ABSTRACT
Acute myeloid leukemia (AML) was confirmed to be associated with hematopoietic insufficiency, as well as abnormal proliferation, differentiation or survival of myeloid progenitors. Multiple studies reported that microRNA-204 (miR-204) and Hepatocyte growth factor (HGF) played important roles in types of cancers. However, the potential molecular regulatory mechanism between miR-204 and HGF in AML remains to be further defined. Real-time PCR (RT-PCR) was adopted to detect the expression of miR-204 and HG. Relative protein levels were detected by western blot assay. The viability, cell cycle, apoptosis, migration, and invasion were analyzed by MTT, flow cytometry, and transwell assays. Moreover, the target relationship between miR-204 and HGF was predicted by MiRcode website and confirmed by luciferase reporter, RNA pull-down, and western blot assays. Our data suggested that miR-204 was downregulated in AML serum samples and cells. MiR-204 overexpression repressed cell proliferation, migration, invasion, and induced cell apoptosis in AML cells. HGF was upregulated in AML samples and cells, and HGF knockdown inhibited the malignancy of AML cells. In addition, HGF was directly targeted by miR-204. HGF overexpression reversed the effects of miR-204 mimic on AML cell proliferation, apoptosis, migration, and invasion. Besides, miR-204 regulated the c-Met signaling by targeting HGF, thereby regulating the downstream protein levels related to cell proliferation, apoptosis, migration, and invasion in AML cells. In conclusion, miR-204 could regulate AML progression through regulating the HGF/c-Met pathway.
Introduction
Acute myeloid leukemia (AML) is the most common acute leukemia in adults with various genetic abnormalities including chromosomal translocations and/or somatic mutations [Citation1], which frequently lead to hematopoietic insufficiency and the abnormal proliferation, differentiation or survival of myeloid progenitors [Citation2–4]. Even with current treatments, about 70% of patients over 65 years or older will die within 1 year after diagnosis [Citation5].
MiRNAs were reported as prognostic biomarkers and relapse indicators in leukemia [Citation6]. Furthermore, miRNAs have been identified as important regulators or biomarkers [Citation7, Citation8] in AML disease development, including cell proliferation, differentiation, and apoptosis. Moreover, miRNAs was confirmed to exert effects on AML therapeutic resistance [Citation9]. For example, miR-139-5p was identified as tumor suppressor, regulating AML cell progression by targeting Tspan3 through PI3K/Akt pathway [Citation10]. It was reported that miR-194-5p/BCLAF1 modulated orchestrates differentiation, cell fate and treatment susceptibility in AML [Citation11]. Besides, miR-155, miR-182-5p and miR-125b were reported to be involved in AML cell progression [Citation12–15].
As for miR-204, several studies suggest that this regulator is not only as the prognostic factor [Citation16,Citation17], but also as a novel biomarker and therapeutic target with an anti-carcinogenic role by targeting BIRC6 in AML [Citation18]. In addition, miR-204 was also reported to be a useful strategy to improve the efficacy of arsenic trioxide [Citation19]. However, the functional role and potential molecular mechanism of miR-204 in AML remain to be further defined.
HGF/c-Met pathway was associated with several cellular processes, such as proliferation, survival, motility, angiogenesis, invasion and metastasis in different tumor cell types, such as melanoma, non-small cell lung cancer and gastrointestinal cancers [Citation20–24]. It has also reported that targeting HGF/c-Met pathway remained an important method for the development of anti-cancer drug [Citation25]. Moreover, HGF mainly produced by mesenchymal cells and was a powerful angiogenesis factor in vivo angiogenesis assay [Citation26]. Aberrant expression of HGF was identified as a critical factor in AML pathogenesis [Citation27], and was confirmed to be a prognostic factor in AML patients [Citation26,Citation28].
However, the relationship between miR-204 and HGF/c-Met pathway in AML is little reported. Thus, this study aimed to explore the potentially-existing regulatory mechanism between miR-204 and HGF/c-Met pathway.
Materials and methods
Samples and cell culture. This study was approved by the Human Ethics Committee of The First Affiliated Hospital of Zhengzhou University. Written informed consent was obtained from each AML patient and healthy volunteers involved in this study. AML patients were diagnosed according to the World Health Organization criteria, and the clinic-pathologic characteristics of 30 AML patients were listed in Supplement Table 1. 30 AML serum samples from AML patients and 30 normal serum samples from healthy volunteers were collected from the First Affiliated Hospital of Zhengzhou University and stored at -80°C until RNA isolation.
