ABSTRACT
Objective
Acute myeloid leukemia (AML) is a common blood cancer associated with poor prognosis and high mortality. In this study, we investigated the role and underlying mechanism of action of circ_0104700 in the pathogenesis of AML.
Methods
Circ_0104700 was screened from the GEO database and detected in AML samples and cell lines. The effect of circ_0104700 on AML was analyzed using a methylcellulose colony assay, CCK-8 assay, and cell cycle and apoptosis analyses. The mechanism was explored using bioinformatic analysis, quantitative reverse transcription-PCR, dual-luciferase reporter assays, northern blotting and western blot analysis in AML cells.
Results
Circ_0104700 expression was higher in AML patients and AML cell lines. Functionally, circ_0104700 depletion attenuated cell viability and induced apoptosis in MV-4-11 and Kasumi-1 cells. Circ_0104700 depletion enhanced the G0/G1-phase proportion but reduced the proportion of S-phase cells in MV-4-11 and Kasumi-1 cells. circ_0104700 served as a competing endogenous RNA of miR-665 and enhanced MCM2 expression by sponging miR-665 in MV-4-11 and Kasumi-1 cells. Silencing circ_0104700 repressed the proliferation and cell cycle and induced apoptosis of MV-4-11 and Kasumi-1 cells by inhibiting miR-665. MCM2 depletion alleviated the proliferation and cell cycle and enhanced the apoptosis of MV-4-11 and Kasumi-1 cells by inactivating JAK/STAT signaling. JAK/STAT signaling was involved in circ_0104700-mediated malignant phenotypes of MV-4-11 and Kasumi-1 cells.
Conclusion
circ_0104700 contributed to AML progression by enhancing MCM2 expression by targeting miR-665. Our findings provide novel potential therapeutic targets for AML, including circ_0104700, miR-665, and MCM2.
Introduction
Acute myeloid leukemia (AML) is lethal malignancy described by deleterious proliferation and a loss of normal differentiation ability [Citation1,Citation2]. AML has been recognized as a treatable cancer with the advancement of bone marrow transplantation, targeted therapies, immunotherapy, and chemotherapy; however, recurrence remains a clinical challenge for AML patients with poor prognosis and outcomes [Citation3–6]. Therefore, there is an urgent need to understand the molecular mechanisms underlying the progression of AML. Circular RNAs (circRNAs) are non-coding RNAs with circular structures that play an essential role in cancer initiation and development [Citation7–9]. Several circular RNAs, such as circ_0004277 and DLEU2, regulate AML [Citation10,Citation11]. Meanwhile, circ_0104700 is a poorly understood circular RNA with the gene symbol HOMER2, and its function in AML remains unknown.
Minichromosome maintenance protein 2 (MCM2) serves as a cell proliferation biomarker and is an oncogene in multiple tumors, such as breast, colon, and lung cancer [Citation12–14]. Several microRNAs (miRNAs), including miR-1296 and miR-195-5p, have been reported to target MCM2 to suppress cancer progression [Citation15,Citation13]. In addition, miR-665 has been identified as a tumor suppressor that inhibits tumorigenesis by modulating different processes, such as epithelial–mesenchymal transition, proliferation, apoptosis, and migration of cancer cells [Citation16–18]. However, the effectS of MCM2 and miR-665 on AML development and their correlation with circ_0104700 have not yet been reported.
In this study, we focused on the role of the circular RNA circ_0104700 in AML development. We elucidated the critical role of circ_0104700 in promoting malignant progression of AML by targeting the miR-665/MCM2 axis.
Materials and methods
AML clinical samples
Between March 2020 and December 2021, peripheral blood mononuclear cells (MCs) were obtained from 26 untreated patients with primary AML and 20 healthy patients at our hospital. The samples were under the written approval by the patients and healthy cases. This study conformed to the experimental guidelines of the World Medical Association and the Ethics Committee of our hospital. Details of the clinical characteristics of AML patients are listed in .
Table 1. Clinical characteristics of newly diagnosed AML patients.
Cell culture and transfection
HS-5, MV-4-11, Kasumi-1, HL-60, and AML2 cell lines were acquired from the American Type Culture Collection (ATCC) and cultured according to ATCC recommendation using culture medium, fetal bovine serum (FBS), and penicillin/streptomycin (Gibco, USA). We synthesized and purchased siRNAs targetingcirc_0104700 and MCM2, pcDNA3.1-MCM2, miR-665 mimic and inhibitor (GenePharma, China). Lipofectamine 3000 (Invitrogen, USA) was used to transfect MV-4-11 and Kasumi-1 cells.
Quantitative reverse transcription-PCR (qRT-PCR)
Total RNAs were isolated using TRIZOL (Invitrogen, USA) according to the manufacturer’s protocol. First-strand cDNA was prepared according to the manufacturer's instructions (TaKaRa, China). RT-qPCR were applied to analyze MCM2, miR-665, and circ_0104700 expression in the AML samples and cell lines. The expression was quantified by the cycle threshold (CT) (2 − ΔΔCT) method relative to GAPDH or U6.
