ABSTRACT
Background
A previous study reported circ_0001821 serves as a new diagnostic biomarker in lung and gastric cancer. However, whether circ_0001821 associates with the development of multiple myeloma is still unclear.
Objective
To investigate the clinical significance and biological function of circ_0001821 in human multiple myeloma.
Methods
The expression of circ_0001821 in bone marrow tissues and cell lines of multiple myeloma was detected by quantitative real-time PCR. The relationship between circ_0001821 expression and clinicopathological characteristics in multiple myeloma patients was determined by the Chi-square test. MTT, Western blotting analysis and flow cytometry were performed to evaluate the impact of circ_0001821 on cell proliferation and apoptosis.
Results
Circ_0001821 expression was increased in bone marrow tissues and cell lines from multiple myeloma compared with that in normal controls, and its expression was related to hemoglobin, bone injury, β2-MG, and globulin. In multiple myeloma patients aged ≥60 years, high circ_0001821 expression displayed lower overall survival (OS) rate compared with low circ_0001821 expression. High circ_0001821 expression was an independent prognostic indicator for the poor OS. Furthermore, the expression of Caspase-3 protein was lower in patients with high circ_0001821 expression than that in those patients with low circ_0001821expression. Overexpression of circ_0001821 led to increased proliferation and arrested apoptosis of multiple myeloma cells, while knockdown of circ_0001821 had the opposite effects.
Conclusion
High expression of circ_0001821 associated with aggressive pathological indicators and predicted a poor prognosis in multiple myeloma. Circ_0001821 played an oncogenic role in multiple myeloma by modulating cell proliferation and apoptosis.
Introduction
Multiple myeloma is one of the common malignant tumors of the blood system, which is originated from human plasma cells in bone marrow tissues and leads to the overproduction of immunoglobulins [Citation1,Citation2]. The incidence rate of multiple myeloma is increasing with the increasing elderly population in China. With the progress of therapeutic strategies over the past few decades, the median survival rate of patients with multiple myeloma has improved significantly; however, the overall survival rate remains unsatisfactory with a five-year survival rate of less than 50% [Citation3–5]. It is well known that the initiation and progression of multiple myeloma are related to genetic and epigenetic regulation as well as the activation of oncogenic genes and/or inactivation of tumor suppressor genes. Although various biomarkers have been considered to be related to multiple myeloma diagnoses in clinical practice, including β2-MG, light chain λ/κ, and β-CTX, their reliability remains controversial [Citation6,Citation7]. Therefore, it is critical to identify novel mechanisms involved in the aggressive progression of multiple myeloma and to enhance the diagnosis and treatment strategies for multiple myeloma patients.
Circular RNAs (circRNAs) are a novel class of long non-coding RNAs that form a covalently closed continuous loop, which makes them much more stable than linear RNA and insusceptible to degradation by Ribonuclease R [Citation8,Citation9]. CircRNAs have been found to be involved in human diseases, and to play key roles in many types of physiological and pathological conditions [Citation10]. In recent years, circRNAs have been identified as oncogenic genes and/or tumor suppressor genes to regulate cell biological behaviors of cancer at the posttranscriptional level [Citation11]. For example, in hepatocellular carcinoma, circRNA circ_5692 suppresses tumor progression by targeting miR-328-5p and upregulation of DAB2IP expression [Citation12]. In breast cancer, circRNA circ_0025202 regulates cell sensitivity to tamoxifen through the miR-182-5p/FOXO3a signaling pathway [Citation13]. In bladder cancer, circRNA cric_SLC8A1 functions as a tumor suppressor by modulating cell migration, invasion, and growth [Citation14]. Nowadays, although increasing data have demonstrated that circRNAs are dysregulated in the development of multiple myeloma and closely associated with clinical prognosis and diagnosis, most circRNAs still remain functionally unspecifically characterized.
A recent research study by Wang et al. reported a circRNA, circ_0001821 and served as a potential biomarker for both lung adenocarcinoma and squamous cell carcinoma [Citation15]. In addition, by comprehensive circular RNA profiling, Kong et al. firstly demonstrated that circ_0001821 serves as a new diagnostic biomarker in gastric cancer [Citation16]. Whether circ_0001821 contributes to the development of multiple myeloma is still unclear. In this study, we investigated the expression profile of circ_0001821 in bone marrow tissues and cell lines of multiple myeloma by quantitative real-time PCR, and identified that upregulation of circ_0001821 expression was an independent prognostic factor for multiple myeloma. We also determined the biological role of circ_0001821 in multiple myeloma cells, and found that circ_0001821 overexpression promoted cell growth, but arrested cell apoptosis, whereas circ_0001821 knockdown had the opposite effects. Our results suggested that circ_0001821 exerted an oncogenic role in the development of multiple myeloma and might be a promising molecular target for multiple myeloma treatment.
