ABSTRACT
Background
Acute myeloid leukemia (AML) is a malignant blood cancer with a poor prognosis and complex pathogenesis. Recently, the critical role of circular RNAs (circRNAs) has been demonstrated in the malignant progression of AML. This study aimed to investigate the functional role and underlying mechanism of circ_0001602 in AML development.
Methods
Quantitative real-time polymerase chain reaction (qRT-PCR) assay was conducted for detecting the expression of circ_0001602, CCND3, microRNA-192-5p (miR-192-5p), and Zinc Finger and BTB Domain-Containing Protein 20 (ZBTB20) mRNA. RNase R assay and Actinomycin D assay were implemented to determine the characteristics of circ_0001602. Cell counting Kit-8 (CCK-8) assay was performed to evaluate cell proliferation. Flow cytometry was employed for assessing cell cycle distribution and apoptosis. Dual-luciferase reporter assay and RIP assay were utilized for confirming the interactions between miR-192-5p and circ_0001602 or ZBTB20.
Results
Circ_0001602 and ZBTB20 were upregulated and miR-192-5p level was reduced in AML tissues and cells. Depletion of circ_0001602 repressed cell proliferation and induced cell cycle arrest and apoptosis in AML cells. Functionally, circ_0001602 was identified to be the sponge of miR-192-5p, and miR-192-5p silence restored the suppressive effects of circ_0001602 knockdown on AML cell progression. Furthermore, ZBTB20 was a target of miR-192-5p, and ZBTB20 overexpression neutralized the miR-192-5p-mediated inhibiting actions on the malignant phenotypes of AML cells. Besides, circ_0001602 could sponge miR-192-5p to positively regulate ZBTB20 expression.
Conclusion
Circ_0001602 contributed to AML cell development at least partially through modulating the miR-192-5p/ZBTB20 axis, which provided new insights for AML treatment.
KEYWORDS:
Introduction
Acute myeloid leukemia (AML) is a form of hematopoietic disease of the bone marrow, featured by the damaged differentiation and indisciplinable proliferation of myeloid progenitor cells [Citation1,Citation2]. As a well-known acute leukemia in grown-up persons, AML comprised varieties of subtypes, and the morbidity of AML rises with increasing age [Citation3,Citation4]. In spite of considerable improvements has been made in the treatment strategies of AML, the prognosis of it is still dissatisfactory [Citation5,Citation6]. Thereby, it is imperative to expound on the pathogenesis of AML and find effective molecular therapeutic targets for AML patients.
Circular RNAs (circRNAs) are a sort of endogenous, noncoding RNAs (ncRNAs) carrying covalent closed-loop structures [Citation7,Citation8]. Lately, diverse circRNAs have been proven to be related to the initiation and advancement of human malignancies [Citation9,Citation10], including AML [Citation11]. For example, circ_0035381 could facilitate the proliferative ability and block the apoptotic ability of AML cells through the miR-582-3p/YWHAZ pathway [Citation12]. Furthermore, circ_0001602 (derived from CCND3 precursor mRNA, located in chr6:42012350-42012461) was previously proved to be visibly upregulated in AML bone marrow specimens [Citation13]. Nonetheless, the role and potential theory of circ_0001602 in AML etiology are indistinct.
MicroRNA (miRNAs) are small ncRNAs that directly modulate the posttranscriptional expression of genes by interacting with the 3’UTRs of targeted mRNAs [Citation14,Citation15]. Accumulating proofs have illustrated that the dysregulation of miRNAs participates in the modulation of tumor progression in diversiform human malignancies [Citation16], including AML [Citation17]. For example, Wang et al. unraveled that miR-9 could accelerate the vicious behaviors of AML cells by adjusting the Hippo/YAP signaling pathway [Citation18]. Shang and his colleagues declared that miR-625-5p had an inhibitory effect on the development of AML cells via targeting SOX12 [Citation19]. Of interest, as a vital tumor-associated miRNA, miR-192 has been verified to act a tumor-suppressive role in leukemia [Citation20,Citation21]. It has been reported that the overexpression of miR-192-5p might induce cell apoptosis and hinder cell proliferation in AML cells [Citation22]. However, whether miR-192-5p is relevant to circ_0001602-mediated AML progression has not been explored. Besides, a previous study manifested that Zinc Finger and BTB Domain-Containing Protein 20 (ZBTB20), an essential transcription factor, was upregulated and related to AML evolution [Citation23]. A recent study indicated that the overexpression of ZBTB20 might boost the malignant growth of AML cells via increasing proliferation, migration, invasion, and glycolysis [Citation24]. These findings implied that ZBTB20 might exert a vital role during AML progression.
Currently, the circRNA-miRNA-mRNA regulatory mechanism that circRNAs, a miRNA sponge, might derepress target mRNA expression via competitively binding to miRNAs [Citation25,Citation26]. Herein, the abundance of circ_0001602 in AML tissue specimens and cells was surveyed. Further, bioinformatics analysis discovered that miR-192-5p has some binding sites with circ_0001602 or ZBTB20. Thus, we aimed to explore whether circ_0001602 might regulate AML cell malignant behaviors via targeting miR-192-5p/ZBTB20.