CCK-8 assay
For the cell viability analysis, MV-4-11 and Kasumi-1 cells (2 × 104 cells/well) were seeded in 96-well plates and incubated for 48 h. Cell Count Kit-8 (CCK-8) solution (Sigma-Aldrich) was added to the culture medium and incubated for 4 h. The absorbance at 450 nm was measured using a microplate reader.
Methylcellulose colony assay
MV-4-11 and Kasumi-1 cells were seeded in triplicate into 24-well plate in semisolid methylcellulose medium (H4100 Methocult, StemCell, Canada) supplemented with a final serum concentration of 10% FBS (Gibco, USA) and 1% penicillin/streptomycin (Sigma, USA) and incubated for 14 days. On day 14, the colonies were counted and evaluated microscopically.
Cell cycle and apoptosis
Cell cycle detection was performed using the staining of RNA enzyme containing iodide (PI, Sigma, USA). Apoptosis measurement was performed using an Annexin V-FITC Apoptosis Detection kit (Sigma, USA). Cell cycle and apoptosis were analyzed using flow cytometry (BD, USA).
Dual luciferase reporter assays
The circ_0104700 sequences containing wild-type or mutant miR-665 binding sites were respectively cloned into pmirGLO dual-luciferase vectors to form the circ_0104700-wild-type vector (circ_0104700-WT) and the circ_0104700-mutated type vector (circ_0104700-Mut). Similarly, the vectors of MCM2-WT and MCM2-Mut were constructed based on the above method. Subsequently, these vectors were co-transfected into MV-4-11 and Kasumi-1 cells with miR-665 mimics or mimics NC utilizing Lipofectamine 3000 (Invitrogen, USA) as per the manufacturer’s procedures. The luciferase activities were detected using the Dual Luciferase Assays (Promega, USA). The data was normalized to the Renilla luciferase activities.
Northern blotting
DIG Northern Starter kit (Roche, Switzerland) was applied in northern blotting. Total RNA samples were denatured in formaldehyde, resolved on a 1% agarose-formaldehyde gel, and then transferred to the Hybond-N + nylon membranes (Beyotime, Jiangsu, China). After crosslinking by ultraviolet irradiation (265 nm; 0.15 J/cm2), the membrances were hybridized with a biotin-labeled DNA probe. At last, RNA signals were measured using a Chemiluminescent Nucleic Acid Detection Module (Thermo, USA).
Western blot analysis
Total proteins were extracted from cells using RIPA buffer (CST, USA) and quantified using a BCA Protein Quantification Kit (Abbkine, USA). Equal proteins were subjected in 10% SDS-PAGE and transferred into PVDF membrane (Millipore, USA). The membrane was incubated with primary antibodies (JAK2, p-JAK2, STAT3, p-STAT3, and GAPDH; CST, USA) at 4°C overnight. The membranes were then incubated with secondary antibodies (CST, USA) for 1.5 h at room temperature. The immunoreactivity was visualized by ECL kits (Sigma, USA).
Bioinformatics analysis
The microarray data of GSE94591, GSE142700, GSE49665, and GSE22187 from GEO database was obtained to analyze the expression of circular RNA, miRNAs, and mRNAs [Citation10,Citation19,Citation20]. The expression of circ_0104700, miR-665, and MCM2 was analyzed in the AML samples from TCGA database. The Circular RNA Interactome online database and Starbase online database were used to predict the circ_0104700-targeted miRNAs and miR-665-targeted genes. KEGG downstream pathway prediction for MCM2 was performed using the GSEA software.
Statistical analysis
Data were assessed by unpaired Student’s t-test and one-way ANOVA using SPSS 18.0 software based on mean ± SD. The correlation of miR-665 with circ_0104700 and MCM2 was analyzed using Pearson’s correlation coefficient. Differences were considered statistically significant at P < 0.05.
Results
Circ-0104700 is highly expressed in AML samples and cells
We selected the circular RNA microarray from the GEO database GSE94591 and identified aberrant expression of circ_0104700 (LogFC > 4, P < 0.001) in the AML patients relative to healthy controls (A). We validated that circ_0104700 was highly expressed in AML samples from TCGA database (B). qRT-PCR assays presented that circ_0104700 expression was higher in AML patients than in normal patients (C). AML cell lines, including MV-4-11, Kasumi-1, HL-60, and AML2, showed elevated expression of circ_0104700 compared tobone marrow stromal HS-5 cells (D). The MV-4-11 and Kasumi- 1 cell lines with the highest circ_0104700 expression were selected for subsequent studies.