Material and methods
Clinical specimen collection
Bone marrow samples were obtained from 115 multiple myeloma patients who underwent surgical punctures between January 2015 and January 2017 in the First Affiliated Hospital of Bengbu Medical College. Patients with multiple myeloma were confirmed by three experienced pathologists. In total, 115 healthy volunteers with a mean of 48.27 years (59 male and 56 female) were treated as normal controls. All tissues were stored immediately at −80°C for further experiments. The 115 patients with multiple myeloma included 66 males and 49 females, and their mean age was 46.33 ± 7.16 years. All patients had not received any clinical therapy including radiation therapy, chemotherapy, and/or other treatments before admission. The clinicopathological features of the 115 patients are shown in . The study was authorized by the First Affiliated Hospital of Bengbu Medical College (approval number: 20170521), and all participants had provided written informed consents.
Table 1. The association of circ_0001821 expression with the clinicopathological characteristics in 115 multiple myeloma patients.
Cell culture
The human multiple myeloma cell lines, including RPMI-8226, NCI-H929, JJN-3, and U266, and a normal peripheral blood mononuclear cell (PBMC), were purchased from the Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). They were incubated with RPMI-1640 medium (Gibco, CA, USA) added with 10% heat-inactivated fetal bovine serum (FBS; Gibco, CA, USA) at 37°C in a humidified incubator containing 5% CO2.
Quantitative real-time PCR
The total RNA from the specimens and cell lines was extracted with the TRIzol reagent (Thermo Fisher Scientific, MA, USA). The eligible RNA was reversely transcribed to synthesize complementary DNA (cDNA) using the PrimeScript RT reagent kit (Takara, Beijing, China). Quantitative real-time PCR was conducted with TB Green® Premix Ex Taq™ II (Tli RNaseH Plus) (Takara) on the ABI PRISM 7000 Fluorescent Quantitative PCR System (Applied Biosystems, CA, USA), according to the manufacturer’s instructions. The amplification protocol was as follows: 95°C for 10 min, 36 cycles of 95°C for 10 s, and 60°C for 45 s. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was regarded as the internal control. The primers for quantitative real-time PCR were ordered from Genecopoeia (Guangzhou, China): circ_0001821 (forward, 5′-TGGAATGTAAGACCCCGACT-3′ reverse, 5′-CCATCTTGAGGGGCATCTTT-3′), GAPDH (forward, 5′-AGAAGGCTGGGGCTCATTTG-3′ reverse, 5′-AGGGGCCATCCACAGTCTTC-3′). The expression levels of circ_0001821 were normalized internally by using the 2−ΔΔCt method of the housekeeping gene GAPDH [Citation17].
Cell transfection
The pcDNA4.0 and pcDNA4.0 expressing circ_0001821 (pcDNA4.0 + circ_0001821) were constructed by Sangon Biotech (Shanghai, China). Small interfering RNA (siRNA) against circ_0001821 (si-circ_0001821) and siRNA negative control (si-NC) were constructed by Genepharma (Shanghai, China). pcDNA4.0 and pcDNA4.0 + circ_0001821 at a final concentration of 2 μg/well were transiently transfected into RPMI-8226 cells with Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific), according to the manufacturer’s instructions. The si-circ_0001821 and si-NC were transiently transfected into the NCI-H929 cell at a final concentration of 50 nM/well. RPMI-8226 and NCI-H929 cells were collected 48 h after transfection for the subsequent experiments. The efficiency of circ_0001821 overexpression and knockdown was evaluated by using quantitative real-time PCR.
3-(4,5-dime-thyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium Bromide (MTT) assay
RPMI-8226 and NCI-H929 cells were seeded into 96-well plates at a density of 7000 cells/well in triplicates and transfected with pcDNA4.0 or pcDNA4.0 + circ_0001821 and si-circ_0001821 or si-NC, respectively. The transfected cells were cultivated at 37°C in a humidified incubator containing 5% CO2, followed by the addition of 15 μl MTT reagent (5 mg/ml; Sigma-Aldrich, CA, USA) at 0, 24, 48, 72, and 96 h after the beginning of cell culture. Cells were then incubated for an additional 4 h and 180 μl dimethylsulfoxide (DMSO; Sigma-Aldrich) was used to dissolve formazan following the removal of the supernatant. The absorbance of each well at 450 nm was observed and recorded by using a microplate reader (Bio-Rad, CA, USA).