AML cell lines Kasumi-1 and HL-60 obtained from bnbio (Beijing, China) were cultured in Roswell Park Memorial Institute 1640 (RPMI1640; Gibco, Grand Island, NY, USA) with 10% fetal bovine serum (FBS; HyClone, Logan, UT, USA) and 50 μg/mL Gentamicin (Gibco) and cultured at 37°C with 5% CO2.
Transfection. MiR-204 mimics (miR-204), miRNA negative control (miR-NC), pcDNA-HGF, short-hairpin RNAs against HGF (shRNA-HGF), pcDNA-control and sh-NC obtained from Syngentech (Beijing, China) were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instruction at a final concentration of 10-100 nM.
RNA extraction and Real-time polymerase chain reaction (RT-PCR). Total RNA from AML serum samples and cell lines was extracted by Trizol reagent (Invitrogen). RNA reverse transcription was performed ultilizing the Prime Script TM RT Master Mix (TaKaRa Bio Technology, Dalian, China). RT-PCR for mRNA was carried out with SYBR Premix Ex Taq II reagent by 7500 RT-PCR System (Applied Biosystems, Foster City, CA, USA). The expression level of miR-204 was calculated using 2−ΔΔCT method normalized to U6, while HGF mRNA expression level was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The primers of miR-204 and U6 were obtained from RiboBio (Guangzhou, China). Other primers were listed: GAPDH forward primer 5′-GCACCGTCAAGGCTGAGAAC-3′ and reverse primer 5′-TGGTGAAGACGCCAGTGGA-3′, HGF forward primer 5′-GGGCACTGTCAATACCAT-3′ and reverse primer 5′-CAGTAGCCAACTCGGATG-3′.
Western blot assay. Kasumi-1 and HL-60 cells were collected and lysed using RIPA cell lysis buffer (Thermo Fisher Scientific, Waltham, MA, USA) with protease inhibitors to extract total protein. The protein concentration was measured by bicinchonininc acid (BCA) method (Thermo Fisher Scientific). Then these protein samples were separated by 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred on the polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA, USA). Subsequently, PVDF membranes were blocked for 1 h with 5% non-fat milk at room temperature, and then were incubated with primary antibodies overnight at 4°C. Antibodies of c-Met, p-Met, c-Myc, Cyclin D1, Bcl-2, Bax, MMP2, MMP9 and GAPDH were purchased from Affinity (Affinity Biosciences, Changzhou, China). Moreover, membranes were incubated with the respective horseradish peroxidase (HRP)-conjugated secondary antibodies, protein signals were visualized with enhanced chemiluminescence (Millipore).
Cell viability assay. Cell viability was measured by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay in this study. Transfected Kasumi-1 and HL-60 cells were seeded in 96-well plates (Corning Costar, Corning, NY, US) for 24 h at the concentration of 2.0 × 103 cells per well. Then MTT (Beyotime, Shanghai, China) reagent was added into each well at 24 h, 48 h and 72 h after transfection, respectively. Cell viability was detected by measuring the absorbance at 490 nm with a microplate reader.
Cell cycle analysis. The cell cycle and apoptosis analysis kit (Beyotime) was utilized for cell cycle analysis based on the manufacturer’s instruction. Transfected AML cells were collected and washed with ice-cold PBS, followed by fixing with 70% ethanol for 12 h at –20°C. Subsequently, the cells were washed with ice-cold PBS and then stained with propidium iodide (PI) solution, which contained 50 µg/mL PI and 200 mg/mL RNase A, in the dark. 30 min later, the cells were analyzed using FACS Caliber flow cytometry (Becton Dickinson, Mountain View, CA, USA).
Cell apoptosis analysis. Transfected AML cell apoptosis rate was evaluated using the fluorescein isothiocyanate (FITC)-Annexin V Apoptosis Detection Kit (Beyotime Institute of Biotechnology, Shanghai, China) by FACS Caliber flow cytometry (Becton Dickinson) according to the manufacturer’s instruction. Transfected AML cells were washed using PBS for three times and stained with Annexin V-FITC and propidium iodide (PI) for 20 min at room temperature in the dark. Then the apoptotic cells were counted using flow cytometry and analyzed using WinMDI software (The Scripps Research Institute, CA, USA)
Transwell migration and invasion assay. For the determination of cell invasion, transwell chambers were coated with 30 μL matrigel, and then were incubated at 37°C for 40 min. Cells were trypsinized and seeded in the chambers at the density of 1 × 106 cells/well at 24 h after transfection. These cells were cultured in RPMI 1640 medium with 2% serum [Citation29]. 600 μL of medium supplemented with 15% FBS was injected into the lower chambers. Then, the cells in the bottom well were collected and counted by trypan blue exclusion assay. For cell migration experiment, matrigel was not used and other steps were same to cell invasion experiment.