Figure 1. Circ_0104700 was highly expressed in AML samples and cells. (A) Analysis of GSE94591 microarray to select the differential circRNA with LogFC > 4 and P < 0.001. (B) TCGA database showing high expression of circ_0104700 in AML. (C) RT-qPCR was used to detect the expression of circ_0104700 in normal cases (n = 20) and AML patients (n = 26). (D) RT-qPCR was used to detect the expression of circ_0104700 in human bone marrow stromal cells (HS-5) and AML cells (MV-4-11, Kasumi-1, HL-60, and AML2). Compared with Control group or HS-5 cells, **P < 0.01.
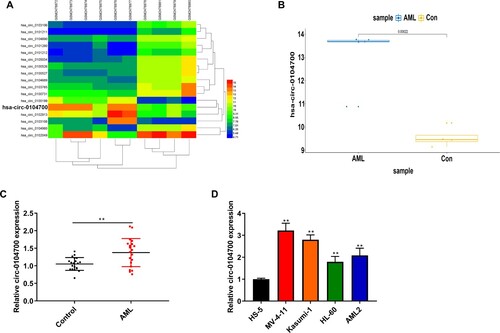
Silencing of circ_0104700 represses proliferation and the cell cycle and induces apoptosis in AML cells
We silenced circ_0104700 expression using two siRNAs and confirmed that these siRNAs reduced circ_0104700 expression in MV-4-11 and Kasumi-1 cells (A). The viability of MV-4-11 and Kasumi-1 cells was attenuated by circ_0104700 depletion (B). Moreover, circ_0104700 depletion decreased the number of MV-4-11 and Kasumi-1 cell colonies (C). The circ_0104700 depletion enhanced the G0/G1-phase proportion but reduced the proportion of MV-4-11 and Kasumi-1 cells in the S-phase (D). Besides, the apoptosis of MV-4-11 and Kasumi-1 cells was induced by circ_0104700 knockdown (E).
Figure 2. Silencing of circ_0104700 repressed proliferation, cell cycle and induced apoptosis in AML cells. (A) Circ_0104700 expression in MV-4-11 and Kasumi-1 cells was detected by RT-qPCR. (B) MV-4-11 and Kasumi-1 cell viabilities was detected by CCK-8 assay. (C) MV-4-11 and Kasumi-1 cell colony ability was detected by colony formation assay. (D) MV-4-11 and Kasumi-1 cell cycle was detected by flow cytometry. (E) MV-4 11 and Kasumi-1 cell apoptosis was detected by flow cytometry. Compared with si-NC, **P < 0.01.
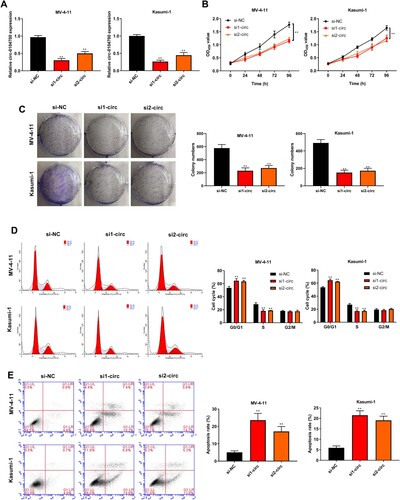
Circ_0104700 negatively regulates miR-665 expression
We predicted circ_0104700-targeted miRNAs using the Circular RNA Interactome online database and identified 52 potential miRNAs. AIntersection analysis of these 52 miRNAs in the miRNA microarray from the GEO databases GSE142700 and GSE49665 identified miR-665, and we synthesized circ_0104700 with the miR-665-binding site mutant (A and B). Treatment with the miR-665 mimic repressed the luciferase activity of wild type circ_0104700 but not of the circ_0104700 mutant in MV-4-11 and Kasumi-1 cells (C). Silencing circ_0104700 up-regulated miR-665 expression in cells (D). Moreover, miR-665 was down-regulated in AML samples from TCGA database (E). The miR-665 expression was lower in AML patients than in normal patients (F). The circ_0104700 and miR-665 expression were negatively correlated in AML patients (G). The binding of circ_0104700 to miR-665 was clearly observed via utilizing northern hybridization analysis of miR-665 (H).
Figure 3. Circ_0104700 negatively regulated miR-665 expression. (A) The predicted targeted miRNAs of circ_0104700 using Circular RNA Interactome online database were intersected with the differentially expressed miRNAs in the microarray analysis of GSE142700 and GSE49665 from GEO database. (B) The binding site of circ_0104700 with miR-665. (C) Luciferase activity was detected by dual luciferase reporter gene assays. (D) miR-665 expression in MV-4-11 and Kasumi-1 cells was detected by RT-qPCR. (E) TCGA database showed the low expression of miR-665 in AML samples. (F) RT-qPCR was used to detect the expression of miR-665 in normal cases (n = 20) and AML patients (n = 26). (G) Correlation analysis of circ_0104700 with miR-665 in AML patients. (H) Level of miR-665 after being pulled down by the circular probe for circ_0104700 and a random probe measured utilizing northern blotting. Compared with Control, si-NC or mimic NC group, **P < 0.01.