Flow cytometry
RPMI-8226 and NCI-H929 cells were seeded into 6-well plates in triplicate at a density of 3.0 × 106 cells/well, and transiently transfected with pcDNA4.0 or pcDNA4.0 + circ_0001821 and si-circ_0001821 or si-NC, respectively. 24 h after cell transfection, cell apoptosis was detected by performing AnnexinV-FITC/PI double staining flow cytometry analysis, according to the manufacturer’s instructions. Briefly, the transfected cells were washed twice with warm PBS, and resuspended with a binding buffer at a final concentration of 1 × 106 cells/ml. Then, 5 μl of AnnexinV-FITC (BD Biosciences, MA, USA) was added to the cells followed by incubation for 30 min at 37°C in dark. Before loading the samples, 2 μl of propidiumiodide (PI; BD Biosciences) was added to each tube. Cell apoptosis rate was determined with a FACSCalibur™ Flow Cytometer (BD Biosciences). The FlowJo software (FlowJo, Ashland, USA) was applied to analyze the flow cytometry data.
Western blotting analysis
The protein from the specimens and cell lines was extracted with the RIPA lysis buffer (Thermo Fisher Scientific) containing 1% protease inhibitor. An equal amount of proteins (35 μg) was then separated on 10% sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred into polyvinylidene difluoride membranes (PVDF; Millipore, MA, USA), followed by blocking with 5% nonfat milk for 1 h at 37°C. Subsequently, the membranes were incubated with the primary antibodies (1:1000 dilution; Abcam, MA, USA) against Caspase-3 (#ab13847) overnight at 4°C. GAPDH (1:2000 dilution; #ab181602) was used as loading control. After being washed with TBST buffer three times, the membranes were incubated with horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibodies (1:5000 dilution; #ab6721) for an additional 2 h at 37°C. The protein signals were detected by the Beyoecl Plus Enhanced Chemiluminescence Kit (Beyotime Biotechnology, Shanghai, China) and analyzed with the ImageJ 142 software (NIH, MD, USA).
Statistical analysis
Statistical analysis was done by using SPSS 25.0 (SPSS, Chicago, USA). All experiments were repeated three times, and data were recorded as the mean ± standard deviation (SD). Graphs were mainly derived from GraphPad Prism 5 (GraphPad, San Diego, USA). The difference with P < 0.05 was regarded as statistically significant. Comparison between two groups was evaluated by Student’s t-test, and comparison among three or more groups was performed with one-way ANOVA followed by the Tukey–Kramer post hoc analysis. The relationship between clinical pathologic feature and circ_0001821 was analyzed with the Chi-square test. Cox regression and Kaplan-Meier method were conducted to analyze the prognostic value of circ_0001821 in multiple myeloma patients.
Results
The expression of circ_0001821 in bone marrow tissues and cell lines of multiple myeloma
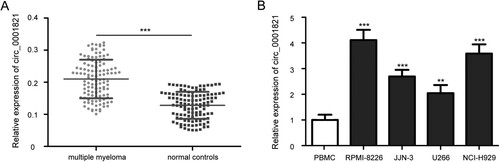
To detect the expression of circ_0001821 in multiple myeloma, we applied quantitative real-time PCR to examine its levels in 115 cases of bone marrow tissues from patients with multiple myeloma and normal controls. Compared with the normal controls, the expression levels of circ_0001821 in multiple myeloma patients were upregulated ((A), P < 0.001). We then analyzed circ_0001821 expression in a normal PBMC cell line, and four multiple myeloma cell lines, including JJN-3, U266, RPMI-8226, and NCI-H929, and found that circ_0001821 was highly expressed in these multiple myeloma cell lines ((B), all P < 0.01). Among these cell lines, RPMI-8226 and NCI-H929 cells had higher circ_0001821 expression than JJN-3 and U266 cells and were chosen for further experiments.
