ABSTRACT
Objective
Myelodysplastic syndromes (MDS) constitute a heterogeneous group of clonal hematological diseases. Previous investigations reported that tumor necrosis factor-alpha (TNF-α) gene polymorphisms were associated with MDS susceptibility, but the results remained controversial. Thus, we conducted a meta-analysis to higher elucidate the correlation between TNF-α gene polymorphisms and MDS susceptibility.
Methods
The PubMed, Cochrane Library, Embase, Chinese National Knowledge Infrastructure (CNKI), and Wan Fang databases were searched for eligible literatures published up to July 2021. Pooled odds ratios (ORs) and 95% confidence intervals (CIs) were applied to evaluate the strength of association.
Results
Eight studies involving 1180 MDS patients and 1387 controls were included in this meta-analysis. For the TNF-α G308A polymorphism, we confirmed that the G allele (G versus A: P = 0.001), GG genotypes (GG versus GA: P = 0.005; GG versus GA + AA: P = 0.002), and GG + AA genotypes (GG + AA versus GA: P = 0.008) was significantly associated with decreased MDS susceptibility according to different genetic models. Furthermore, the G308A polymorphism was significantly correlated with decreased occurrence risk of MDS in the Caucasian population as compared with Asians in the above four genetic models (P < 0.05). However, no significant association was observed between the TNF-α G238A polymorphism and MDS risk.
Conclusion
This research showed that TNF-α G308A polymorphism might be a potential biomarker in early clinical screening of MDS, which would contribute to improving the individualized prevention of MDS patients in clinic.
Introduction
Myelodysplastic syndromes (MDS) are a type of heterogeneous clonal blood disease that originates from hematopoietic stem cells. It is characterized by low hematopoietic function, multilineage dysplasia and decrease of peripheral blood cells [Citation1]. Previous studies reported that the incidence of MDS ranged from 5.3 to 13.1 per 100,000 and the incidence of patients with 65 years of age or older was between 75 and 162 per 100,000 in the United States [Citation2–4]. Furthermore, the number of patients with MDS was estimated at 60,000–170,000 and still projected to grow [Citation4], indicating a relatively series situation we are now facing. However, although this disease was extensively investigated previously and many theories regarding its etiology and pathogenesis were proposed [Citation5,Citation6], its early clinical screening and prevention remained poor due to the complexity and diversity of its pathogenic mechanisms and further novel related biomarkers are still needed to be identified [Citation6–8].
Tumor necrosis factor alpha (TNF-α) is a pleiotropic inflammatory cytokine and was demonstrated to be involved in inducing the apoptosis of hematopoietic cells in the process of ineffective hematopoiesis in MDS and eventually affected the development of MDS [Citation9,Citation10]. Furthermore, it was also demonstrated that the levels of serum TNF-α expression significantly increased in MDS patients [Citation11,Citation12]. Therefore, this cytokine was suggested to be one of the pathogenic factors to induce MDS.
Human TNF-α is encoded by the TNF-α gene that is located in human chromosome 6p21.3 within the major histocompatibility complex (MHC) region [Citation13]. It was recently suggested that several TNF-α gene polymorphisms located in the promoter region significantly influenced the expression of TNF-α [Citation14–20] and were hence associated with MDS susceptibility [Citation9,Citation15–17,Citation19,Citation21]. Therefore, these polymorphisms in the promoter region of TNF-α gene are very likely to act as potential biomarkers for MDS prevention, among which the TNF-α −308G > A and −238G > A single nucleotide polymorphisms (SNPs) were the most frequently investigated SNPs regarding their associations with MDS susceptibility [Citation9,Citation15–23]. However, previous studies reported conflicting results about the associations between gene mutation of these SNPs and MDS. For instance, it was shown that the allele of TNF-α G308A was associated with MDS susceptibility [Citation9,Citation19], while several other studies indicated no significant association between this polymorphism and the risk of MDS [Citation18,Citation22,Citation23]. In addition, similar inconsistent results regarding the TNF-α G238A gene polymorphism were also observed [Citation9,Citation17,Citation23]. Therefore, it is a high degree of uncertainty for the accurate influence of TNF-α gene polymorphisms on MDS susceptibility. The inconsistent and inconclusive results were probably attributed to the relatively small sample size of each individual study and the ethnic diversity of research populations, as well as the differences in detecting methodologies. Nevertheless, although a meta-analysis briefly investigated the correlation between TNF-α G308A polymorphism and the risk of bone marrow failure syndrome [Citation20], the detailed discussing and analyzing regarding the correlation between TNF-α gene polymorphisms and MDS were unfortunately missing.