Quantitation of HGF by enzyme-linked immunosorbent assay (ELISA). Total protein was extracted from AML cells. HGF ELISA kit (eBioscience, CA, USA) was used to determine the concentration of HGF according to instruction of manufacturer.
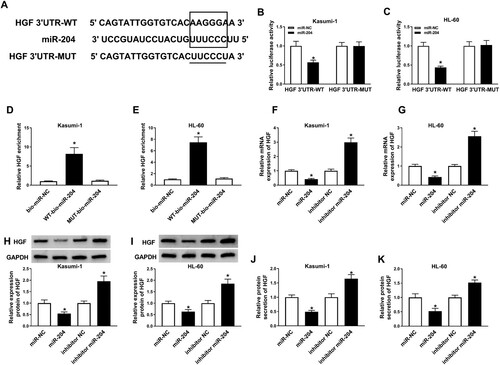
Luciferase reporter assay. By bioinformatics databases, there was miR-204 putative binding sequences on HGF, then the putative binding sequence was cloned into the downstream of firefly luciferase gene of pGL3 vectors (Promega Corporation, Fitchburg, WI, USA), named as HGF 3’UTR-WT. The mutant potential binding site was also cloned into the pGL3 vector, named as HGF 3’UTR-MUT. Kasumi-1 and HL-60 cells were seeded into the 24-well plate for 48 h after co-transfection with 100 ng HGF 3’UTR-WT or HGF 3’UTR-MUT, 50 nM miR-204 or miR-NC, 50 nM of Renilla luciferase reporter vector (pRL-TK, Promega Corporation) using the Lipofectamine 2000 (Invitrogen). Finally, and the relative luciferase activity was examined using Dual Luciferase Reporter Assay Kit (Promega Corporation).
RNA pull-down assay. AML cells were transfected with 20 nmol/L bio-miR-NC, WT-bio-miR-204, or MUT-bio-miR-204 for 24 h. Then, the cell lysates were incubated with streptavidin-coated magnetic beads (Invitrogen). Afterward, the biotin-coupled RNA complex was eluted with Trizol reagent (Invitrogen), and the abundance of HGF mRNA in bound fractions analyzed by qRT-PCR.
Statistical analysis. GraphPad 7.0 software was used in statistical analyses. All data were shown as the mean ± standard deviation (SD) of at least 3 individual experiments. The differences between two or more groups were estimated with the Student’s t-test or one-way analysis of variance. P less than 0.05 was considered as statistically significant.
Results
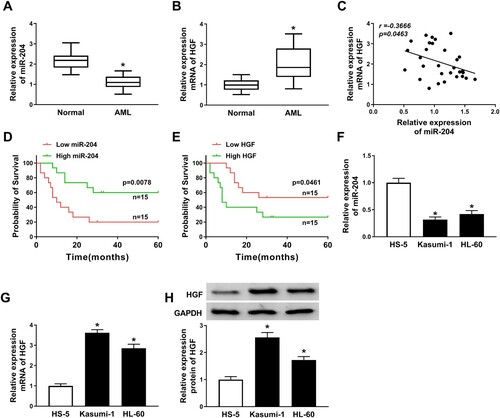
MiR-204 was downregulated and HGF was upregulated in AML samples and cell lines. In order to determine the roles of miR-204 and HGF in AML, the relative expression of miR-204 and HGF was measured. The result showed that miR-204 expression level in AML serum samples was abnormally lower than that in normal samples, while HGF mRNA expression level was higher than that in normal samples ((A,B)). Besides, miR-204 was negatively correlated with HGF level ((C)). To analyze the correlation between serum miR-204 and HGF and the clinical characteristics of AML patients, 30 AML patients were divided into high (n = 15) and low miR-204 group (n = 15), as well as the high (n = 15) and low HGF group (n = 15) based on the median miR-204 and HGF expression level. As shown in Supplement Table 1, there was no significant associate of miR-204 expression or HGF expression with patients’ age, gender, WBC count, Cytogenetics and FAB classification of AML (Supplement Table 1). However, we found that high miR-204 (P = 0.0078) and low HGF expression (P = 0.0461) was highly correlated with favorable prognosis ((D,E)). Similarly, decreased miR-204 and increased mRNA and protein expression of HGF were observed in Kasumi-1 and HL-60 cells compared with the HS-5 cells ((F–H)). These results suggested that miR-204 and HGF plated crucial roles in AML.
