?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Objectives
This meta-analysis examined the prognostic role of brain and acute leukemia, cytoplasmic (BAALC), Ecotropic virus integration site-1 (EVI1) and Wilms’ tumor 1 (WT1) genes at different time-points during conventional chemotherapy.
Methods
A systematic search of publications indexed in the electronic databases from January 1988 to October 2020 was performed. Over 7525 cases of AML from 25 studies were involved.
Results
At diagnosis, overexpression of either BAALC or EVI1 had a negative impact on complete remission achievement (Summary Odds ratios [SORs] for BAALC = 0.32; SORs for EVI1 = 0.49) and survival outcome. The summary hazard ratios of overall survival (OS) and disease-free survival (DFS) were 1.97 and 2.04 for BAALC and 1.33 and 1.86 for EVI1, respectively. The prognostic value of pretreatment WT1 levels was heterogeneous while subgroup analyses unveiled that overexpressed WT1 may correlate with a favorable outcome (summary hazard ratio [SHR] for OS = 0.42). Both WT1 and BAALC played a role in prognosis assessment at post-induction and the diagnostic performance of WT1 transcript reduction was superior to the absolute WT1 level. Post-consolidation WT1 overexpression consistently indicated an increased risk of relapse, while the combined HR for RFS was statistically insignificant (SHR = 4.22).
Conclusion
These findings confirm the application of BAALC and EVI1 at diagnosis, WT1 after induction chemotherapy in AML patients throughout conventional chemotherapy.
1. Background
Acute myeloid leukemia (AML) is a heterogeneous hematologic malignancy, which occurs mostly in adults. Cytogenetic abnormalities and response to initial treatment have a major influence on prognostic assessment.
However, nearly 45% of AML patients have normal cytogenetics and the absence of cytogenetic markers poses significant challenges for risk stratification and minimal residual disease monitoring (MRD) [Citation1]. MRD denotes the presence of residual leukemia cells far below the morphology-based 5% blast threshold and multiparameter flow cytometry (MFC) and real-time quantitative polymerase chain reaction (qPCR) are two methods currently widely applied. Nevertheless, a paucity of residual leukemia-specific markers in AML makes the prognostication lag behind compared with acute lymphoblastic leukemia [Citation2]. With the goal of improving outcome prediction and identifying early relapse, the prognostic value and sequential qPCR MRD monitoring of prognostic markers at diagnosis and during chemotherapy are widely investigated.
Significant effort has gone into identifying potential prognostic markers to stratify AML patients [Citation3,Citation4]. Recently, a variety of novel molecular abnormalities including mutations (e.g. NPM1, FLT3-ITD, CEBPA) and abnormal genes expression (e.g. EVI1, WT1, BAALC, MN1, ERG, and BCL2) have been identified. Overexpression of pathogenetic genes such as brain and acute leukemia, cytoplasmic (BAALC), Ecotropic virus integration site-1 (EVI1), and Wilms’ tumor 1 (WT1) have been reported to be able to differentiate AML patients with a high or low risk of relapse during therapy [Citation5,Citation6]. BAALC gene, which is located on chromosome 8q22.3, encodes a protein involved in the development of neuroectoderm and hematopoiesis [Citation7]. EVI1 gene, also known as MDS1 and EVI1 complex locus (MECOM) oncogene, located at chromosome 3q26 encodes a nuclear DNA binding protein with two zinc-finger domains [Citation8,Citation9]. WT1 is a tumor suppressor gene mapped on chromosome 11p13, encoding a zinc-finger transcription factor [Citation10]. These genes play important roles in normal hematopoiesis by regulating cell growth and differentiation [Citation11–13] and they are found to be highly expressed in CD34-positive cells while their levels rapidly decrease during differentiation [Citation14–16]. Meanwhile, several studies in the biology of disease have revealed their dysregulated expression might contribute to leukemogenesis and each of these three genes has been proposed as prognostic markers in AML [Citation17].
Even though numerous studies have described and evaluated their impact on prognosis at diagnosis, after induction and post-remission chemotherapy, the extent of the correlation between gene expression levels and risk of relapse has remained controversial and the optimal testing time for each gene over the course of conventional chemotherapy varies widely between studies. Since BAALC expression has been demonstrated highly correlated to MN1 expression and ERG expression [Citation17], in this meta-analysis, we mainly focused on investigating the prognostic importance of BAALC, EVI1, and WT1 in acute myeloid leukemia and ascertained the optimal time-points for gene expression monitoring during conventional chemotherapy.