Based on this background, we conducted a comprehensive meta-analysis based on current available studies to evaluate the associations between the abovementioned TNF-α SNPs and MDS susceptibility, expecting to provide reliable evidence-based data for the clinical risk assessment and prevention of MDS.
Materials and methods
Literature search
We performed the Population-Intervention-Comparator-Outcome-Study Design (PICOS) framework to structure the research question and its corresponding literature search. A systematically searched relevant literatures was conducted in PubMed, Cochrane library, Embase, Chinese National Knowledge Infrastructure (CNKI) and Wan Fang database, covering all studies published up to July 2021. The search keywords were Tumor necrosis factor-alpha (TNF-α), Myelodysplastic syndromes (MDS), Single nucleotide polymorphism (SNP) and polymorphism. The search strategies for PubMed as an example was provided in Supplementary Material Table S1. Only published studies with full-text articles were included. In addition, we also checked for relevant publication based on all eligible studies’ bibliographies. All articles involved were reviewed by two independent authors.
Inclusion and exclusion criteria
Studies included in this meta-analysis contained the following inclusion criteria: (a) studies investigating the association between the TNF-α polymorphisms and risk of MDS, (b) published in English or Chinese, (c) case–control or cohort studies, and (d) the preliminary studies provide available data that can be used to calculate the odds ratios (ORs) and 95% confidence intervals (CIs). Studies were excluded under the following criteria (a) systematic reviews, meta-analyses, case reports and obviously irrelevant studies (b) studies lacking full text or not indicating the available data or essential information.
Date extraction
Two authors independently searched the literatures, selected the studies and collected required information and data from each study using a standardized form. Meanwhile, if there were inconsistent results, the differences were resolved through discussion between the two authors or by asking a third author. The information and data were extracted from the original studies, including first author’s name, year of publication, country, ethnicity of study population, TNF-α typing methods, source of controls, sample size and genotype distribution in both case and control groups, and P value for Hardy-Weinberg equilibrium.
Quality assessment
The quality of the included studies was assessed by the Newcastle-Ottawa quality assessment scale (NOS) [Citation24]. The score for assessing quality was based on three aspects, including the selection of the study groups, the comparability of the groups, and exposure in the preliminary researches. The total scores ranged from 0 to 9. Studies with a NOS score of 5 or higher were rated as high-quality researches, while those with a score below 5 were low-quality studies.
Ethical statement
All outcomes and analyses were based on previous ethically approved studies. Thus, no further ethics approval and patient consent were required.
Statistical analysis
We used the Review Manager 5.4 software (Cochrane Collaboration, Oxford, UK) for statistical analysis. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated to investigate the strength of the association between TNF-α gene polymorphism and MDS risk. Genotype frequencies were analyzed by using the allelic, homozygote codominant, heterozygote codominant, dominant, recessive and over-dominant models in this meta-analysis. We calculated the heterogeneities across studies and between subgroups by using Chi-square test and I2 statistic. The random-effect model was applied if the heterogeneity was considered statistically important (I2 > 50% or P < 0.10); otherwise, the fixed-effect model was used. Meanwhile, we also performed subgroups analysis to evaluate the types of ethnicity specific effects in the meta-analysis. Cumulative analysis was applied to investigate changes in estimates of overall effects over time. Sensitive analysis was carried out to test the stability of the results. Funnel plots were used to visually assess possible publications bias.