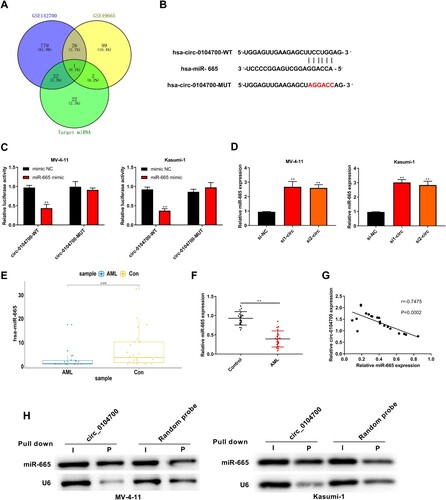
Silencing circ_0104700 inhibits proliferation and the cell cycle and induces apoptosis in AML cells by regulating miR-665
Next, we confirmed that the depletion of circ_0104700 enhanced miR-665 expression, which was blocked by the miR-665 inhibitor (A). The MV-4-11 and Kasumi-1 cell viability and colony number were reduced by circ_0104700 depletion but were elevated by the miR-665 inhibitor, and the miR-665 inhibitor rescued the circ_0104700 depletion-induced inhibition of cell viability and colony formation ability (B and 4C). The circ_0104700 depletion enhanced the G0/G1-phase proportion but reduced the proportion of S-phase cells in MV-4-11 and Kasumi-1 cells, which was reversed by the miR-665 inhibitor (D). In addition, the apoptosis of MV-4-11 and Kasumi-1 cells was increased by circ_0104700 knockdown, whereas the miR-665 inhibitor reversed this effect and attenuated the function of circ_0104700 knockdown in the cells (E).
Figure 4. Silencing circ_0104700 inhibited the proliferation, cell cycle and induced apoptosis in AML cells by regulating miR-665. (A) miR-665 expression in MV-4-11 and Kasumi-1 cells was detected by qRT-PCR. (B) MV-4-11 and Kasumi-1 cell viability was detected by CCK-8 assay. (C) MV-4-11 and Kasumi-1 cell colony ability was detected by colony formation assay. (D) MV-4-11 and Kasumi-1 cell cycle was detected by flow cytometry. (E) MV-4 11 and Kasumi-1 cell apoptosis was detected by flow cytometry. Compared with si-NC + inhibitor NC group, *P < 0.05, **P < 0.01; compared with si1-circ group, #P < 0.05, ##P < 0.01; compared with inhibitor group, &P < 0.05, &&P < 0.01.
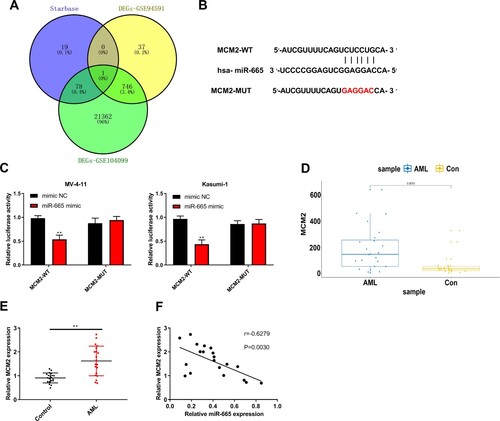
MiR-665 negatively regulates MCM2 expression
We predicted the miR-665-targeted genes using Starbase (A). An overlap analysis of these genes with microarray profiling from the GEO databases GSE94591 and GSE22187 identified MCM2, and we synthesized MCM2 with the miR-665-binding site mutant (A and B). Luciferase activity of wild type MCM2 was attenuated by the miR-665 mimic in MV-4-11 and Kasumi-1 cells (C). MCM2 expression was elevated in AML samples from TCGA database (D). AML patients showed a higher expression of MCM2 relative to normal cases (E). MCM2 and miR-665 expression were negatively correlated in patients with AML (F).
Figure 5. MiR-665 negatively regulated MCM2 expression. (A) The predicted targeted genes of miR-665 using Starbase online database were intersected with the differentially expressed genes in the microarray analysis of GSE94591 and GSE104099 from GEO database. (B) The binding site of MCM2 with miR-665. (C) Luciferase activity was detected by dual luciferase reporter gene assays. (D) TCGA database showed the high expression of MCM2 in AML samples. (E) RT-qPCR was used to detect the expression of MCM2 in normal cases (n = 20) and AML patients (n = 26). (F) Correlation analysis of MCM2 with miR-665 in AML patients. Compared with Control or miminc NC group, **P < 0.01.