2. Methods
We specifically sought observational studies including EVI1, Ecotropic virus integration site-1, MECOM, WT1, Wilms’ tumor gene, BAALC, Brain and acute leukemia, cytoplasmic, acute myeloid leukemia, minimal residual disease, MRD, dynamic changes, time-points as keywords. The titles and abstracts of identified references were screened and studies that fulfilled predefined inclusion criteria below were selected.
2.1. Data sources and searches
We conducted a systematic search on PubMED, EMBASE, ISI Web of Science, the Cochrane Central Register of Controlled Trials, Scopus, and Registers of ongoing trials from January 1988 through October 2020. We also searched bibliographic databases for relevant reviews and meta-analyses, as well as gray literature databases for any relevant unpublished research. If needed, we contacted the original authors for additional information.
2.2. Study selection
Two investigators independently evaluated each study’s abstract according to prespecified inclusion criteria. We included high-quality retrospective and prospective cohort studies which assessed the prognostic value of BAALC, EVI1 and WT1 expression levels in acute myeloid leukemia at different time-points. Study inclusion criteria were the following: (1) Studies were conducted among patients diagnosed with AML, including de no AML, AML with MDS-related changes and therapy-related myeloid neoplasms; (2) Duration of follow-up assessments should be more than 2 years; (3) Data on survival analysis including disease-free survival (DFS), overall survival (OS), relapse-free survival (RFS), or Kaplan-Meier curves were provided. (4) During follow-up, AML patients who underwent allogeneic hematopoietic stem cell transplantation were censored at the time of HSCT. Publications were excluded if insufficient information was provided or if they focused exclusively on children. When a single study was illustrated in multiple reports, only the most recent publication was chosen for further analysis.
2.3. Data extraction
One investigator extracted study design information, baseline patient characteristics, WT1, EVI1 and BAALC expression levels at different time-points, treatment details, disease outcome, and survival data from all included studies into a combined evidence table. A second investigator evaluated these data for accuracy. We specified the OS as the primary outcome measure, DFS and RFS as the secondary outcome measure. The prognostic value of WT1, EVI1 and BAALC overexpression in patients with AML was assessed by hazard ratio (HR) and 95% confidence intervals (CIs) of OS, DFS and RFS obtained from Cox multivariate analyses; the association between gene overexpression and complete remission rate was expressed in odds ratios (ORs). If there were only Kaplan-Meier curves available on published papers and raw data from the authors were unattainable, KM data was retrieved using DigitizeIt software extracted from the coordinates of the published Kaplan-Meier curves and calculations were made based on the method of Tierney et al. [Citation18].
2.4. Quality assessment
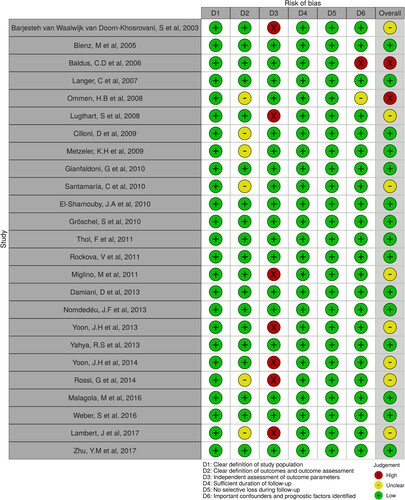
Two investigators independently appraised each observational study’s quality according to the risk of bias assessment checklist proposed by MOOSE [Citation19]. Risks of bias were designated as high, unclear, or low. Studies at high risk of bias were further evaluated by sensitivity analyses, in which one study was removed at a time and the others analyzed to examine the potential impact of bias on cumulative results. All studies at higher risk of bias that could invalidate the cumulative results were excluded. When disagreements occurred, we resolved them through discussion.
2.5. Data synthesis and analysis
To investigate the inter-study heterogeneity, the chi-squared test was used to determine differences with a P value < 0.10 considered statistically significant and statistic was adopted to quantify inconsistency across studies. Cut-points between high and low expression levels were derived from the criteria specified by the individual publications. Here, we performed a cumulative meta-analysis to estimate the prognostic value of WT1, EVI1 and BAALC expression levels at diagnosis, following completion of induction chemotherapy and sequential monitoring. Odds ratios less than 1 (greater than) correspond to lower (higher) odds of achieving CR for gene overexpression and hazard ratios greater than (less than) 1 correspond to shorter DFS and OS for gene overexpression, relating to the increased (decreased) risk of death. A random-effects model was employed if moderate or high heterogeneity was observed by
values (
close to 50% or above); otherwise, a fixed-effects model was used. Subgroup analyses were performed to discover possible sources of heterogeneity. Stata software, version 15 (Stata Corp, TX, USA) was employed for statistical analyses.
