of A. mongholicus.
Author contributions
Conceptualization, methodology, investigation, and writing – original draft preparation: Jiamei Wang, Zhen Wang, and Panpan Wang; software: Jianhao Wu and Xiaozhuang Zhang; validation: Lingyang Kong; formal analysis: Lengleng Ma; resources: Weichao Ren; data curation: Weili Liu and Yanli Guo; project administration and funding acquisition: Wei Ma and Xiubo Liu. All authors have read and agreed to the published version of the manuscript.
Supplementary File S1.doc
Download MS Word (13.5 KB)Supplementary Table S1.doc
Download MS Word (18 KB)Supplementary Figure.doc
Download MS Word (134.4 KB)Supplementary Table S2.doc
Download MS Word (27.5 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The transcriptome data were deposited at the NCBI database under accession number PRJNA1064679.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15592324.2024.2355740
Additional information
Funding
References
- Wang P, Wang Z, Zhang Z, Cao H, Kong L, Ma W, Ren W. A review of the botany, phytochemistry, traditional uses, pharmacology, toxicology, and quality control of the Astragalus memeranaceus. Front Pharmacol. 2023;14:1242318. doi:10.3389/fphar.2023.1242318.
- Yuan YM, Gao JW, Shi Z, Huang P, Lu YS, Yao MC, Huang M. Herb-drug pharmacokinetic interaction between radix astragali and pioglitazone in rats. J Ethnopharmacol. 2012;144(2):300–9. doi:10.1016/j.jep.2012.09.012.
- Dong M, Li J, Yang D, Li M, Wei J. Biosynthesis and pharmacological activities of flavonoids, Triterpene Saponins and polysaccharides derived from Astragalus membranaceus. Molecules. 2023;28(13):5018. doi:10.3390/molecules28135018.
- Chen Z, Liu L, Gao C, Chen W, Vong CT, Yao P, Yang Y, Li X, Tang X, Wang S. et al. Astragali radix (Huangqi): A promising edible immunomodulatory herbal medicine. J Ethnopharmacol. 2020;258:112895. doi:10.1016/j.jep.2020.112895.
- Fu J, Wang Z, Huang L, Zheng S, Wang D, Chen S, Zhang H, Yang S. Review of the botanical characteristics, phytochemistry, and pharmacology of Astragalus membranaceus (Huangqi). Phytother Res. 2014;28:1275–1283. doi:10.1002/ptr.5188.
- Zhang CH, Yang X, Wei JR, Chen NM, Xu JP, Bi YQ, Yang M, Gong X, Li Z-Y, Ren K. et al. Ethnopharmacology, phytochemistry, pharmacology, toxicology and clinical applications of radix astragali. Chin J Integr Med. 2021;27(3):229–240. doi:10.1007/s11655-019-3032-8.
- Guo Z, Lou Y, Kong M, Luo Q, Liu Z, Wu J. A systematic review of phytochemistry, pharmacology and pharmacokinetics on astragali radix: implications for astragali radix as a personalized medicine. Int J Mol Sci. 2019;20(6):1463. doi:10.3390/ijms20061463.
- Ma XQ, Shi Q, Duan JA, Dong TT, Tsim KW. Chemical analysis of Radix Astragali (Huangqi) in China: a comparison with its adulterants and seasonal variations. J Agric Food Chem. 2002;50(17):4861–4866. doi:10.1021/jf0202279.
- Bowman JL, Smyth DR. CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development. 1999;126(11):2387–2396. doi:10.1242/dev.126.11.2387.
- Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews GN, Bowman JL. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development. 1999;126(18):4117–4128. doi:10.1242/dev.126.18.4117.
- Li Z, Li G, Cai M, Priyadarshani S, Aslam M, Zhou Q, Huang X, Wang X, Liu Y, Qin Y. Genome-wide analysis of the YABBY transcription factor family in pineapple and functional identification of AcYABBY4 involvement in salt stress. Int J Mol Sci. 2019;20(23):5863. doi:10.3390/ijms20235863.