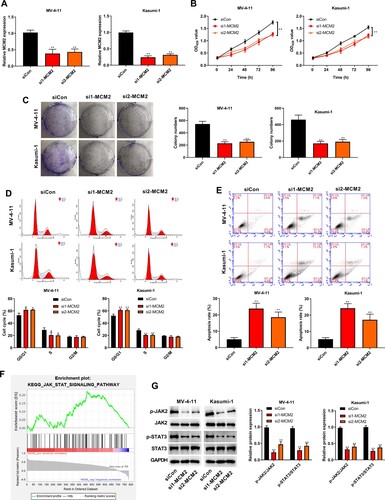
Silencing of MCM2 alleviates the proliferation and cell cycle of AML cells and induces apoptosis by regulating the JAK/STAT signaling
Next, we depleted MCM2 expression using two siRNAs and validated siRNA efficiency in MV-4-11 and Kasumi-1 cells (A). The viability of MV-4-11 and Kasumi-1 cells was reduced by MCM2 silencing (B). Similarly, the silence of MCM2 reduced the colony number of MV-4-11 and Kasumi-1 cells (C). MCM2 depletion increased the G0/G1-phase proportion but decreased the proportion of cells in the S-phase in MV-4-11 and Kasumi-1 cells (D). The apoptosis of MV-4-11 and Kasumi-1 cells was promoted by MCM2 knockdown (E). Moreover, we identified the JAK/STAT signaling pathway downstream of MCM2 by KEGG pathway analysis using GSEA (F). Phosphorylation of JAK2 and STAT3 was suppressed by MCM2 knockdown in MV-4-11 and Kasumi-1 cells (G).
Figure 6. Silencing of MCM2 alleviated the proliferation and cell cycle of AML cells and induces apoptosis by regulating the JAK/STAT signaling. (A) MCM2 expression in MV-4-11 and Kasumi-1 cells was detected by RT-qPCR. (B) MV-4-11 and Kasumi-1 cell viabilities was detected by CCK-8 assay. (C) MV-4-11 and Kasumi-1 cell colony ability was detected by colony formation assay. (D) MV-4-11 and Kasumi-1 cell cycle was detected by flow cytometry. (E) MV-4 11 and Kasumi-1 cell apoptosis was detected by flow cytometry. (F) KEGG downstream pathway prediction for MCM2 using GSEA software. (G) Western blot analysis was used to detected the expression of JAK/STAT signaling-related proteins in MV-4-11 and Kasumi-1 cells. Compared with siCon group, *P < 0.05, **P < 0.01.
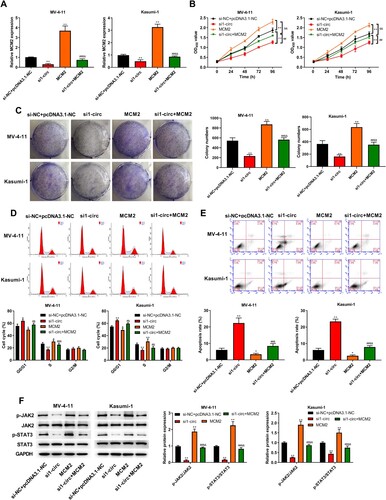
Silencing circ_0104700 inhibits proliferation and the cell cycle and induces apoptosis in AML cells by regulating MCM2
The expression of MCM2 was reduced by circ_0104700 siRNA, while the expression of MCM2 was rescued by MCM2 overexpression plasmid (A). MV-4-11 and Kasumi-1 cell viability and colony number were decreased by circ_0104700 depletion, but were increased by MCM2 overexpression, and MCM2 overexpression rescued the circ_0104700 depletion-induced suppression of cell viability and colony formation ability (B and 7C). The circ_0104700 depletion induced the G0/G1-phase proportion but attenuated the S-phase proportion of MV-4-11 and Kasumi-1 cells, which was reversed by MCM2 overexpression (D). Apoptosis of MV-4-11 and Kasumi-1 cells was enhanced by circ_0104700 knockdown, and MCM2 overexpression reversed this effect and attenuated the function of circ_0104700 knockdown in the cells (E). Meanwhile, the circ_0104700 depletion repressed JAK2 and STAT3 phosphorylation in MV-4-11 and Kasumi-1 cells and MCM2 overexpression rescued this phenotype (F).
Figure 7. Silencing circ_0104700 inhibited proliferation, cell cycle and induced apoptosis in AML cells by regulating MCM2. (A) MCM2 expression in MV-4-11 and Kasumi-1 cells was detected by RT-qPCR. (B) MV-4-11 and Kasumi-1 cell viabilities was detected by CCK-8 assay. (C) MV-4-11 and Kasumi-1 cell colony ability was detected by colony formation assay. (D) MV-4-11 and Kasumi-1 cell cycle was detected by flow cytometry. (E) MV-4 11 and Kasumi-1 cell apoptosis was detected by flow cytometry. (F) Western blot analysis was used to detected the expression of of JAK/STAT signaling-related proteins in MV-4-11 and Kasumi-1 cells. Compared with si-NC + pcDNA3.1-NC group, *P < 0.05, **P < 0.01; compared with si1-circ group, #P < 0.05, ##P < 0.01; compared with MCM2 group, &P < 0.05, &&P < 0.01.