Results
Study selection and qualitative assessment
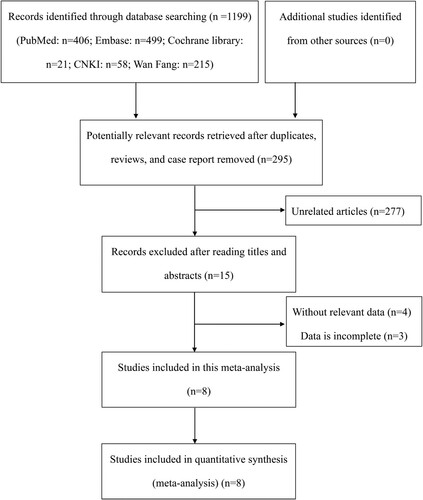
Initially, we performed a search in the electronic databases and screened 295 articles after excluding duplicate publications, reviews, and case reports (). After reading titles and abstracts, 277 unrelated articles were excluded. Then, we reviewed the full text of the remaining 15 articles. Four articles without relevant data about the relationship between TNF-α gene polymorphism and MDS susceptibility and three studies with incomplete data were excluded. Eventually, 7 eligible case–control studies and 1 cohort study were included according to the present inclusion and exclusion criteria, involving 1180 patients and 1387 controls [Citation9,Citation15–19,Citation22,Citation23].
summarized the main characteristics of the 8 included studies. Among these studies, there were 8 comparisons for TNF-α G308A polymorphism and 4 for TNF-α G238A polymorphism. Furthermore, 7 of the included studies were conducted in Caucasian and 1 in Asian. The genetic polymorphisms were assayed by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), polymerase chain reaction-sequence specific primer (PCR-SSP) or TaqMan. Meanwhile, for the source of control group, there were 7 population-based studies and 1 hospital-based study. The NOS score used to assess the quality of case–control studies ranged from 6 to 8 and the cohort study had a quality score of 5.
Table 1. Main characteristic of studies in the meta-analysis.
Association between TNF-α G308A polymorphism and MDS
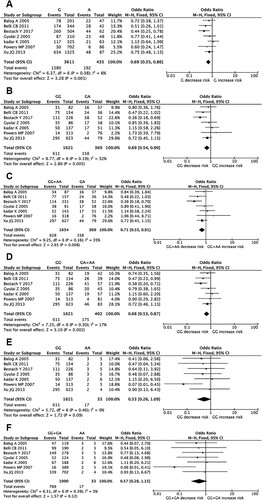
Seven studies involving 786 cases and 1237 controls evaluated the association between TNF-α G308A polymorphism and MDS risk (). When all eligible studies were collected into the meta-analysis, we found that the G allele significantly decreased MDS susceptibility in allelic model (G allele versus A allele, OR = 0.69, 95% CI = 0.55-0.86, P = 0.001, I2 = 6%) ((A)). Meanwhile, the GG genotype significantly decreased MDS susceptibility in heterozygote codominant model (GG versus GA, OR = 0.69, 95% CI = 0.54-0.90, P = 0.005, I2 = 32%) ((B)) and the GG + AA genotypes played same role in over-dominant model (GG + AA versus GA, OR = 0.71, 95% CI = 0.55-0.91, P = 0.008, I2 = 35%) ((C)). In addition, the GG genotype also obviously decreased MDS susceptibility in dominant model (GG versus GA + AA, OR = 0.68, 95% CI = 0.53-0.87, P = 0.002, I2 = 17%) ((D)). However, no significant association was observed in homozygote codominant and recessive models ((E,F)). Additionally, we found no obvious heterogeneity among included studies in all genetic models. As for the included cohort study, the reported results also indicated that high expression of TNF-α G308A genotypes was independently associated with MDS risk, which was consistent with the results of the pooled meta-analysis based on the case–control studies.
Figure 2. Forest plots of pooled ORs and 95% CI for evaluating the association between TNF-α G308A polymorphism and MDS susceptibility in six different genetic models: (A) allelic (G versus A), (B) heterozygous (GG versus GA), (C) over-dominant (GG + AA versus GA), (D) recessive (GG versus GA + AA), (E) homozygous (GG versus AA), (F) dominant (GG + GA versus AA).