Figure 1. The expression of circ_0001821 in tissues and cell lines of multiple myeloma. (A) The expression levels of circ_0001821 were measured in bone marrow tissues from 115 multiple myeloma patients and normal controls by using quantitative real-time PCR. (B) Comparison of circ_0001821 expression in a normal peripheral blood mononuclear cell (PBMC) and four multiple myeloma cell lines (RPMI-8226, NCI-H929, JJN-3 and U266), and circ_0001821 was upregulated in the four multiple myeloma cell lines. **P < 0.01, ***P < 0.001.
The relationship between circ_0001821 expression and clinicopathological characteristics in patients with multiple myeloma
According to the circ_0001821 expression levels, we obtained the median value of circ_0001821 in bone marrow tissues of multiple myeloma as a cutoff value and divided the patients into two groups: the high circ_0001821 expression (n = 58) and the low circ_0001821 expression (n = 57). We further analyzed the relationship between circ_0001821 expression and the clinicopathological characteristics and indicated that expression of circ_0001821 was correlated with hemoglobin (P < 0.001), bone injury (P = 0.017), β2-MG (P = 0.020), and globulin (P = 0.009) (), but had no association with other factors, including age, sex, M protein, and light chain.
The prognostic value of circ_0001821 in patients with multiple myeloma
The Kaplan-Meier analysis revealed that the multiple myeloma patients with high circ_0001821 expression displayed a lower overall survival (OS) rate compared with those patients with low circ_0001821 expression ((A), P = 0.031). Furthermore, due to different treatments for patients with multiple myeloma aged <60 years and ≥60 years, we also analyzed the prognosis of patients in the two groups based on circ_0001821 expression. Interestingly, a similar result was found in multiple myeloma patients aged ≥60 years ((B), P = 0.024). However, in patients with multiple myeloma aged <60 years, there was no significant difference in the OS rate between the high and low circ_0001821 expression groups ((C), P = 0.452). In addition, multivariate Cox regression analyses suggested that high circ_0001821 expression was an independent prognosticator of poor OS in patients with multiple myeloma (, P = 0.046).
Figure 2. Upregulated circ_0001821 was associated with poor patient outcome. (A) The OS rate of multiple myeloma patients with low and high circ_0001821 expression was analyzed by the using Kaplan-Meier survival method, and the survival curves were evaluated with the log-rank test. (B) In patients with multiple myeloma aged ≥60 years, high circ_0001821 expression group displayed a shorter OS rate compared with the low circ_0001821 expression group. (C) In patients with multiple myeloma aged <60 years, there was no significant difference in the OS rate between the high and low circ_0001821 expression groups.
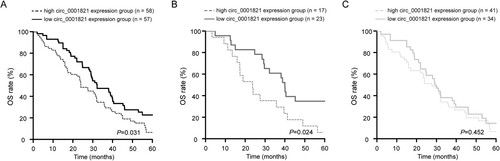
Table 2. Univariate and multivariate Cox regression analyses of clinicopathological characteristics as overall survival predictor in multiple myeloma patients.
Circ_0001821 overexpression and silencing in multiple myeloma cell lines
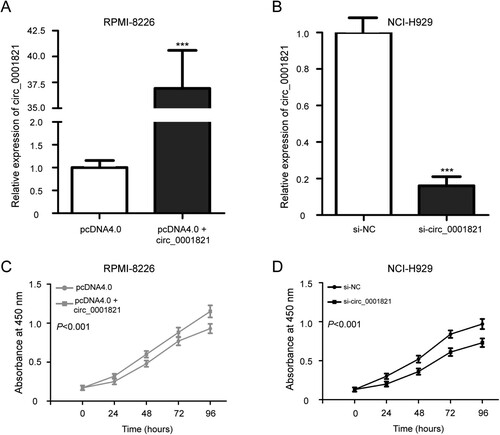
To investigate the potential role of circ_0001821 in the development of multiple myeloma, RPMI-8226 and NCI-H929 cells were chosen and cultured in 6-well plates, and then transfected with pcDNA4.0 or pcDNA4.0 + circ_0001821 and si-circ_0001821 or si-NC, respectively. The analysis of transfection efficacy demonstrated that the circ_0001821 expression was effectively increased in the RPMI-8226 cells after the transfection of pcDNA4.0 + circ_0001821 compared with the pcDNA4.0 group ((A), P < 0.001). Compared with the si-NC group, si-circ_0001821 led to downregulated expression of circ_0001821 in NCI-H929 cells ((B), P < 0.001).