Figure 1. Downregulation of miR-204 and upregulation of HGF were observed in AML samples and cell lines. (A–B) The relative expression of miR-204 and HGF mRNA in AML and normal serum samples was analyzed by RT-PCR assay. (C) The relationship between miR-204 and HGF was analyzed. (D) Kaplan-Meier analysis displayed the correlation between miR-204 expression and 5-year overall survival of AML patients. (E) Kaplan-Meier analysis revealed the correlation between HGF expression and 5-year overall survival of AML patients. (F–G) RT-PCR assay was used to detect the expression of miR-204 and HGF in AML cell lines and HS-5 cells. (H) The HGF protein expression was measured by western blot assay. *P < 0.05.
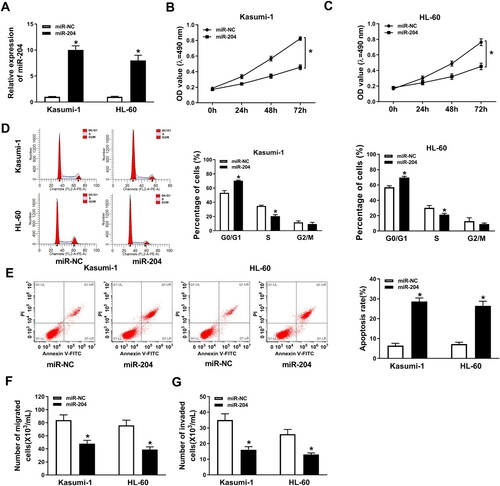
Overexpression of miR-204 inhibited AML progression. To explore the function of miR-204 in AML cell progression, proliferation, migration, invasion and apoptosis of AML cells with overexpressed miR-204 were detected. We obtained a successful overexpression efficiency of miR-204 in AML cells ((A)). The result showed that miR-204 overexpression significantly inhibited cell proliferation ((B,C)). Moreover, miR-204 mimic elevated the number of AML cells in the G0/G1 phase and decreased the number of AML cells in the S phase, manifesting that miR-204 overexpression repressed cell cycle progression in AML cells ((D)). However, cell apoptosis rate in AML cells was accelerated by miR-204 overexpression ((E)). Furthermore, miR-204 overexpression remarkably constrained cell migration and invasion ((F,G)). These results suggested that miR-204 overexpression repressed cell proliferation, migration and invasion, and induced cell apoptosis in AML cells.
Figure 2. MiR-204 overexpression inhibited AML progression. (A) RT-PCR assay confirmed the efficiency of miR-204 in AML cells. (B–C) Cell proliferation in AML cells transfected with miR-204 or miR-NC was measured by MTT assay. (D and E) Flow cytometry method was employed to detect cell cycle and apoptosis in AML cells transfected with miR-204 or miR-NC. (F–G) Transwell assay was used to determine cell migration and invasion in AML cells transfected with miR-204 or miR-NC. *P < 0.05.
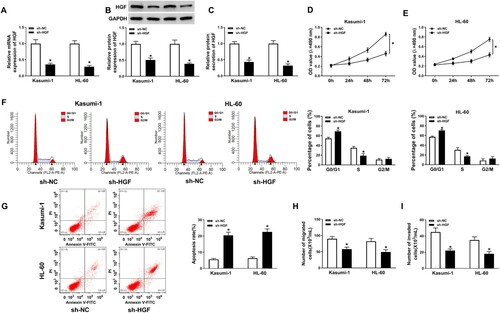
Knockdown of HGF promoted AML progression. To investigate the role of HGF in AML, proliferation, migration, invasion and apoptosis of AML cells with HGF knockdown were examined. The mRNA and protein level of HGF was suppressed by HGF knock in both Kasumi-1 and HL-60 cells ((A–C)). HGF knockdown significantly inhibited the proliferation and cell cycle progression of AML cells ((D–F)). However, cell apoptosis rate was promoted by HGF knockdown in AML cells ((G)). Furthermore, HGF deletion inhibited cell migration and invasion in AML cells ((H,I)). These results indicated that HGF knockdown inhibited cell proliferation, migration and invasion, and promoted cell apoptosis in AML cells.