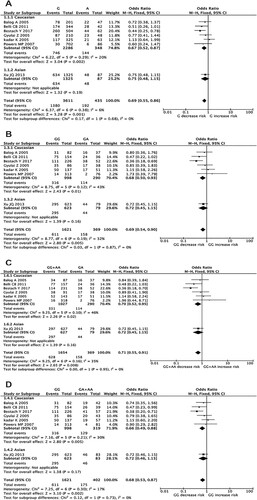
In further subgroup analyses, positive association results were also observed. As for different ethnicities, the G allele obviously decreased the risk of MDS in Caucasians (G versus A, OR = 0.67, 95% CI = 0.52-0.87, P = 0.002; (A) and Supplementary Table S2). Meanwhile, the GG genotype also decreased MDS susceptibility significantly in Caucasian population in heterozygote codominant model (GG versus GA, OR = 0.68, 95% CI = 0.50-0.93, P = 0.01; (B) and Supplementary Table S2) and the GG + AA genotypes played same role in over-dominant model (GG + AA versus GA, OR = 0.70, 95% CI = 0.52-0.95, P = 0.02; (C) and Supplementary Table S2). As for dominant model, the GG genotype also decreased MDS susceptibility obviously in Caucasian population (GG versus GA + AA, OR = 0.66, 95% CI = 0.49-0.88, P = 0.005; (D) and Supplementary Table S2). However, no associations were observed in other genetic models (Supplementary Table S2).
Figure 3. Forest plots of pooled ORs and 95% CI for evaluating the association between TNF-α G308A polymorphism and MDS susceptibility according to different ethnicities in four different genetic models: (A) allelic (G versus A), (B) heterozygous (GG versus GA), (C) over-dominant (GG + AA versus GA), (D) recessive (GG versus GA + AA).
In addition, we found that the GG + AA genotypes decreased the risk of mild MDS significantly as compared with that of severe MDS in over-dominant model (GG + AA versus GA, OR = 0.39, 95% CI = 0.17-0.88, P = 0.02, I2 = 0%) (Supplementary Figure S1C). As compared with health controls, the GG + AA genotypes also decreased the risk of mild MDS obviously in the over-dominant model (GG + AA versus GA, OR = 0.49, 95% CI = 0.26-0.92, P = 0.03, I2 = 0%) (Supplementary Figure S2C). Meanwhile, no significant associations were observed between the TNF-α G308A polymorphism and severe MDS as compared with health controls in all genetic models (Supplementary Figure S3).
Association between TNF-α G238A polymorphism and MDS
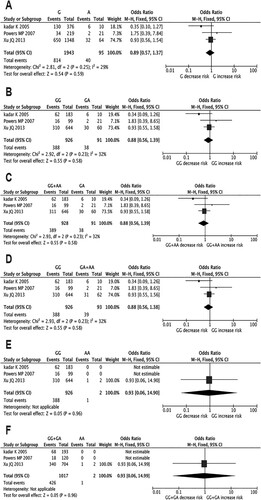
Three studies (428 cases and 592 controls) evaluated the association between TNF-α G238A polymorphism and MDS risk. We did not find any associations between the risk of MDS and genetic polymorphism in all genetic models (). Meanwhile, we also found no obvious heterogeneity among included studies in all genetic models. As for the subgroup analyses, no significant associations were observed according to different ethnicities (Caucasian and Asian) in all genetic models (Supplementary Table S2). However, the results of included cohort study showed that high-expressing TNF-α G238A genotypes was independently associated with MDS risk, which was inconsistent with the results of the pooled meta-analysis based on the case–control studies.
Figure 4. Forest plots of pooled ORs and 95% CI for evaluating the association between TNF-α G238A polymorphism and MDS susceptibility in six different genetic models: (A) allelic (G versus A), (B) heterozygous (GG versus GA), (C) over-dominant (GG + AA versus GA), (D) recessive (GG versus GA + AA), (E) homozygous (GG versus AA), (F) dominant (GG + GA versus AA).
Cumulative analysis and sensitivity analysis
Results of cumulative analysis revealed a consistent trend toward correlation after initial discovery about association between the TNF-α gene polymorphisms and susceptibility of MDS (Supplementary Figures S4 and S5). The overall effects of TNF-α G308A reached statistical significance by 2013 in allelic model (G allele versus A allele, OR = 0.76, 95% CI = 0.59–0.96, P = 0.02, Supplementary Figure S4A) and dominant model (GG versus GA + AA, OR = 0.76, 95% CI = 0.58–1.00, P < 0.05, Supplementary Figure S4B) and had not been materially changed since 2013, with 1 more case–control study and 706 more participants. Furthermore, the overall effects of TNF-α G308A heterozygote codominant model (GG versus GA, OR = 0.69, 95% CI = 0.54-0.90, P = 0.005) (Supplementary Figure S4C) and over-dominant model (GG + AA versus GA, OR = 0.71, 95% CI = 0.55-0.91, P = 0.008) (Supplementary Figure S4D) also achieved statistical significance by 2017. As for other genetic models of G308A and all genetical models of G238A, the overall effects were not statistically significant. These results were consistent with the main outcomes from the present meta-analysis.