Figure 3. Circ_0001821 stimulated the cell proliferation of multiple myeloma cells. (A) RPMI-8226 cells were treated with pcDNA4.0 and pcDNA4.0 + circ_0001821 for 48 h, and quantitative real-time PCR was applied to determine circ_0001821 expression. (B) The expression level of circ_0001821 was decreased in NCI-H929 cells after treatment with si-circ_0001821 compared with cells transfected with si-NC. (C) MTT assay showed that pcDNA4.0 + circ_0001821 promoted the cell proliferation in RPMI-8226 cells. (D) Knockdown of circ_0001821 reduced cell proliferation in NCI-H929 cells compared to the si-NC group. ***P < 0.001.
The impact of circ_0001821 expression on cell proliferation of multiple myeloma
We further determined the biological function of circ_0001821 on cell proliferation of multiple myeloma cell lines. The MTT assay indicated that RPMI-8226 cells incubated with the pcDNA4.0 + circ_0001821 resulted in a significant promotion in cell proliferation compared to the growth in the pcDNA4.0 group ((C), P < 0.001), while the NCI-H929 cells incubated with the si-circ_0001821 exhibited reduced cell proliferation compared to the si-NC group ((D), P < 0.001).
The effect of circ_0001821 expression on cell apoptosis of multiple myeloma
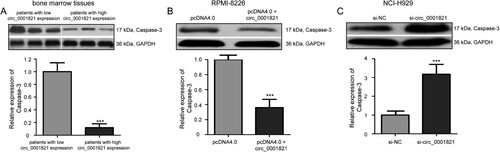
To investigate the effect of circ_0001821 on multiple myeloma cell apoptosis, we performed the AnnexinV-FITC/PI double staining flow cytometry analysis and found that circ_0001821 overexpression inhibited cell apoptosis in RPMI-8226 cells compared with the pcDNA4.0 group ((A), P < 0.001). (B) shows that circ_0001821 knockdown had the opposite effect on cell apoptosis in NCI-H929 cells (P < 0.001). Since Caspase-3 plays a key role in cell apoptosis, the expression of Caspase-3 protein in bone marrow tissues of multiple myeloma was detected by the Western blotting analysis. The result showed that the expression of Caspase-3 protein was lower in patients with high circ_0001821 expression than that in those patients with low circ_0001821 expression ((A), P < 0.001). Furthermore, we explored the effect of circ_0001821 on protein expression levels of Caspase-3. We found that the expression level of Caspase-3 protein was downregulated in the pcDNA4.0 + circ_0001821-treated RPMI-8226 cells compared with the pcDNA4.0-transfected cells ((B), P < 0.001), while Caspase-3 protein level was increased in NCI-H929 cells after treatment with si-circ_0001821 compared with cells treated with si-NC ((C), P < 0.001).
Figure 4. Circ_0001821 suppressed the cell apoptosis of multiple myeloma cells. (A) Cell apoptotic rate was detected with flow cytometry analysis in circ_0001821-overexpressed RPMI-8226 cells. Overexpression of circ_0001821 led to inhibit RPMI-8226 cell apoptosis. (B) NCI-H929 cells incubated with the si-circ_0001821 exhibited increased cell apoptosis compared to the si-NC group. ***P < 0.001.
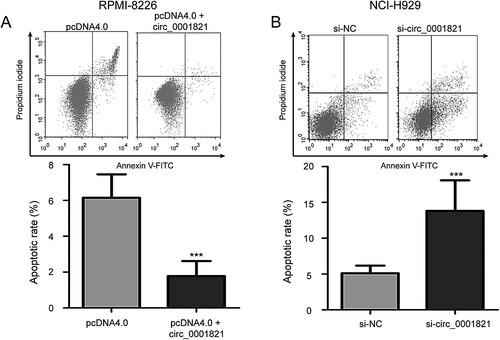
Figure 5. Circ_0001821 reduced Caspase-3 protein expression in multiple myeloma cells. (A) The Western blotting analysis of the expression level of Caspase-3 protein in bone marrow tissues with high and low circ_0001821 expression. (B) pcDNA4.0 + circ_0001821 could decrease Caspase-3 protein in RPMI-8226 cells compared with the si-NC group. (C) Caspase-3 protein expression was increased in NCI-H929 cells after treatment with si-circ_0001821 compared with cells treated with si-NC. ***P < 0.001.