Discussion
The molecular mechanisms underlying AML development are complex. Emerging evidence indicates that abnormally expressed circular RNAs are essential regulators of AML development via various mechanisms [Citation7–9]. Circular RNA PVT1 is abnormally expressed in AML patients and regulates the proliferation and apoptosis of AML cells [Citation21]. Circ_100290 represses apoptosis and enhances the proliferation of AML cells by targeting miR-203 in AML cells [Citation22]. Circ_0000370 promotes AML progression by modulating the miR-1299/S100A7A axis [Citation23]. CircPAN3 enhances drug resistance in AML cells by regulating autophagy [Citation24]. Circ_0009910 depletion suppresses cell growth of AML by inducing miR-20a-5p expression [Citation25]. CircPAN3 regulates drug resistance via the miR-153-5p/miR-183-5p/XIAP axis in AML cells [Citation26]. Moreover, the function of circ_0104700, a circular RNA with the gene symbol of HOMER2, has not yet been reported. Here, we screened aberrantly expressed circ_0104700 in AML samples form the GEO and TCGA databases, and validated that circ_0104700 expression was elevated in AML patients compared to normal cases. We also confirmed that circ_0104700 expression was enhanced in AML cell lines, including MV-4-11, Kasumi-1, HL-60 and AML2, which showed elevated circ_0104700 expression relative to bone marrow stromal HS-5 cells. In the functional investigation, we identified that silencing circ_0104700 repressed proliferation and cell cycle and induced apoptosis in AML cells. These data indicated that circ_0104700 plays a critical role in AML progression by modulating cell proliferation, apoptosis, and the cell cycle, enriching our understanding of the function of circular RNAs in AML pathogenesis.
Previous studies have revealed a wide range of mechanisms involving miRNAs in AML. It has been reported that miR-29b/Sp1 signaling regulates AML malignant progression and stemness by modulating fucosylation through Wnt/β-catenin signaling [Citation27]. MiR-34c-5p contributes to AML eradication by enhancing senescence via targeting RAB27B-mediated exosome shedding [Citation28]. Long non-coding RNA SNHG5 modulates chemoresistance by targeting miR-32/DNAJB9 signaling in AML cells [Citation29]. Meanwhile, miR-665 plays a critical role in the development of cancer, and many reports have identified that miR-665 serves as a tumor suppressor. It has been reported that miR-665 represses progression and epithelial–mesenchymal transition by regulating CRIM1 in gastric cancer [Citation16]. lncRNA linc00462 contributes to the invasiveness of pancreatic cancer cells via miR-665/TGFBR1/TGFBR2/SMAD2/3 signaling [Citation30]. The expression of miR-665 is associated with tumor metastasis and breast cancer prognosis by regulating NR4A3 [Citation31]. CircABCC2 modulates the development of hepatocellular cancer by targeting miR-665 [Citation32]. LINC00565 reduces apoptosis and contributes to cell proliferation by regulating miR-665/AKT3 signaling in gastric cancer [Citation17]. The lncRNA BCAR4 targets miR-665/STAT3 signaling to maintain the stemness of colorectal cancer stem cells and enhance tumorigenicity [Citation33]. Long non-coding RNA RHPN1-AS1 contributes to the migration and proliferation of ovarian cancer cells by miR-665/Akt3 axis [Citation18]. The up-regulation of CircRNA_100876 contributes to the gastric cancer cell invasion, migration, and growth by the miR-665/YAP axis [Citation34].
Moreover, MCM2 is a well-investigated oncogene and a proliferation biomarker of cancer progression. It has been found that the suppression of MCM2 contributes to the carboplatin sensitivity of ovarian cancer cells [Citation35]. MCM2 can be used to measure breast cancer proliferation in a manner similar to Ki-67 [Citation14]. MCM2 is correlated with the diagnosis and prognosis of breast cancer [Citation36]. MCM2 serves as a potential marker for pancreatic cancer prognosis and diagnosis [Citation37]. Inhibition of MCM2 function is an innovative therapeutic strategy for malignant breast cancer [Citation38]. Up-regulation of MCM2 contributes to the regulation of the cell cycle and proliferation of lung squamous cell carcinoma cells [Citation39]. Suzuki et al. [Citation40] have confirmed that MCM2 is upregulated in myelodysplastic syndromes and associated with bone marrow cell apoptosis and peripheral cytopenia. Mechanistically, we found that circ_0104700 sponges miR-665 to enhance MCM2 expression, resulting in the activation of JAK/STAT signaling in AML cells. Circ_0104700 depletion reduced proliferation and cell cycle progression and enhanced apoptosis of AML cells by inhibiting miR-665. MCM2 depletion repressed proliferation and cell cycle progression and enhanced apoptosis of AML cells by inactivating JAK/STAT signaling. MCM2/AK/STAT signaling is involved in circ_0104700-mediated malignant phenotypes of AML cells. Our findings present a new mechanism involving miR-665/MCM2/JAK/STAT signaling by which circ_0104700 promotes AML progression.