In addition, the results of sensitivity showed that no changes of meta-analysis results were observed in any comparisons (Supplementary Figures S6 and S7), which indicated that our findings were statistically reliable. Of note, the heterogeneity was decreased by eliminating each included study.
Publication bias
Potential publication bias in current study was evaluated with funnel plots. The shape of the funnel plots was obviously symmetrical (Supplementary Figures S8 and S9), indicating no significant publication bias of the study.
Discussion
The present study is a relatively comprehensive meta-analysis elucidating the association between TNF-α gene polymorphism and MDS susceptibility. The results demonstrated that the G allele of TNF-α G308A polymorphisms was associated with decreased MDS risk. These findings provided further solid evidence regarding the role of TNF-α polymorphisms during the development of MDS and the potential application of these TNF-α gene loci as biomarkers in early clinical screening and prevention of MDS.
TNF-α has various significant roles in the regulation of the hematopoietic microenvironment [Citation25]. Previous studies demonstrated that this molecule was a proinflammatory cytokine and its expression level was associated with the severity of anemia. Furthermore, population with high TNF-α expression also showed more severe thrombocytopenia [Citation16, Citation26]. Actually, the severity of anemia and thrombocytopenia would eventually affect the progress of MDS, which strengthened the association between TNF-α and MDS. The underlying mechanisms of TNF-α in the hematopoietic microenvironment were reported in many studies. It was reported that TNF-α mediated negative feedback regulation of endogenous erythropoietin (EPO), inhibition of hematopoietic function and apoptosis of intramedullary hematopoietic progenitor cells, suggesting a potential effect on the development and progress of MDS [Citation13, Citation18, Citation25, Citation27–29]. Recently, the significant correlation between the level of serum TNF-α expression and MDS was also demonstrated [Citation11, Citation12]. As the expression of TNF-α was continuously suggested to be influenced by certain functional TNF-α gene polymorphisms located in the promoter region [Citation14–20], these polymorphisms might eventually affect the hematopoietic microenvironment of the pathogenesis of MDS.
It should be mentioned that one previous meta-analysis also reported the correlation between the TNF-α G308A polymorphism and MDS risk [Citation20]. However, its results showed that no significant association was observed. In this previous study, only four genetic models were employed, while the allelic and over-dominant gene models were missing. As a matter of fact, the allelic model is the most ideal and widely employed model to directly reflect the effect of base mutation on the expression and function of an allele. In addition, the over-dominant model is also a commonly employed genetic model in previous reports, based on which the heterozygote superiority profile of a certain polymorphism would be better clarified. Therefore, we added these two gene models for further exploring the association between TNF-α gene polymorphisms and MDS risk in the present study. As a result, we found that the G allele in allelic model and the GG + AA genotypes in over-dominant model significantly decreased the MDS susceptibility, respectively. These findings for the first time confirmed that the G allele of G308A polymorphism was associated with decreased risk of MDS according to the allelic model. Furthermore, that the GA genotype in the over-dominant model increased the risk of MDS also indicated heterozygote superiority of this polymorphism and the heterozygotes of G308A polymorphism (GA genotype) might exhibit more elastic and broader gene expression profiles in terms of specific stimuli or cyto-specificity [Citation30,Citation31]. In this regard, the heterozygote superiority of this polymorphism should be paid more attention in future studies regarding its association with risk of diseases.
According to previous investigations, the A allele of G308A polymorphism demonstrated a 6-9-fold higher transcriptional activation than the common G allele [Citation32,Citation33]. Furthermore, it was demonstrated that the G allele decreased the binding affinity of nuclear factor to the promoter to reduce TNF-α transcription activity and mRNA expression, which regulated the expression level of TNF-α in the hematopoietic microenvironment and ultimately affected the development of MDS [Citation22,Citation25,Citation34–36]. Meanwhile, in our study, the G allele of TNF-α G308A polymorphisms was found to be correlated with decreased significantly risk of MDS, especially in the Caucasian population, which was consistent with the above report. Nevertheless, owing to the complicated effects of TNF-α on the pathogenesis of MDS, further extensive functional investigations are required to clarify the exact underlying mechanisms of how this SNP affects TNF-α expression and influences the development of MDS.