Figure 3. HGF knockdown suppressed AML progression. (A) HGF mRNA expression in Kasumi-1 and HL-60 cells transfected with sh-HGF or sh-NC was detected by RT-PCR. (B-C) HGF protein level in Kasumi-1 and HL-60 cells transfected with sh-HGF or sh-NC was measured by western blot assay and ELISA. (D–E) MTT assay was applied to check cell proliferation in Kasumi-1 and HL-60 cells transfected with sh-HGF or sh-NC. (F–G) The cell cycle and apoptosis of Kasumi-1 and HL-60 cells transfected with sh-HGF or sh-NC were measured by flow cytometry assay. (H–I) Transwell assay was performed to detect cell migration and invasion in AML cells transfected with sh-HGF or sh-NC. *P < 0.05.
MiR-204 directly interacted with HGF. To determine whether miR-204 could interact with HGF in AML, the relationship between miR-204 and HGF was predicted by MiRcode Tools bioinformatic website and confirmed by dual-luciferase, RNA pull-down, and western blot assays. MiRcode predicted that HGF had the binding sites with miR-204 ((A)). Furthermore, the luciferase activity in AML cells co-transfected with HGF 3’UTR-WT and miR-204 was remarkably decreased, while there was no significant change in AML cells transfected with HGF 3’UTR-MUT and miR-204 ((B,C)). Also, the abundance of HGF mRNA was higher in the WT-bio-miR-204 group than bio-miR-NC and MUT-bio-miR-204 groups ((D,E)). Moreover, the mRNA and protein level of HGF were decreased in miR-204 mimic-transfected AML cells and elevated in miR-204 inhibitor-transfected AML cells ((F–I)). Analogously, miR-204 mimic decreased the protein level of HGF in the culture medium of AML cells, but miR-204 inhibitor played an opposite influence ((J,K)). These results revealed that HGF was directly targeted by miR-204.
Figure 4. HGF was a target of miR-204. (A)The possible interaction sites between miR-204 and HGF was predicted by MiRcode Tools (http://mircode.org/). (B–E) The target relationship between miR-204 and HGF was confirmed by dual-luciferase assay and RNA pull-down assay. (D–I) The mRNA and protein levels of HGF in AML cells transfected with miR-NC, miR-204, inhibitor miR-204 or inhibitor NC was measured by RT-PCR, western blot assay, and ELISA. *P < 0.05.
HGF overexpression promoted cell proliferation, migration, invasion, and inhibited cell apoptosis in AML cells. Supplement Figure 1(A) showed a successful overexpression efficiency of pcDNA-HGF in AML cells. Similarly, the protein level of HGF was upregulated in pcDNA-HGF-transfected AML cells and the culture medium of pcDNA-HGF-transfected AML cells (Supplement Figure 1(B,C)). Also, HGF overexpression significantly promoted the proliferation and cell cycle progression in AML cells (Supplement Figure 1(D–G)). On the contrary, overexpressed HGF expression dramatically suppressed cell apoptosis (Supplement Figure 1(H)). In addition, the abilities of migration and invasion were facilitated by HGF overexpression in AML cells (Supplement Figure 1(I,J)). All these results revealed that HGF overexpression promoted AML progression.
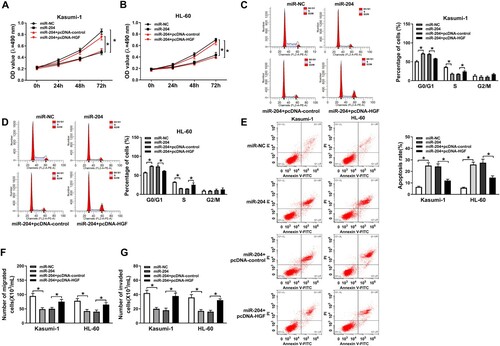
Overexpression of HGF recovered effects of miR-204 on cell proliferation, apoptosis, migration and invasion in AML cells. The data displayed that the inhibitive effect of miR-204 mimic on the proliferation and cell cycle progression of AML cells was restored by HGF overexpression ((A–D)). Furthermore, HGF overexpression reversed the promoting effect of miR-204 upregulation on cell apoptosis in AML cells ((E)). Moreover, HGF upregulation abolished the inhibiting effect of miR-204 overexpression on cell migration and invasion ((F,G)). These results demonstrated that miR-204 regulated AML cell proliferation, apoptosis, migration and invasion by targeting HGF.