3. Results
3.1. Study characteristics
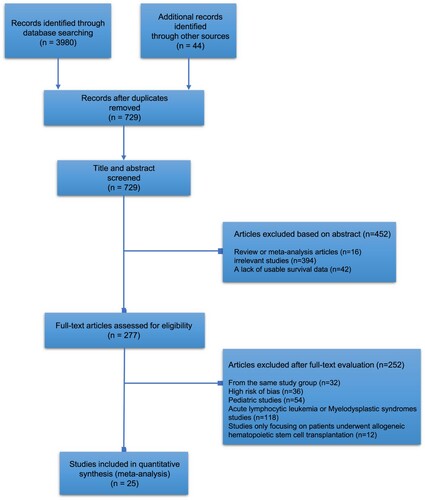
We screened 729 abstracts, reviewed 277 full-text articles, and included 25 articles encompassing 7525 AML cases for full analysis. The PRISMA flow diagram is demonstrated in . All 25 included studies were cohort studies wherein treatment consisted of conventional chemotherapy regimens having a similar backbone, including intensive chemotherapy with cytarabine/anthracycline. summarizes study characteristics, including the year of publication, country, cases, maximum follow-up months, gender and prognostic markers studied. Of the 25 included studies, 10 studies [Citation6,Citation20–27] assessed the prognostic value of WT1 expression levels in AML. Thirteen studies [Citation16,Citation26–37] evaluated the prognostic importance of BAALC and 5 studies [Citation38–41] examined the efficacy of EVI1 gene expression to predict survival outcome in AML patients. The number of cases ranged from 38 cases to 1382 cases. The maximum follow-up months range from 24 months to 224 months. In terms of the study population, four studies excluded patients with acute promyelocytic leukemia (M3), seven studies only included cytogenetically normal acute myeloid leukemia (CN-AML) patients, and two studies specifically analyzed patients with intermediate-risk AML. The quality assessment of each included study is presented in .
Table 1. Features of included studies in this meta-analysis.
There was no statistical evidence of publication bias found in studies that evaluate the expression level of molecular markers at diagnosis, after induction and after consolidation chemotherapy judged from funnel plots and Egger’s test. Given that statistics for heterogeneity demonstrated there may have been heterogeneity for WT1 expression level at diagnosis presenting adjusted overall survival HR (
= 86.3%), the random-effects model was employed for OS at diagnosis.
3.2. Risk stratification at diagnosis
summarizes the adjusted estimates for complete remission from three molecular markers. Eleven studies [Citation16,Citation28–36] for BAALC gene and 3 studies [Citation39–41] for EVI1 gene evaluated the prognostic value of gene expression level at diagnosis on CR rate. The pooled adjusted odds ratios for BAALC gene overexpression and EVI1 gene overexpression were 0.32 (95%CI, 0.23–0.42) and 0.49 (95%CI, 0.35–0.63), respectively, which proved the overexpression of BAALC gene and EVI1 gene were both negative predictors of achieving CR. One study [Citation21] for WT1 gene assessed the CR rate in patients between WT1 high-expressers and low-expressers and showed no statistically significant effect on CR rate (P = 0.07).
Figure 3. Forest plots depicting odds ratios (ORs) for complete remission (A) and hazard ratios (HRs) for overall survival (B) and disease-free survival (C) at diagnosis with pooling of results by each prognostic marker. ORs less than 1 (greater than) correspond to lower (higher) odds of achieving CR for gene overexpression and HRs greater than (less than) 1 correspond to shorter DFS and OS for gene overexpression. CI: confidence interval.
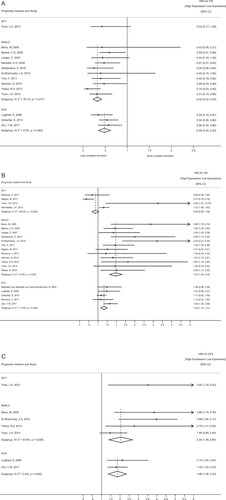
The usefulness of BAALC and EVI1 gene expression levels at diagnosis for stratifying AML patients was observed. The summary HRs for OS and DFS of BAALC overexpression at diagnosis were 1.97 (95%CI, 1.62–2.32) and 2.04 (95%CI, 1.39–2.69). EVI1 overexpression at diagnosis showed similar unfavorable results with a summary HR for OS of 1.33 (95%CI, 1.14–1.51) and a summary HR for DFS of 1.86 (95%CI, 1.36–2.35). We found few consistent results for OS across 4 studies of WT1 expression level at diagnosis. Although the HR for OS reported for WT1 overexpression at diagnosis was greater than 1 corresponding to an increased risk in 2 studies [Citation21,Citation22], the opposite was observed in another 2 studies [Citation26,Citation27].