Discussion
Although circRNAs have no protein-coding capacity at all, they have recently been reported to play a pivotal role in various physiological and pathological activities, including cell differentiation, viability, migration, apoptosis, metastasis, and carcinogenesis [Citation18]. Some circRNAs have been reported as new diagnostic biomarkers, effective prognosticators, and promising therapeutic targets in human cancers including multiple myeloma [Citation19,Citation20]. For example, Yu et al found that circRNA circ_MYBL2 is a novel tumor suppressor and potential biomarker in multiple myeloma [Citation21]. Song et al. indicated that circRNA circ_0007841 is upregulated in multiple myeloma, and may act as a novel potential biomarker and drug resistance for multiple myeloma [Citation22]. Gao et al showed that circRNA circ_0007841 acts as an oncogene to facilitate the proliferation, cell cycle, and motility and suppresses the apoptosis of multiple myeloma cells by sponging of miR-338-3p [Citation23]. However, the clinical application of circRNAs is limited by their unknown function.
We demonstrated for the first time that circ_0001821 was increased in bone marrow tissues and cell lines of multiple myeloma. Moreover, the circ_0001821 expression was found to be related to hemoglobin, bone injury, β2-MG, and globulin in multiple myeloma patients. These data prompted that circ_0001821 is associated with aggressive pathological indicators and might have a pivotal role in the development of multiple myeloma.
As a new identified circRNA, circ_0001821 is located in human genome location (chr8_128902834_128903244_+, hg19/Human) with 410 bp length of its mature transcript [Citation24]. Interestingly, it has been reported that circ_0001821 expression is associated with aggressive phenotypes in lung adenocarcinoma and squamous cell carcinoma [Citation15]. A subsequent study by Kong et al. found that circ_0001821 is a new and promising potential biomarker for the diagnosis of gastric cancer [Citation16]. In our study, we also found that high circ_0001821 expression predicted a poor prognosis in multiple myeloma patients aged≥60 years. However, there was no significant difference in the OS rate between the high and low circ_0001821 expression groups in patients with multiple myeloma aged <60 years. The reason for this phenomenon might be that circ_0001821 expression is related to the treatment of multiple myeloma. In addition, the multivariate Cox regression analysis indicated circ_0001821 as an independent prognosticator for poor OS in patients with multiple myeloma. These findings prompted that circ_0001821 might be a new prognostic marker for multiple myeloma.
As the Western blotting analysis indicated that the expression of Caspase-3 protein was lower in multiple myeloma patients with high expression of circ_0001821 than that in those patients with low expression of circ_0001821, which prompted us to speculate whether circ_0001821 expression is related to cell apoptosis in multiple myeloma. Functionally, we then assessed the role of circ_0001821 on the proliferation and apoptosis in multiple myeloma cells. Consistent with previous report [Citation15], we showed that circ_0001821 overexpression enhanced cell proliferation, but inhibited cell apoptosis in multiple myeloma cells, whereas its knockdown had opposite effects. These results suggested that upregulated circ_0001821 acts as an oncogenic factor in multiple myeloma by modulating cell proliferation and apoptosis.
However, the study has some limitations: (1) bone marrow tissues of multiple myeloma are relatively small, (2) primary CD138+ cells are not isolated from bone marrow tissues, (3) in vitro cell functional experiments of circ_0001821 overexpression and knockdown may not sufficiently reflect the effect of circ_0001821 in patients with multiple myeloma, (4) the molecular mechanism of circ_0001821 in the development of multiple myeloma is unclear. Thus, we will continue to explore the function and mechanism of circ_0001821 in multiple myeloma from the above aspects in the future study.
High expression of circ_0001821 was found in multiple myeloma and predicted a poor prognosis. Increased circ_0001821 plays an oncogenic role in multiple myeloma by regulating cell proliferation and apoptosis. This study indicated that circ_0001821 might be a new biomarker for the prognosis of multiple myeloma and represent a promising therapeutic strategy for multiple myeloma treatment.
Acknowledgements
The authors thank Hui Zeng from Department of Hematology, the First Affiliated Hospital of Jinan University for experimental assistance.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:https://doi.org/10.3322/caac.21590.
- Rollig C, Knop S, Bornhauser M. Multiple myeloma. Lancet. 2015;385(9983):2197–2208. doi:https://doi.org/10.1016/S0140-6736(14)60493-1.
- Joshua DE, Bryant C, Dix C, et al. Biology and therapy of multiple myeloma. Med J Aust. 2019;210(8):375–380. doi:https://doi.org/10.5694/mja2.50129.