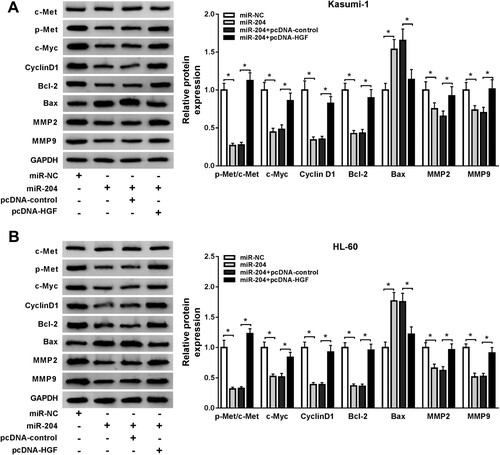
Figure 5. HGF overexpression reversed the effects of miR-204 overexpression on AML cell proliferation, apoptosis, migration, and invasion. (A–B) MTT assay was used to examine cell proliferation in AML cells transfected with miR-NC, miR-204, miR-204+pcDNA-control or miR-204+pcDNA-HGF. (C–D) The cell cycle and apoptosis of AML cells transfected with miR-NC, miR-204, miR-204+pcDNA-control, or miR-204+pcDNA-HGF were assessed by flow cytometry. (F–G) Transwell assay was used to determine cell migration and invasion in AML cells transfected with miR-NC, miR-204, miR-204+pcDNA-control, or miR-204+pcDNA-HGF. *P < 0.05.
MiR-204 regulated HGF/c-Met pathway in AML cells. To further explore the underlying mechanism of miR-204/HGF axis in AML, we detected the expression of Met. Moreover, cell proliferation-related proteins including Cyclin D1 and c-Myc [Citation30], cell apoptosis-related proteins including Bcl-2 and Bax [Citation31,Citation32], and tumor metastasis-related proteins including MMP9 and MMP2 were also detected. Our data showed that miR-204 mimic significantly suppressed the protein levels of c-Myc, Cyclin D1, Bcl-2, MMP-2, MMP-9 and p-Met/c-Met, and promoted the protein level of Bax in AML cells, but these effects caused by miR-204 mimic were reversed by HGF overexpression ((A,B)). These data indicated that miR-204 could cell proliferation, apoptosis, and metastasis in AML cells by regulating the HGF/c-Met pathway.
Discussion
AML is a common hematopoietic malignancy with various genetic abnormalities. MiR-204 and HGF have been reported to play important roles in AML. However, the molecular regulatory mechanism between miR-204 and HGF has not been elucidated. In this study, our data indicated that miR-204 could regulate AML progression through the HGF/c-Met pathway.
MiR-204 originates from the sixth intron of the transient receptor potential melastatin 3 (TRPM3) gene. Most of studies have revealed that miR-204 functioned as a tumor-suppressive gene in various human cancers. The low expression of miR-204 was observed in AML samples and cells in this study, which was consistent with the previous researches [Citation16,Citation33]. HGF was isolated from rat platelets and plasma of patients with hepatic abnormalities. HGF was confirmed to be involved in the progression of AML [Citation34]. Kentsis A and his colleagues indicated that HGF functioned as a vital element in AML pathogenesis, and aberrant expression of HGF induced the autocrine activation Met in most AML samples and cells [Citation35]. Moreover, Kim JG et al. suggested that HGF might be related to the low survival of AML patients [Citation36]. Thus, HGF might be a crucial factor in AML. Consistent with the results in the previous study [Citation37], we found that the mRNA and protein level of HGF were increased in AML serum samples and cells, suggesting that HGF might play a promoting role in AML. In addition, we found that the expression of miR-204 or HGF was closely linked to the prognosis of AML patients. In this study, the data also showed that miR-204 mimic suppressed cell proliferation, migration and invasion, and promoted cell apoptosis in AML cells. Meanwhile, HGF knockdown played an inhibitive role in AML progression. Furthermore, HGF was a target of miR-204, and HGF upregulation reversed the effects of miR-204 overexpression on AML progression. Besides, miR-204 regulated HGF/c-Met pathway and the expression of proliferation, apoptosis and metastasis-related proteins in AML cells.