As for TNF-α G238A polymorphism, it is characterized by a mutation from guanine (G) to adenine (A) at position 238 in the promoter region [Citation37]. Previous studies demonstrated that the polymorphism of TNF-α G238A changed the transcriptional activity of TNF-α and affected its expression level, which eventually influenced the pathological process of MDS [Citation17,Citation38]. However, no significant correlation between this SNP and MDS susceptibility was observed in our study, which might be due to the relatively limited number of original studies and sample sizes included. Therefore, further large-scale clinical studies with lager sample sizes from different ethnic populations are required to confirm the relationship between TNF-α G238A polymorphism and MDS risk.
Although we performed a relatively comprehensive analysis of the association between TNF-α gene polymorphisms and MDS risk, some limitations still need to be addressed. First, our meta-analysis only focused on the articles from English and Chinese databases, which might lead to a potential language bias. Second, the sample size of certain SNPs in the present study was relatively limited, which might cause false positives and underpowered results. Third, only published studies were included in the meta-analysis, the unpublished ones in remote countries might be missed. Fourth, most of the studies involved Caucasian populations and further large-scale studies were required from different ethnic populations. Finally, lack of original data limited our further assessment of other confounding factors, such as age, gender, environment factors as well as lifestyle, which might affect the risk of MDS.
In conclusion, our meta-analysis demonstrated that the G allele, GG genotypes and GG + AA genotypes of TNF-α G308A polymorphisms were associated with decreased MDS risk in the allelic, heterozygote codominant, dominant and over-dominant model. Furthermore, the TNF-α G308A polymorphism was also associated with the severity of MDS. However, the TNF-α G238A polymorphism had no significant influence on the risk of MDS. These findings showed that the G308A polymorphisms in the TNF-α gene might act as biomarker, which would contribute to early clinical screening and prevention of MDS.
Supplemental Material
Download PDF (8.3 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Nishino HT, Chang CC. Myelodysplastic syndromes: clinicopathologic features, pathobiology, and molecular pathogenesis. Arch Pathol Lab Med. 2005;129(10):1299–1310. doi:https://doi.org/10.5858/2005-129-1299-MSCFPA.
- Ma X, Does M, Raza A, et al. Myelodysplastic syndromes: incidence and survival in the United States. Cancer. 2007;109(8):1536–1542. doi:https://doi.org/10.1002/cncr.22570.
- Rollison DE, Howlader N, Smith MT, et al. Epidemiology of myelodysplastic syndromes and chronic myeloproliferative disorders in the United States, 2001-2004, using data from the NAACCR and SEER programs. Blood. 2008;112(1):45–52. doi:https://doi.org/10.1182/blood-2008-01-134858.
- Cogle CR. Incidence and burden of the myelodysplastic syndromes. Curr Hematol Malig Rep. 2015;10(3):272–281. doi:https://doi.org/10.1007/s11899-015-0269-y.
- Hasserjian RP. Myelodysplastic syndrome updated. Pathobiology. 2019;86(1):7–13. doi:https://doi.org/10.1159/000489702.
- Haferlach T. The molecular pathology of myelodysplastic syndrome. Pathobiology. 2019;86(1):24–29. doi:https://doi.org/10.1159/000488712.
- Wang C, Yang Y, Gao S, et al. Immune dysregulation in myelodysplastic syndrome: clinical features, pathogenesis and therapeutic strategies. Crit Rev Oncol Hematol. 2018;122:123–132. doi:https://doi.org/10.1016/j.critrevonc.2017.12.013.
- Gill H, Leung AY, Kwong YL. Molecular and cellular mechanisms of myelodysplastic syndrome: implications on targeted therapy. Int J Mol Sci. 2016;17(4):440. doi:https://doi.org/10.3390/ijms17040440.
- Xu JQ, Wang JY, Qin TJ, et al. [Relationship between polymorphisms of tumor necrosis factor alpha gene and primary myelodysplastic syndromes]. Zhonghua Xue Ye Xue Za Zhi. 2013;34(10):873–876. doi:https://doi.org/10.3760/cma.j.issn.0253-2727.2013.10.010.