3.3. Prognostic value at post-induction
Six studies investigated the predictive impact of WT1 expression after induction chemotherapy on OS and DFS (). Three of these studies [Citation23,Citation25,Citation42] provided data on WT1 level and reported that patients with higher WT1 expression had poor OS (Summary hazard ratio [SHR] = 1.91; 95% CI, 1.16–2.66) and DFS (SHR = 3.41; 95% CI, 0.44–6.38). Likewise, four studies [Citation20,Citation21,Citation23,Citation43] evaluated the prognostic value of WT1 transcript reduction at post-induction and implicated that a greater decrement ratio of WT1 expression after induction compared with the pretreatment level was associated with improved DFS and OS. The results showed summary HR for OS of 0.33 (95% CI, 0.10–0.56) and summary HR for DFS of 0.43 (95% CI, 0.28–0.58) in favor of patients with greater WT1 transcript reduction. One of these studies [Citation23] specifically compared the diagnostic performance of both WT1 level and WT1 transcript reduction after induction and confirmed that WT1 decrement ratio was the better prognostic measurement with higher specificity, positive predictive value, and positive likelihood ratio.
Figure 4. Forrest plots depicting hazard ratios (HRs) and 95% confidence intervals (CI) for overall survival (A) and disease-free survival (B) after induction chemotherapy by each prognostic marker.
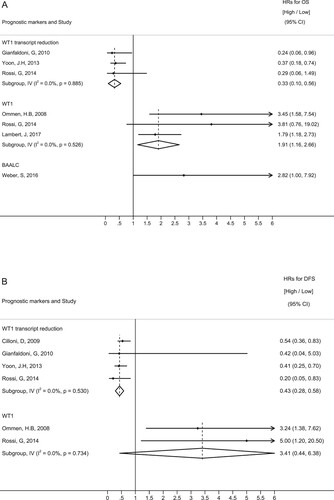
In one study [Citation37] of BAALC, the adjusted OS for BAALC high expression compared with low expression after induction chemotherapy was 2.82 (95%CI, 1.00–7.92), indicating an increased risk of death.
3.4. Sequential monitoring post-consolidation and post-intensification
Relapse-free survival was calculated from the date of complete remission to that of relapse or death, whichever occurred first. Our 2 included studies [Citation24,Citation43] showed a consistent trend that post-consolidation WT1 level was associated with worse RFS (Figure S1), even though the combined RFS was not statistically significant (SHR = 4.22; 95% CI, 0.52–7.93) [Citation24,Citation43]. After intensification, one study [Citation24] found that WT1 overexpression in peripheral blood was an independent predictive factor to impair RFS and another study [Citation22] similarly showed high WT1 expression in bone marrow had inferior RFS.
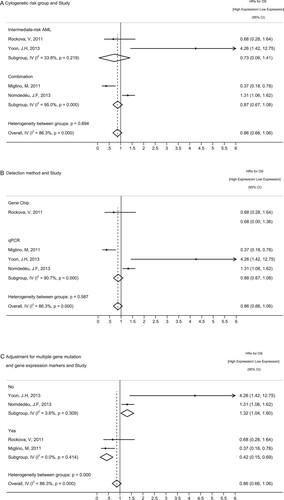
3.5. Subgroup analyses
Since statistical heterogeneity was present in the meta-analysis of WT1 expression level at diagnosis ( = 86.3%, p < 0.001), we employed subgroup analyses to further explore the influence of potential sources of heterogeneity including cytogenetic risk groups, detection method as well as baseline characteristics of the study population. Most subgroup estimates appeared to be consistent with each other and did not indicate heterogeneity. However, as presented in , the summary HR for OS of studies [Citation26,Citation27] with covariates adjusted for multiple (more than three) gene mutations and overexpressed genes was 0.42 (95%CI, 0.15–0.69,
= 0.0%, p = 0.414) opposite to those [Citation21,Citation22] only adjusted with less than two other genes exemplified as FLT3-ITD and NPM1 mutation (SHR = 1.32; 95% CI, 1.04–1.60,
= 3.6%, p = 0.309). This result suggests that the favorable prognostic value of WT1 gene overexpression at diagnosis seems to be overshadowed, as it is often associated with several unfavorable gene mutations and gene expression markers, which indicates that WT1 overexpression alone is not suitable to serve as a prognostic marker at diagnosis.