c-Met, HOXA10, and MEIS1 have been reported to be the targets of miR-204 [Citation33,Citation38]. In the present study, HGF was confirmed as the target of miR-204 and exerted a promoting effect on AML progression. Furthermore, lncRNA was reported to be associated with therapeutic resistance of AML [Citation9,Citation39]. LncRNA can function as miRNA sponges to exert effects in various diseases. Regrettably, we did not explore the upstream regulatory mechanism of miR-204, which will be further studied in the future. Moreover, the lack of immunofluorescence analysis was a limitation in the present study. Another limitation of our study is that the number of samples is smaller, which might lead to over-fitting and negatively influence the diagnostic performance. Thus further analysis is warranted with a larger number of samples to verify the results obtained in current research.
In summary, our findings confirmed that miR-204 exerted an inhibitory effect on AML progression through regulating the HGF/c-Met pathway, providing a novel regulatory mechanism between miR-204 and HGF in AML.
Supplemental Material
Download MS Word (165.3 KB)Supplemental Material
Download MS Word (19.1 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Prada-Arismendy J, Arroyave JC, Rothlisberger S. Molecular biomarkers in acute myeloid leukemia. Blood Rev. 2017;31:63–76.
- Meyer SC, Levine RL. Translational implications of somatic genomics in acute myeloid leukaemia. Lancet Oncol. 2014;15:e382–e394.
- Saadatpour A, Guo G, Orkin SH, et al. Characterizing heterogeneity in leukemic cells using single-cell gene expression analysis. Genome Biol. 2014;15:525.
- Lowenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med. 1999;341:1051–1062.
- Meyers J, Yu Y, Kaye JA, et al. Medicare fee-for-service enrollees with primary acute myeloid leukemia: an analysis of treatment patterns, survival, and healthcare resource utilization and costs. Appl Health Econ Health Policy. 2013;11:275–286.
- Gomwalk NE, Umoh UJ, Gosham LT, et al. Influence of climatic factors on rotavirus infection among children with acute gastroenteritis in Zaria, northern Nigeria. J Trop Pediatr. 1993;39:293–297.
- Wang X, Chen H, Bai J, et al. MicroRNA: an important regulator in acute myeloid leukemia. Cell Biol Int. 2017;41:936–945.
- Wallace JA, O’Connell RM. MicroRNAs and acute myeloid leukemia: therapeutic implications and emerging concepts. Blood. 2017;130:1290–1301.
- Zebisch A, Hatzl S, Pichler M, et al. Therapeutic resistance in acute myeloid leukemia: the role of non-coding RNAs. Int J Mol Sci. 2016;17:2080.
- Zhang R, Tang P, Wang F, et al. Tumor suppressor miR-139-5p targets Tspan3 and regulates the progression of acute myeloid leukemia through the PI3 K/Akt pathway. J Cell Biochem. 2019;120:4423–4432.
- Dell’Aversana C, Giorgio C, D’Amato L, et al. MiR-194-5p/BCLAF1 deregulation in AML tumorigenesis. Leukemia. 2017;31:2315–2325.
- Sadras T, Kok CH, Perugini M, et al. MiR-155 as a potential target of IL-3 signaling in primary AML cells. Leuk Res. 2017;57:57–59.
- Liu J, Guo B, Chen Z, et al. MiR-125b promotes MLL-AF9-driven murine acute myeloid leukemia involving a VEGFA-mediated non-cell-intrinsic mechanism. Blood. 2017;129:1491–1502.
- Wallace JA, Kagele DA, Eiring AM, et al. MiR-155 promotes FLT3-ITD-induced myeloproliferative disease through inhibition of the interferon response. Blood. 2017;129:3074–3086.
- Zhang S, Zhang Q, Shi G, et al. MiR-182-5p regulates BCL2L12 and BCL2 expression in acute myeloid leukemia as a potential therapeutic target. Biomed Pharmacother. 2018;97:1189–1194.
- Butrym A, Rybka J, Baczynska D, et al. Low expression of microRNA-204 (miR-204) is associated with poor clinical outcome of acute myeloid leukemia (AML) patients. J Exp Clin Cancer Res. 2015;34:68.
- Butrym A, Lacina P, Kuliczkowski K, et al. Genetic variation of the gene coding for microRNA-204 (miR-204) is a risk factor in acute myeloid leukaemia. BMC Cancer. 2018;18:107.