In summary, we concluded that circ_0104700 contributed to AML progression by enhancing MCM2 expression through targeting miR-665. Our findings providenovel potential therapeutic targets for AML, including circ_0104700, miR-665, and MCM2.
Declarations
Ethics approval
The protocol of this research has been approved by the Ethics Committee of The Affiliated Qingdao Central Hospital of Qingdao University. All patients have signed written informed consent.
Consent to participate
Not applicable.
Availability of data and material
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Authors’ contributions
Conception and design: Kang Chen; Perform research: Kang Chen and Xiaohan Ning; Data analysis and interpretation: Xiaohong Yan and Lingling Song; Manuscript writing: Kang Chen; Final approval of manuscript: All authors.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Fang H, Yabe M, Zhang X, et al. Myelodysplastic syndrome with t(6;9)(p22;q34.1)/DEK-NUP214 better classified as acute myeloid leukemia? A multicenter study of 107 cases. Mod Pathol. 2021;34(6):1143–1152.
- Libbrecht C, Xie HM, Kingsley MC, et al. Menin is necessary for long term maintenance of meningioma-1 driven leukemia. Leukemia. 2021;35(5):1405–1417.
- Ma D, Liu P, Wang P, et al. PKC-beta/Alox5 axis activation promotes Bcr-Abl-independent TKI-resistance in chronic myeloid leukemia. J Cell Physiol. 2021;236(9):6312–6327.
- Maiti A, DiNardo CD, Daver NG, et al. Triplet therapy with venetoclax, FLT3 inhibitor and decitabine for FLT3-mutated acute myeloid leukemia. Blood Cancer J. 2021;11(2):25.
- Pautas C, Raffoux E, Lambert J, et al. Outcomes following hematopoietic stem cell transplantation in patients treated with standard chemotherapy with or without gemtuzumab ozogamicin for acute myeloid leukemia. Bone Marrow Transplant. 2021;56(6):1474–1477.
- Vial JP, Lechevalier N, Lacombe F, et al. Unsupervised flow cytometry analysis allows for an accurate identification of minimal residual disease assessment in Acute Myeloid Leukemia. Cancers (Basel). 2021;13(4):629.
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691.
- Kristensen LS, Hansen TB, Veno MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565.
- Lei M, Zheng G, Ning Q, et al. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19(1):30.
- Li W, Zhong C, Jiao J, et al. Characterization of hsa_circ_0004277 as a new biomarker for Acute Myeloid Leukemia via circular RNA profile and bioinformatics analysis. Int J Mol Sci. 2017;18(3):597.
- Wu DM, Wen X, Han XR, et al. Role of circular RNA DLEU2 in Human Acute Myeloid Leukemia. Mol Cell Biol. 2018;38(20):e00259-18.
- Cheung CHY, Hsu CL, Chen KP, et al. MCM2-regulated functional networks in lung cancer by multi-dimensional proteomic approach. Sci Rep. 2017;7(1):13302.
- Wang L, Guo J, Zhou J, et al. NF-kappaB maintains the stemness of colon cancer cells by downregulating miR-195-5p/497-5p and upregulating MCM2. J Exp Clin Cancer Res. 2020;39(1):225.
- Yousef EM, Furrer D, Laperriere DL, et al. MCM2: an alternative to Ki-67 for measuring breast cancer cell proliferation. Mod Pathol. 2017;30(5):682–697.
- Majid S, Dar AA, Saini S, et al. Regulation of minichromosome maintenance gene family by microRNA-1296 and genistein in prostate cancer. Cancer Res. 2010;70(7):2809–2818.
- Wu KZ, Zhang CD, Zhang C, et al. miR-665 suppresses the Epithelial-Mesenchymal transition and progression of gastric cancer by targeting CRIM1. Cancer Manag Res. 2020;12:3489–3501.
- Hu J, Ni G, Mao L, et al. LINC00565 promotes proliferation and inhibits apoptosis of gastric cancer by targeting miR-665/AKT3 axis. Onco Targets Ther. 2019;12:7865–7875.
- Zhao J, Yang T, Ji J, et al. RHPN1-AS1 promotes cell proliferation and migration via miR-665/Akt3 in ovarian cancer. Cancer Gene Ther. 2020;28(1-2):33–41.
- Rommer A, Steinleitner K, Hackl H, et al. Overexpression of primary microRNA 221/222 in acute myeloid leukemia. BMC Cancer. 2013;13:364.