Figure 5. Subgroup analyses of WT1 levels among OS analyses at diagnosis for (A) Cytogenetic risk group: In two studies, survival analyses were exclusively conducted in AML patients cytogenetically classified into intermediate-risk categories, while the other two studies analyzed the complete cohort of AML patients combining favorable, intermediate, and adverse risk categories. (B) Detection method: Gene chip versus qPCR (C) Adjustment for multiple gene mutations and gene expression markers in multivariate survival analysis: ‘Yes’ indicates studies with covariates of overall survival adjusted for multiple (more than three) gene mutations and overexpressed genes. ‘No’ indicates studies only adjusted with less than two other genes. HR: hazard ratios, CI: confidence intervals.
4. Discussion
Recent developments in genomics technology, such as genome sequencing and gene expression analysis, have made the identification of numerous genetic abnormalities in AML patients on a large scale in parallel possible. WT1, EVI1, and BAALC gene overexpression are well-known risk factors that provide important prognostic information in AML. Even though the most common method for risk stratification was the detection of fusion genes resulting from chromosomal translocations [Citation44], a relative paucity of identifiable molecular and phenotypical markers in approximately 45% of AML patients poses significant challenges for prognostic assessment over the course of treatment, particularly for those cases without cytogenetic abnormalities [Citation45]. Thus, the expression level analysis of these genes has been included in the initial diagnostics in clinical practice. Nevertheless, there are no consensus recommendations for the measurement and application of gene expression analysis at different time-points in chemotherapy workup for AML [Citation46]. Hence, our meta-analysis not only intends to examine the prognostic value of WT1, EVI1, and BAALC overexpression but also to evaluate the optimal timing of assessment for individual gene tests.
This meta-analysis summarizes the findings of 25 observational studies, encompassing a total of 7525 AML cases, incorporating data from independent studies for a specified gene and integrating the relevant studies at different time-points to assess their diagnostic utility. At diagnosis, overexpression of either BAALC gene or EVI1 gene identifies patients at a lower rate of complete remission and shorter survival compared to patients with lower expression, which is in line with the results of previous meta-analyses either for the BAALC gene or EVI1 gene [Citation47,Citation48]. Surprisingly, we found that high WT1 expression may have a positive effect on survival outcome after excluding the effect of adverse gene mutations and gene expression markers by subgroup analyses, which varied from previous reports [Citation49]. Nevertheless, WT1 overexpression alone at diagnosis is not suitable to serve as a prognostic marker since its prognostic value is easily affected by other prognostic markers. Meanwhile, the evaluation of WT1 decremental response comparable to absolute WT1 expression level after induction chemotherapy was helpful in predicting outcome and guiding post-remission potential therapy. Similarly, high BAALC expression shortly following induction chemotherapy correlated with shorter overall survival. Addressing post-consolidation level for follow-up assessment, overexpressed WT1 indicated an increased risk of relapse.
In recent years, the expression levels of several genes over the course of AML therapy became the subject of research interest, exemplified as EVI1, BAALC, ERG, as well as WT1. EVI1 was shown to be overexpressed in both adult and pediatric AML patients with and without cytogenetic abnormalities involving chromosome 3q [Citation50]. Aberrant expression of EVI1 was found to be closely related to monosomy of chromosome 7 and chromosome 11q23 aberrations, both of which are proven to be correlated to unfavorable outcome [Citation40]. EVI1 overexpression and favorable risk cytogenetics were virtually exclusive. The poor effect also remains significant in intermediate-risk AML and in AML with normal karyotypes [Citation38]. The BAALC expression differed with AML FAB subtype distribution, with high BAALC expressing patients correlated with the more immature FAB subtypes M0 and M1, and less frequently with the monocytic differentiated FAB M4 and M5 [Citation16]. Pretreatment BAALC expression levels were associated with the mutation status of FLT3-ITD, NPM1wt, KMT2A-PTDs and CEBPA [Citation51]. Notably, there were significant pairwise correlations between ERG, BAALC, and MN1 transcript levels, especially between BAALC and MN1 [Citation30]. Elevated WT1 expression levels can be detected in both peripheral blood and bone marrow during relapse [Citation52]. Nevertheless, Ommen et al. [Citation42] provided data suggesting that relapses were identified significantly sooner in BM relative to PB and the intervening lag phase could last up to two months in some cases.