- Hideshima T, Mitsiades C, Tonon G, et al. Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat Rev Cancer. 2007;7(8):585–598. doi:https://doi.org/10.1038/nrc2189.
- Landgren O, Rajkumar SV. New developments in diagnosis, prognosis, and assessment of response in multiple myeloma. Clin Cancer Res. 2016;22(22):5428–5433. doi:https://doi.org/10.1158/1078-0432.CCR-16-0866.
- Levin A, Hari P, Dhakal B. Novel biomarkers in multiple myeloma. Transl Res. 2018;201:49–59. doi:https://doi.org/10.1016/j.trsl.2018.05.003.
- Gupta N, Sharma A. Emerging biomarkers in multiple myeloma: a review. Clin Chim Acta. 2020;503:45–53. doi:https://doi.org/10.1016/j.cca.2019.12.026.
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi:https://doi.org/10.1038/s41576-019-0158-7.
- Wu Z, Sun H, Li J, et al. Circular RNAs in leukemia. Aging (Albany NY). 2019;11(13):4757–4771. doi:https://doi.org/10.18632/aging.102091.
- Chen X, Yang T, Wang W, et al. Circular RNAs in immune responses and immune diseases. Theranostics. 2019;9(2):588–607. doi:https://doi.org/10.7150/thno.29678.
- Kristensen LS, Hansen TB, Veno MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. doi:https://doi.org/10.1038/onc.2017.361.
- Liu Z, Yu Y, Huang Z, et al. CircRNA-5692 inhibits the progression of hepatocellular carcinoma by sponging miR-328-5p to enhance DAB2IP expression. Cell Death Dis. 2019;10(12):900. doi:https://doi.org/10.1038/s41419-019-2089-9.
- Sang Y, Chen B, Song X, et al. CircRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR-182-5p/FOXO3a axis in breast cancer. Mol Ther. 2019;27(9):1638–1652. doi:https://doi.org/10.1016/j.ymthe.2019.05.011.
- Lu Q, Liu T, Feng H, et al. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol Cancer. 2019;18(1):111. doi:https://doi.org/10.1186/s12943-019-1040-0.
- Wang C, Tan S, Liu WR, et al. RNA-seq profiling of circular RNA in human lung adenocarcinoma and squamous cell carcinoma. Mol Cancer. 2019;18(1):134. doi:https://doi.org/10.1186/s12943-019-1061-8.
- Kong S, Yang Q, Tang C, et al. Identification of hsa_circ_0001821 as a novel diagnostic biomarker in gastric cancer via comprehensive circular RNA profiling. Front Genet. 2019;10:878. doi:https://doi.org/10.3389/fgene.2019.00878.
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: https://doi.org/10.1038/nprot.2008.73.
- Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. doi:https://doi.org/10.1186/s12943-017-0663-2.
- de Acha OP, Rossi M, Gorospe M. Circular RNAs in blood malignancies. Front Mol Biosci. 2020;7:109. doi:https://doi.org/10.3389/fmolb.2020.00109.
- Zhang Y, Pisano M, Li N, et al. Exosomal circRNA as a novel potential therapeutic target for multiple myeloma-related peripheral neuropathy. Cell Signal. 2021;78:109872. doi:https://doi.org/10.1016/j.cellsig.2020.109872.
- Yu S, Ai L, Wei W, et al. CircRNA circ-MYBL2 is a novel tumor suppressor and potential biomarker in multiple myeloma. Hum Cell. 2021;34(1):219–228. doi:https://doi.org/10.1007/s13577-020-00441-8.
- Song Y, Hu N, Song X, et al. Hsa_Circ_0007841 enhances multiple myeloma chemotherapy resistance through upregulating ABCG2. Technol Cancer Res Treat. 2020;19:1533033820928371. doi:https://doi.org/10.1177/1533033820928371.
- Gao M, Li C, Xiao H, et al. Hsa_circ_0007841: a novel potential biomarker and drug resistance for multiple mmyeloma. Front Oncol. 2019;9:1261. doi:https://doi.org/10.3389/fonc.2019.01261.
- Chen S, Ding J, Wang Y, et al. RNA-seq profiling of circular RNAs and the oncogenic role of circPVT1 in cutaneous squamous cell carcinoma. Onco Targets Ther. 2020;13:6777–6788. doi:https://doi.org/10.2147/OTT.S252233.