- Nuño-Ayala M, Guillén N, Arnal C, et al. Cystathionine β-synthase deficiency causes infertility by impairing decidualization and gene expression networks in uterus implantation sites. Physiol Genomics. 2012;44(14):702–716.
- Hu J, Han Q, Gu Y, et al. Circular RNA PVT1 expression and its roles in acute lymphoblastic leukemia. Epigenomics. 2018;10(6):723–732.
- Fan H, Li Y, Liu C, et al. Circular RNA-100290 promotes cell proliferation and inhibits apoptosis in acute myeloid leukemia cells via sponging miR-203. Biochem Biophys Res Commun. 2018;507(1–4):178–184.
- Zhang L, Bu Z, Shen J, et al. A novel circular RNA (hsa_circ_0000370) increases cell viability and inhibits apoptosis of FLT3-ITD-positive acute myeloid leukemia cells by regulating miR-1299 and S100A7A. Biomed Pharmacother. 2020;122:109619.
- Shang J, Chen WM, Liu S, et al. CircPAN3 contributes to drug resistance in acute myeloid leukemia through regulation of autophagy. Leuk Res. 2019;85:106198.
- Ping L, Jian-Jun C, Chu-Shu L, et al. Silencing of circ_0009910 inhibits acute myeloid leukemia cell growth through increasing miR-20a-5p. Blood Cells Mol Dis. 2019;75:41–47.
- Shang J, Chen WM, Wang ZH, et al. CircPAN3 mediates drug resistance in acute myeloid leukemia through the miR-153-5p/miR-183-5p-XIAP axis. Exp Hematol. 2019;70:42–54 e43.
- Liu B, Ma H, Liu Q, et al. MiR-29b/Sp1/FUT4 axis modulates the malignancy of leukemia stem cells by regulating fucosylation via Wnt/beta-catenin pathway in acute myeloid leukemia. J Exp Clin Cancer Res. 2019;38(1):200.
- Peng D, Wang H, Li L, et al. miR-34c-5p promotes eradication of acute myeloid leukemia stem cells by inducing senescence through selective RAB27B targeting to inhibit exosome shedding. Leukemia. 2018;32(5):1180–1188.
- Wang D, Zeng T, Lin Z, et al. Long non-coding RNA SNHG5 regulates chemotherapy resistance through the miR-32/DNAJB9 axis in acute myeloid leukemia. Biomed Pharmacother. 2020;123:109802.
- Zhou B, Guo W, Sun C, et al. Linc00462 promotes pancreatic cancer invasiveness through the miR-665/TGFBR1-TGFBR2/SMAD2/3 pathway. Cell Death Dis. 2018;9(6):706.
- Zhao XG, Hu JY, Tang J, et al. miR-665 expression predicts poor survival and promotes tumor metastasis by targeting NR4A3 in breast cancer. Cell Death Dis. 2019;10(7):479.
- Bai N, Peng E, Xia F, et al. CircABCC2 regulates hepatocellular cancer progression by decoying MiR-665. J Cancer. 2019;10(17):3893–3898.
- Ouyang S, Zhou X, Chen Z, et al. LncRNA BCAR4, targeting to miR-665/STAT3 signaling, maintains cancer stem cells stemness and promotes tumorigenicity in colorectal cancer. Cancer Cell Int. 2019;19:72.
- Lin X, Huang C, Chen Z, et al. CircRNA_100876 is upregulated in Gastric Cancer (GC) and promotes the GC cells’ growth, migration and invasion via miR-665/YAP1 signaling. Front Genet. 2020;11:546275.
- Deng M, Sun J, Xie S, et al. Inhibition of MCM2 enhances the sensitivity of ovarian cancer cell to carboplatin. Mol Med Rep. 2019;20(3):2258–2266.
- Issac MSM, Yousef E, Tahir MR, et al. MCM2, MCM4, and MCM6 in breast cancer: clinical utility in diagnosis and prognosis. Neoplasia. 2019;21(10):1015–1035.
- Deng Y, Ma H, Hao J, et al. MCM2 and NUSAP1 are potential biomarkers for the diagnosis and prognosis of pancreatic cancer. Biomed Res Int. 2020;2020:8604340.
- Abe S, Yamamoto K, Kurata M, et al. Targeting MCM2 function as a novel strategy for the treatment of highly malignant breast tumors. Oncotarget. 2015;6(33):34892–34909.
- Wu W, Wang X, Shan C, et al. Minichromosome maintenance protein 2 correlates with the malignant status and regulates proliferation and cell cycle in lung squamous cell carcinoma. Onco Targets Ther. 2018;11:5025–5034.
- Suzuki S, Kurata M, Abe S, et al. Overexpression of MCM2 in myelodysplastic syndromes: association with bone marrow cell apoptosis and peripheral cytopenia. Exp Mol Pathol. 2012;92(1):160–166.