Our analysis has several limitations. The major challenge is that the included studies were observational studies rather than prospective randomized clinical trials. In addition, the limited number of studies in each pooled analysis made it difficult to demonstrate a more robust summary survival estimate. Thirdly, reporting of the adverse events of interest was not uniform. Finally, as is common the case with meta-analysis, a considerable effect of heterogeneity such as study settings, different treatment strategies, and cut-off points chosen to define gene overexpression may have a negative impact on the accuracy of our study. Despite these limitations, this study is the first meta-analysis focusing on the optimal time-points for gene expression markers over the course of AML chemotherapy and provides validated information on time-point selection of gene expression analysis in clinical use.
5. Conclusions
In conclusion, measurement of BAALC, EVI1, and WT1 at several time-points from diagnosis to post-intensification in conventional chemotherapy serves as an important predictor for disease outcome. Our results support the use of BAALC expression and EVI1 expression at diagnosis for AML risk stratification and underscore the importance of WT1 transcript reduction after induction chemotherapy in risk-adapted post-induction therapy. Sequential WT1 level monitoring after consolidation and after intensification may add extra information to predict relapse. Future studies of gene expression profiling markers are recommended to evaluate their optimal timing of assessment in conventional chemotherapy and HSCT.
Code availability
Stata software, version 15 (Stata Corp, TX, USA).
Supplemental Material
Download PDF (98.8 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Additional information
Funding
References
- Gupta M, Mahapatra M, Saxena R. Cytogenetics’ impact on the prognosis of acute myeloid leukemia. J Lab Physicians. 2019;11(2):133–137.
- Rubnitz JE, Kaspers GJL. How I treat pediatric acute myeloid leukemia. Blood. 2021;138(12):1009–1018.
- Lussana F, Caprioli C, Stefanoni P, et al. Molecular detection of minimal residual disease before allogeneic stem cell transplantation predicts a high incidence of early relapse in adult patients with NPM1 positive acute myeloid leukemia. Cancers (Basel). 2019;11(10):1455.
- Picharski GL, Andrade DP, Fabro ALMR, et al. The impact of Flt3 gene mutations in acute promyelocytic leukemia: a meta-analysis. Cancers (Basel). 2019;11(9):1311.
- Brand J, van Vliet MH, de Best L, et al. A standardized microarray assay for the independent gene expression markers in AML: EVI1 and BAALC. Exp Hematol Oncol. 2013;2(1):7.
- Cilloni D, Gottardi E, De Micheli D. Quantitative assessment of WT1 expression by real time quantitative PCR may be a useful tool for monitoring minimal residual disease in acute leukemia patients Leukemia. Int J Lab Hematol. 2002;16(10):2115–2121.
- Eid MA, Attia M, Abdou S, et al. BAALC and ERG expression in acute myeloid leukemia with normal karyotype: impact on prognosis. Int J Lab Hematol. 2010;32(2):197–205.
- Balgobind BV, Lugthart S, Hollink IH, et al. EVI1 overexpression in distinct subtypes of pediatric acute myeloid leukemia. Leukemia. 2010;24(5):942–949.
- Glass C, Wilson M, Gonzalez R, et al. The role of EVI1 in myeloid malignancies. Blood Cells Mol Dis. 2014;53(1-2):67–76.
- Marjanovic I, Karan-Djurasevic T, Ugrin M, et al. Use of Wilms Tumor 1 gene expression as a reliable marker for prognosis and minimal residual disease monitoring in acute myeloid leukemia with normal karyotype patients. Clin Lymphoma Myeloma Leuk. 2017;17(5):312–319.
- Marjanovic I, Karan-Djurasevic T, Kostic T, et al. Expression pattern and prognostic significance of EVI1 gene in adult acute myeloid leukemia patients with normal karyotype. Indian J Hematol Blood Transfus. 2020;36(2):292–299.
- Rashed RA, Kadry DY, El Taweel M, et al. Relation of BAALC and ERG gene expression with overall survival in acute myeloid leukemia cases. Asian Pac J Cancer Prev. 2015;16(17):7875–7882.
- Gaiger A, Schmid D, Heinze G, et al. Detection of the WT1 transcript by RT-PCR in complete remission has no prognostic relevance in de novo acute myeloid leukemia. Leukemia. 1998;12(12):1886–1894.