- Uversky VN, El-Baky NA, El-Fakharany EM, et al. Functionality of intrinsic disorder in tumor necrosis factor-α and its receptors. FEBS J. 2017;284(21):3589–3618. doi:https://doi.org/10.1111/febs.14182.
- Gersuk GM, Beckham C, Loken MR, et al. A role for tumour necrosis factor-alpha, Fas and Fas-ligand in marrow failure associated with myelodysplastic syndrome. Br J Haematol. 1998;103(1):176–188. doi:https://doi.org/10.1046/j.1365-2141.1998.00933.x.
- Selleri C, Sato T, Anderson S, et al. Interferon-gamma and tumor necrosis factor-alpha suppress both early and late stages of hematopoiesis and induce programmed cell death. J Cell Physiol. 1995;165(3):538–546. doi:https://doi.org/10.1002/jcp.1041650312.
- El-Tahan RR, Ghoneim AM, El-Mashad N. TNF-α gene polymorphisms and expression. Springerplus. 2016;5(1):1508. doi:https://doi.org/10.1186/s40064-016-3197-y.
- Kennedy JA, Ebert BL. Clinical implications of genetic mutations in myelodysplastic syndrome. J Clin Oncol. 2017;3(9):968–974. doi:https://doi.org/10.1200/JCO.2016.71.0806.
- Bestach Y, Nagore VP, Flores MG, et al. Influence of TNF and IL6 gene polymorphisms on the severity of cytopenias in Argentine patients with myelodysplastic syndromes. Ann Hematol. 2017;96(8):1287–1295. doi:https://doi.org/10.1007/s00277-017-3036-4.
- Belli CB, Bestach Y, Sieza Y, et al. The presence of -308A TNFα is associated with anemia and thrombocytopenia in patients with myelodysplastic syndromes. Blood Cells Mol Dis. 2011;47(4):255–258. doi:https://doi.org/10.1016/j.bcmd.2011.09.003.
- Powers MP, Nishino H, Luo Y, et al. Polymorphisms in TGFbeta and TNFalpha are associated with the myelodysplastic syndrome phenotype. Arch Pathol Lab Med. 2007;131(12):1789–1793. doi:https://doi.org/10.5858/2007-131-1789-PITATA.
- Balog A, Borbényi Z, Gyulai Z, et al. Clinical importance of transforming growth factor-beta but not of tumor necrosis factor-alpha gene polymorphisms in patients with the myelodysplastic syndrome belonging to the refractory anemia subtype. Pathobiology. 2005;72(3):165–170. doi:https://doi.org/10.1159/000084121.
- Parnes A, Nikiforow S, Berliner N, et al. Single nucleotide polymorphisms in the human TNF gene are associated with anaemia and neutropenia in a cohort of patients with de novo myelodysplastic syndrome. Br J Haematol. 2010;150(6):700–701. doi:https://doi.org/10.1111/j.1365-2141.2010.08254.x.
- Chen W, Zhu H, Yu L, et al. TNF-α -308 G > A polymorphism and risk of bone marrow failure syndrome: A meta-analysis. Gene. 2015;565(1):1–8. doi:https://doi.org/10.1016/j.gene.2015.04.038.
- Gowans D, O'Sullivan A, Rollinson S, et al. Allele and haplotype frequency at human leucocyte antigen class I/II and immunomodulatory cytokine loci in patients with myelodysplasia and acute myeloid leukaemia: in search of an autoimmune aetiology. Br J Haematol. 2002;117(3):541–545. doi:https://doi.org/10.1046/j.1365-2141.2002.03452.x.
- Gyulai Z, Balog A, Borbényi Z, et al. Genetic polymorphisms in patients with myelodysplastic syndrome. Acta Microbiol Immunol Hung. 2005;52(3-4):463–475. doi:https://doi.org/10.1556/AMicr.52.2005.3-4.15.
- Kádár K, Demeter J, Andrikovics H, et al. TNF-alpha promoter gene polymorphism in patients with myelodysplastic syndrome. Acta Haematol. 2005;113(4):262–264. doi:https://doi.org/10.1159/000084681.