- Huang J, Chen GZ, Ahmad S, Wang Q, Tu S, Shi XL, Hao Y, Zhou YZ, Lan SR, Liu ZJ. et al. Identification, molecular characteristics, and evolution of YABBY gene family in Melastoma dodecandrum. IJMS. 2023;24(4):4174. doi:10.3390/ijms24044174.
- Yang T, He Y, Niu S, Zhang Y. A YABBY gene CRABS CLAW a (CRCa) negatively regulates flower and fruit sizes in tomato. Plant Sci. 2022;320:111285. doi:10.1016/j.plantsci.2022.111285.
- Sun L, Wei YQ, Wu KH, Yan JY, Xu JN, Wu YR, Li GX, Xu JM, Harberd NP, Ding ZJ, et al. Restriction of iron loading into developing seeds by a YABBY transcription factor safeguards successful reproduction in Arabidopsis. Mol Plant. 2021 Oct 4;14(10):1624–1639. doi:10.1016/j.molp.2021.06.005.
- Hou H, Lin Y, Hou X. Ectopic expression of a Pak-choi YABBY Gene, BcYAB3, causes leaf curvature and flowering stage delay in Arabidopsis thaliana. Genes. 2020;11(4):370. doi:10.3390/genes11040370.
- Lee JY, Baum SF, Oh SH, Jiang CZ, Chen JC, Bowman JL. Recruitment of CRABS CLAW to promote nectary development within the eudicot clade. Development. 2005;132(22):5021–5032. doi:10.1242/dev.02067.
- Eckardt NA. YABBY genes and the development and origin of seed plant leaves. Plant Cell. 2010;22(7):2103. doi:10.1105/tpc.110.220710.
- Sarojam R, Sappl PG, Goldshmidt A, Efroni I, Floyd SK, Eshed Y, Bowman JL. Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell. 2010;22(7):2113–2130. doi:10.1105/tpc.110.075853.
- Villanueva JM, Broadhvest J, Hauser BA, Meister RJ, Schneitz K, Gasser CS. INNER NO OUTER regulates abaxial- adaxial patterning in Arabidopsis ovules. Genes Dev. 1999;13(23):3160–3169. doi:10.1101/gad.13.23.3160.
- Yang H, Shi G, Li X, Hu D, Cui Y, Hou J, Yu D, Huang F. Overexpression of a soybean YABBY gene, GmFILa, causes leaf curling in Arabidopsis thaliana. BMC Plant Biol. 2019;19(1):234. doi:10.1186/s12870-019-1810-2.
- Chen Y, Fang T, Su H, Duan S, Ma R, Wang P, Wu L, Sun W, Hu Q, Zhao M. et al. A reference-grade genome assembly for Astragalus mongholicus and insights into the biosynthesis and high accumulation of triterpenoids and flavonoids in its roots. Plant Commun. 2023;4(2):100469. doi:10.1016/j.xplc.2022.100469.
- Liao B, Hu H, Xiao S, Zhou G, Sun W, Chu Y, Meng X, Wei J, Zhang H, Xu J. et al. Global pharmacopoeia genome database is an integrated and mineable genomic database for traditional medicines derived from eight international pharmacopoeias. Sci China Life Sci. 2022;65(4):809–817. doi:10.1007/s11427-021-1968-7.
- Chen C, Wu Y, Li J, Wang X, Zeng Z, Xu J, Liu Y, Feng J, Chen H, He Y. et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol Plant. 2023;16(11):1733–1742. doi:10.1016/j.molp.2023.09.010.
- Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H. et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49. doi:10.1093/nar/gkr1293.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura KMX, Battistuzzi FU. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi:10.1093/molbev/msy096.
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi:10.1093/nar/gkp335.
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–327. doi:10.1093/nar/30.1.325.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262.