- Steinleitner K, Rampetsreiter P, Köffel R, et al. EVI1 and MDS1/EVI1 expression during primary human hematopoietic progenitor cell differentiation into various myeloid lineages. Anticancer Res. 2012;32(11):4883–4889.
- Svedberg H, Richter J, Gullberg U, et al. Forced expression of the Wilms tumor 1 (WT1) gene inhibits proliferation of human hematopoietic CD34 + progenitor cells. Leukemia. 2001;15(12):1914–1922.
- Langer C, Ruppert AS, Radmacher MD, et al. High BAALC expression associates with other molecular prognostic markers, poor outcome and a distinct gene expression signature in cytogenetically normal acute myeloid leukemia (CN AML): a Cancer and Leukemia Group B (CALGB) study. J Clin Oncol. 2007;25(18):7013.
- Thol F, Yun H, Sonntag AK, et al. Prognostic significance of combined MN1, ERG, BAALC, and EVI1 (MEBE) expression in patients with myelodysplastic syndromes. Ann Hematol. 2012;91(8):1221–1231.
- Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007:8–16.
- Stroup DF, Berlin JA, Morton SC, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA. 2000;283(15):2008–2012.
- Gianfaldoni G, Mannelli F, Ponziani V, et al. Early reduction of WT1 transcripts during induction chemotherapy predicts for longer disease free and overall survival in acute myeloid leukemia. Haematologica. 2010;95(5):833–836.
- Yoon JH, Kim HJ, Shin SH, et al. Serial measurement of WT1 expression and decrement ratio until hematopoietic cell transplantation as a marker of residual disease in patients with cytogenetically normal acute myelogenous leukemia. Biology of blood and marrow transplantation. Leukemia. 2013;19(6):958–966.
- Nomdedéu JF, Hoyos M, Carricondo M, et al. Bone marrow WT1 levels at diagnosis, post-induction and post-intensification in adult de novo AML. Leukemia. 2013;27(11):2157–2164.
- Rossi G, Minervini MM, Melillo L, et al. Predictive role of minimal residual disease and log clearance in acute myeloid leukemia: a comparison between multiparameter flow cytometry and Wilm's tumor 1 levels. Ann Hematol. 2014;93(7):1149–1157.
- Malagola M, Skert C, Borlenghi E, et al. Postremission sequential monitoring of minimal residual disease by WT1 Q-PCR and multiparametric flow cytometry assessment predicts relapse and may help to address risk-adapted therapy in acute myeloid leukemia patients. Cancer Med. 2016;5(2):265–274.
- Rockova V, et al. Early detection of WT1 minimal residual disease predicts outcome in acute myeloid leukemia and identify patients with high risk of relapse independently of allogeneic stem cell transplantation. Blood. 2017;130(Suppl. 1):29.
- Rockova V, Abbas S, Wouters BJ, et al. Risk stratification of intermediate-risk acute myeloid leukemia: Integrative analysis of a multitude of gene mutation and gene expression markers. Leuk Lymphoma. 2011;118(4):1069–1076.
- Miglino M, Colombo N, Pica G, et al. WT1 overexpression at diagnosis may predict favorable outcome in patients with de novo non-M3 acute myeloid leukemia. Leuk Lymphoma. 2011;52(10):1961–1969.
- Bienz M, Ludwig M, Mueller BU, et al. Risk assessment in patients with acute myeloid leukemia and a normal karyotype. Clin Cancer Res. 2005;11(4):1416–1424.
- Baldus CD, Thiede C, Soucek S, et al. BAALC expression and FLT3 internal tandem duplication mutations in acute myeloid leukemia patients with normal cytogenetics: prognostic implications. J Clin Oncol. 2006;24(5):790–797.
- Metzeler KH, Dufour A, Benthaus T, et al. ERG expression is an independent prognostic factor and allows refined risk stratification in cytogenetically normal acute myeloid leukemia: a comprehensive analysis of ERG, MN1, and BAALC transcript levels using oligonucleotide microarrays. J Clin Oncol. 2009;27(30):5031–5038.
- Santamaría C, Chillón MC, García-Sanz R, et al. BAALC is an important predictor of refractoriness to chemotherapy and poor survival in intermediate-risk acute myeloid leukemia (AML). Ann Hematol. 2010;89(5):453–458.
- El-Sharnouby JA, Ahmed LMS, Taha AM, et al. Prognostic significance of CEBPA mutations and BAALC expression in acute myeloid leukemia patients with normal karyotype. Eur J Gen Med. 2010;7(1):17–28.