- The Newcastle-Ottawa Scale (NOS) for assessing the quality of non-randomised studies in meta-analyses [EB/OL]. http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp.
- Moldoveanu E, Moicean A, Vidulescu C, et al. Apoptotic rate in patients with myelodisplastic syndrome treated with modulatory compounds of pro-apoptotic cytokines. J Cell Mol Med. 2003;7(3):313–321. doi:https://doi.org/10.1111/j.1582-4934.2003.tb00232.x.
- Kiss C, Benko I, Kovács P. Leukemic cells and the cytokine patchwork. Pediatr Blood Cancer. 2004;42(2):113–121. doi:https://doi.org/10.1002/pbc.10436.
- Stifter G, Heiss S, Gastl G, et al. Over-expression of tumor necrosis factor-alpha in bone marrow biopsies from patients with myelodysplastic syndromes: relationship to anemia and prognosis. Eur J Haematol. 2005;75(6):485–491. doi:https://doi.org/10.1111/j.1600-0609.2005.00551.x.
- Serio B, Selleri C, Maciejewski JP. Impact of immunogenetic polymorphisms in bone marrow failure syndromes. Mini Rev Med Chem. 2011;11(6):544–552. doi:https://doi.org/10.2174/138955711795843356.
- Camara-Lemarroy CR, Salas-Alanis JC. The role of tumor necrosis factor-α in the pathogenesis of vitiligo. Am J Clin Dermatol. 2013;14(5):343–350. doi:https://doi.org/10.1007/s40257-013-0039-3.
- Ma HJ, Fu SC, Xiao A, et al. The associations of CYP19A1 rs700518 polymorphism with bone mineral density and risk of osteoporosis: a meta-analysis. Gynecol Endocrinol. 2020;36(7):626–631. doi:https://doi.org/10.1080/09513590.2020.1727431.
- Huebner C, Browning BL, Petermann I, et al. Genetic analysis of MDR1 and inflammatory bowel disease reveals protective effect of heterozygous variants for ulcerative colitis. Inflamm Bowel Dis. 2009;15(12):1784–1793. doi:https://doi.org/10.1002/ibd.21019.
- Kim JH, Jeon YJ, Rah H, et al. Tumor necrosis factor-alpha promoter polymorphisms are associated with idiopathic primary ovarian insufficiency in Korean women. Fertil Steril. 2012;98(5):1260–1265. e52. doi:https://doi.org/10.1016/j.fertnstert.2012.07.1111.
- Kampman O, Anttila S, Illi A, et al. Interaction of tumor necrosis alpha - G308A and epidermal growth factor gene polymorphisms in early-onset schizophrenia. Eur Arch Psychiatry Clin Neurosci. 2005;255(4):279–283. doi:https://doi.org/10.1007/s00406-004-0560-8.
- Li Y, Lin Y. Tumor necrosis factor alpha-308G/A polymorphism and the risk of multiple myeloma: a meta-analysis of pooled data from twelve case-control studies. Turk J Haematol. 2019;36(2):72–80. doi:https://doi.org/10.4274/tjh.galenos.2019.2018.0238.
- Abraham LJ, Kroeger KM. Impact of the -308 TNF promoter polymorphism on the transcriptional regulation of the TNF gene: relevance to disease. J Leukoc Biol. 1999;66(4):562–566. doi:https://doi.org/10.1002/jlb.66.4.562.
- Wang X, Zhang H, Cao X, et al. Gene-disease association study of tumor necrosis factor-α G-308A gene polymorphism with risk of major depressive disorder: a systematic review and meta-analysis. Brain Behav. 2020;10(6):e01628. doi:https://doi.org/10.1002/brb3.1628.
- Kaluza W, Reuss E, Grossmann S, et al. Different transcriptional activity and in vitro TNF-alpha production in psoriasis patients carrying the TNF-alpha 238A promoter polymorphism. J Invest Dermatol. 2000;114(6):1180–1183. doi:https://doi.org/10.1046/j.1523-1747.2000.00001.x.
- Tian X, Ma P, Sui C, et al. Comprehensive assessment of the association between tumor necrosis factor alpha G238A polymorphism and liver cancer risk. Tumour Biol. 2014;35(1):103–109. doi:https://doi.org/10.1007/s13277-013-1012-8.