- Romanova MA, Maksimova AI, Pawlowski K, Voitsekhovskaja OV. YABBY genes in the development and evolution of land plants. Int J Mol Sci. 2021;22(8):4139. doi:10.3390/ijms22084139.
- Zhang T, Li C, Li D, Liu Y, Yang X. Roles of YABBY transcription factors in the modulation of morphogenesis, development, and phytohormone and stress responses in plants. J Plant Res. 2020;133(6):751–763. doi:10.1007/s10265-020-01227-7.
- Kayani SI, Shen Q, Ma Y, Fu X, Xie L, Zhong Y, Tiantian C, Pan Q, Li L, Rahman SU. et al. The YABBY family transcription factor AaYABBY5 directly targets cytochrome P450 monooxygenase (CYP71AV1) and Double-Bond Reductase 2 (DBR2) involved in Artemisinin Biosynthesis in Artemisia Annua. Front Plant Sci. 2019;10:1084. doi:10.3389/fpls.2019.01084.
- Kong L, Sun J, Jiang Z, Ren W, Wang Z, Zhang M, Liu X, Wang L, Ma W, Xu J. Identification and expression analysis of YABBY family genes in Platycodon grandiflorus. Plant Signal Behav. 2023 Dec 31;18(1):2163069. doi:10.1080/15592324.2022.2163069. PMID: 36681901; PMCID: PMC9870009.
- Zhang S, Wang L, Sun X, Li Y, Yao J, van Nocker S, Wang X. Genome-wide analysis of the YABBY gene family in grapevine and functional characterization of VvYABBY4. Front Plant Sci. 2019;10:1207. doi:10.3389/fpls.2019.01207.
- Zhao Y, Liu C, Ge D, Yan M, Ren Y, Huang X, Yuan Z. Genome-wide identification and expression of YABBY genes family during flower development in Punica granatum L. Gene. 2020;752:144784. doi:10.1016/j.gene.2020.144784.
- Zeng D, Si C, Teixeira da Silva JA, Dai G, Duan J, He C. Characterization of YABBY genes in Dendrobium officinale reveals their potential roles in flower development. Protoplasma. 2023;260(2):483–495. doi:10.1007/s00709-022-01790-x.
- Zhao S, Zhang Y, Tan M, Jiao J, Zhang C, Wu P, Feng K, Li L. Identification of YABBY transcription factors and their function in ABA and salinity response in Nelumbo nucifera. Plants (Basel). 2023;12(2):380. doi:10.3390/plants12020380.
- Gross T, Broholm S, Becker A. CRABS CLAW acts as a bifunctional transcription factor in flower development. Front Plant Sci. 2018 June 20;9:835. doi:10.3389/fpls.2018.00835.
- Orashakova S, Lange M, Lange S, Wege S, Becker A. The CRABS CLAW ortholog from California poppy (Eschscholzia californica, Papaveraceae), EcCRC, is involved in floral meristem termination, gynoecium differentiation and ovule initiation. Plant Journal. 2009;58(4):682–693. doi:10.1111/j.1365-313X.2009.03807.x.
- Yamaguchi T, Nagasawa N, Kawasaki S, Matsuoka M, Nagato Y, Hirano HY. The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa [W]. Plant Cell. 2004 Feb;16(2):500–509. doi:10.1105/tpc.018044. Epub 2004 Jan 16. PMID: 14729915; PMCID: PMC341919.
- Simon MK, Skinner DJ, Gallagher TL, Gasser CS. Integument development in Arabidopsis depends on interaction of YABBY protein INNER NO OUTER with coactivators and corepressors. Genetics. 2017;207(4):1489–1500. doi:10.1534/genetics.117.300140.
- Zheng Q, Zhao X, Huang Y, Zhang MM, He X, Ke S, Li Y, Zhang C, Ahmad S, Lan S, et al. Genome-wide identification of the YABBY gene family in Dendrobium Orchids and its expression patterns in Dendrobium chrysotoxum. Int J Mol Sci. 2023 June;24(12):10165. doi:10.3390/ijms241210165.