- Thol F, Damm F, Ludeking A, et al. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J Clin Oncol. 2011;29(21):2889–2896.
- Yoon JH, Kim HJ, Shin SH, et al. Implication of higher BAALC expression in combination with other gene mutations in adult cytogenetically normal acute myeloid leukemia. Leuk Lymphoma. 2014;55(1):110–120.
- Damiani D, Tiribelli M, Franzoni A, et al. BAALC overexpression retains its negative prognostic role across all cytogenetic risk groups in acute myeloid leukemia patients. Am J Hematol. 2013;88(10):848–852.
- Yahya RS, Sofan MA, Abdelmasseih HM, et al. Prognostic implication of BAALC gene expression in adult acute myeloid leukemia. Clin Lab. 2013;59(5-6):621–628.
- Weber S, Haferlach T, Alpermann T, et al. Feasibility of BAALC gene expression for detection of minimal residual disease and risk stratification in normal karyotype acute myeloid leukaemia. Br J Haematol. 2016;175(5):904–916.
- Barjesteh van Waalwijk van Doorn-Khosrovani S, Erpelinck C, van Putten WL, et al. High EVI1 expression predicts poor survival in acute myeloid leukemia: a study of 319 de novo AML patients. Blood. 2003;101(3):837–845.
- Lugthart S, Van Drunen E, Van Norden Y, et al. High EVI1 levels predict adverse outcome in acute myeloid leukemia: prevalence of EVI1 overexpression and chromosome 3q26 abnormalities underestimated. Blood. 2008;111(8):4329–4337.
- Gröschel S, Lugthart S, Schlenk RF, et al. High EVI1 expression predicts outcome in younger adult patients with acute myeloid leukemia and is associated with distinct cytogenetic abnormalities. J Clin Oncol. 2010;28(12):2101–2107.
- Zhu YM, Wang PP, Huang JY, et al. Gene mutational pattern and expression level in 560 acute myeloid leukemia patients and their clinical relevance. J Transl Med. 2017;15(1):178.
- Ommen HB, Nyvold CG, Braendstrup K, et al. Relapse prediction in acute myeloid leukaemia patients in complete remission using WT1 as a molecular marker: development of a mathematical model to predict time from molecular to clinical relapse and define optimal sampling intervals. Br J Haematol. 2008;141(6):782–791.
- Cilloni D, Renneville A, Hermitte F, et al. Real-time quantitative polymerase chain reaction detection of minimal residual disease by standardized WT1 assay to enhance risk stratification in acute myeloid leukemia: a European LeukemiaNet study. J Clin Oncol. 2009;27(31):5195–5201.
- Voso MT, Ottone T, Lavorgna S, et al. MRD in AML: the role of new techniques. Front Oncol. 2019;9:655.
- de Boer EN, Johansson LF, de Lange K. Detection of fusion genes to determine minimal residual disease in leukemia using next-generation sequencing. Clin Chem. 2020;66(8):1084–1092.
- Hokland P, Ommen HB. Towards individualized follow-up in adult acute myeloid leukemia in remission. Blood. 2011;117(9):2577–2584.
- Wu X, Wang H, Deng J, et al. Prognostic significance of the EVI1 gene expression in patients with acute myeloid leukemia: a meta-analysis. Annals of Hematology. 2019;98(11):2485–2496.
- Xiao SJ, Shen JZ, Huang JL, et al. Prognostic significance of the BAALC gene expression in adult patients with acute myeloid leukemia: a meta-analysis. Mol Clin Oncol. 2015;3(4):880–888.
- Yi-Ning Y, Xiao-rui W, Chu-xian Z, et al. Prognostic significance of diagnosed WT1 level in acute myeloid leukemia: a meta-analysis. Ann Hematol. 2015;94(6):929–938.
- Hinai AA, Valk PJ. Review: aberrant EVI1 expression in acute myeloid leukaemia. Br J Haematol. 2016;172(6):870–878.
- Weber S, Alpermann T, Dicker F, et al. BAALC expression: a suitable marker for prognostic risk stratification and detection of residual disease in cytogenetically normal acute myeloid leukemia. Blood Cancer J. 2014;4(1):e173.
- Rossi G, Minervini MM, Carella AM, et al. 2020. Wilms’ tumor gene (WT1) expression and minimal residual disease in acute myeloid leukemia. In: van den Heuvel-Eibrink MM, editor. Wilms tumor. Brisbane: Codon Publications Copyright: The Authors; 2016.