- Wang Z, Luo H, Fang Z, et al. MiR-204 acts as a potential therapeutic target in acute myeloid leukemia by increasing BIRC6-mediated apoptosis. BMB Rep. 2018;51:444–449.
- Wang Z, Fang Z, Lu R, et al. MicroRNA-204 potentiates the sensitivity of acute myeloid leukemia cells to arsenic trioxide. Oncol Res. 2019: 1035–1042.
- Noriega-Guerra H, Freitas VM. Extracellular matrix influencing HGF/c-MET signaling pathway: impact on cancer progression. Int J Mol Sci. 2018;19:3300.
- Bradley CA, Salto-Tellez M, Laurent-Puig P, et al. Targeting c-MET in gastrointestinal tumours: rationale, opportunities and challenges. Nat Rev Clin Oncol. 2017;14:562–576.
- Demkova L, Kucerova L. Role of the HGF/c-MET tyrosine kinase inhibitors in metastasic melanoma. Mol Cancer. 2018;17:26.
- Cao HH, Cheng CY, Su T, et al. Quercetin inhibits HGF/c-Met signaling and HGF-stimulated melanoma cell migration and invasion. Mol Cancer. 2015;14:103.
- Stabile LP, Rothstein ME, Gubish CT, et al. Co-targeting c-Met and COX-2 leads to enhanced inhibition of lung tumorigenesis in a murine model with heightened airway HGF. J Thorac Oncol. 2014;9:1285–1293.
- Yap TA, Sandhu SK, Alam SM, et al. HGF/c-MET targeted therapeutics: novel strategies for cancer medicine. Curr Drug Targets. 2011;12:2045–2058.
- Verstovsek S, Kantarjian H, Estey E, et al. Plasma hepatocyte growth factor is a prognostic factor in patients with acute myeloid leukemia but not in patients with myelodysplastic syndrome. Leukemia. 2001;15:1165–1170.
- Kentsis A, Reed C, Rice KL, et al. Autocrine activation of the MET receptor tyrosine kinase in acute myeloid leukemia. Nat Med. 2012;18:1118–1122.
- Kim JG, Sohn SK, Kim DH, et al. Clinical implications of angiogenic factors in patients with acute or chronic leukemia: hepatocyte growth factor levels have prognostic impact, especially in patients with acute myeloid leukemia. Leuk Lymphoma. 2005;46:885–891.
- Liu P, Li J, Lu H, et al. Thalidomide inhibits leukemia cell invasion and migration by upregulation of early growth response gene 1. Leuk Lymphoma. 2009;50:109–113.
- Venneti S, Thompson CB. Metabolic modulation of epigenetics in gliomas. Brain Pathol. 2013;23:217–221.
- Merino D, Kelly GL, Lessene G, et al. BH3-Mimetic drugs: blazing the trail for new cancer medicines. Cancer Cell. 2018;34:879–891.
- Chao DT, Korsmeyer SJ. BCL-2 family: regulators of cell death. Annu Rev Immunol. 1998;16:395–419.
- Garzon R, Garofalo M, Martelli MP, et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci U S A. 2008;105:3945–3950.
- Edwards DK, Watanabe-Smith K, Rofelty A, et al. CSF1R inhibitors exhibit antitumor activity in acute myeloid leukemia by blocking paracrine signals from support cells. Blood. 2019;6:588–599.
- Kentsis A, Reed C, Rice KL, et al. Autocrine activation of the MET receptor tyrosine kinase in acute myeloid leukemia. Nat Med. 2012;7:1118–1122.
- Kim JG, Sohn SK, Kim DH, et al. Clinical implications of angiogenic factors in patients with acute or chronic leukemia: hepatocyte growth factor levels have prognostic impact, especially in patients with acute myeloid leukemia. Leuk Lymphoma. 2005;6:885–891.
- Guo JR, Li W, Wu Y, et al. Hepatocyte growth factor promotes proliferation, invasion, and metastasis of myeloid leukemia cells through PI3K-AKT and MAPK/ERK signaling pathway. Am J Transl Res. 2016;8:3630–3644.
- Liu H, Li SR, Si Q. Regulation of miRNAs on c-met protein expression in ovarian cancer and its implication. Eur Rev Med Pharmacol Sci. 2017;21:3353–3359.
- Fernandes Q. MicroRNA: defining a new niche in leukemia. Blood Rev. 2017;31:129–138.