Materials and methods
Clinical samples
The bone marrow specimens from 52 AML patients and 34 healthy individuals were obtained from Longyan First Affiliated Hospital of Fujian Medical University. All participants signed written informed consent and did not accept any preoperative therapy. Patients with other severe diseases or initiated therapies within 3 months prior to admission were excluded. Meanwhile, the healthy controls showed normal physiological functions in systemic physiological exams carried out at the aforementioned hospital. This research obtained ratification from the Ethics Committee of Longyan First Affiliated Hospital of Fujian Medical University. The diagnosis of AML was made in accordance with the World Health Organization classification of Tumors of Hematopoietic and Lymphoid Tissues 2017 [Citation27].
Cell culture
Human bone marrow stromal cell line (HS-5) was obtained from Bluefbio (Shanghai, China). AML cell line (THP-1, HL-60, and NB4) and HEK 293 T cell line were obtained from Procell (Wuhan, China). HS-5 cells and HEK 293 T cells were fostered in DMEM medium (Procell) replenished with 10% FBS (Procell) and 1% penicillin–streptomycin (P/S; Beyotime, Shanghai, China). HL60 cells were fostered in IMDM medium (Procell) plus 20% FBS and 1% P/S. THP-1 and NB4 cells were fostered in RPMI-1640 medium (Procell) plus 10% FBS and 1% P/S. The above-mentioned cells were cultivated under 37°C in a moist incubator containing 5% CO2.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA extraction from bone marrow samples and cell lines was performed utilizing Triquick Reagent (Trizol Substitute; Solarbio, Beijing, China), and cDNA was synthetic from RNA by M-MLV Reverse Transcriptase Kit (Promega, Madison, WI, USA) for circ_0001602, CCND3 and ZBTB20 or PrimeScript miRNA RT–PCR Kit (Takara, Dalian, China) for miR-192-5p. Then, qRT-PCR was executed employing SYBR Premix Ex Taq II kit (Takara) on a PCR system. Meanwhile, qRT-PCR was carried out at the condition: 95.0°C for 1 min, and 39 circles of 95.0°C for 10s, and 60°C for 30 s. Relative expression was calculated by exploiting the 2−ΔΔCt strategy after normalization with internal contrast GAPDH (for circ_0001602, CCND3, and ZBTB20) or U6 (for miR-192-5p). The sequences of specific primers were listed in .
Table 1. Primer sequences used for qRT-PCR.
Ribonuclease R (RNase R) assay
Total RNA (2 μg) isolated from AML cells was subjected to the treatment of RNase R (3 U/μg; Epicenter, Madison, WI, USA) or not for 20 min under 37°C. Next, the relative RNA abundances of circ_0001602 and linearizing CCND3 were inspected through qRT-PCR analysis. The RNA not incubated with RNase R served as the negative control (Mock).
Actinomycin D assay
Transcription inhibitor Actinomycin D (2 μg/mL; Abcam, Cambridge, UK) was incubated with THP-1 and HL-60 cells for 0, 4, 8, 12, and 24 h to prevent transcription. Then, the RNAs were isolated from cells, and qRT-PCR analysis was conducted for examining the RNA abundances of circ_0001602 and linear CCND3 mRNA.
Cell transfection
To silence circ_0001602, circ_0001602 small interference RNAs (si-circ_0001602#1 and si-circ_0001602#2) were established, with si-NC as a matched control. For miR-192-5p overexpression or inhibition, miR-192-5p mimic or inhibitor (miR-192-5p mimic or in-miR-192-5p) was constructed, with mimic NC or in-miR-NC as the negative control. To upregulate ZBTB20 expression, the ZBTB20 overexpression vector (ZBTB20) was established through introducing the sequence of ZBTB20 into the pcDNA3.1 vector (pcDNA), with empty pcDNA as a control. Above designated plasmids or oligonucleotides were all commercially acquired from GenePharma (Shanghai, China) and transduced into THP-1 and HL-60 cells through Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA).
Cell counting kit-8 (CCK-8) assay
To identify the viability of AML cells, transduced cells were planted in 96-well plates (1 × 104 cells/well) and cultivated overnight in a 5% CO2 incubator at 37°C. At 0, 24, 48, or 72 h, 10 μL CCK-8 (5 mg/mL; Solarbio) was supplemented into each well and maintained for another 2 h. At last, the absorption was determined at 450 nm using a microplate reader.
Flow cytometry analysis
Flow cytometry was utilized for detecting the cycle distribution and apoptosis of AML cells. After being transfected for 48 h, cells were harvested. To analyze cycle distribution, transduced cells were gathered and washed with PBS, followed by the fixation of 70% ethanol at 4°C for 24 h. And the cells were subsequently incubated with propidine iodide (PI; Solarbio) in darkness at 37°C for 30 min after being resuspended in a binding buffer. Through utilizing a flow cytometer, the distributed proportions of AML cells at different stages were monitored.
For the exploration of apoptosis, Annexin V-FITC Apoptosis Detection kit (Beyotime) was utilized. Briefly, transduced cells were gathered and resuspended in binding buffer, and later subjected to the dyeing of Annexin V-FITC and PI in darkness for 15 min. The percentage of apoptotic cells was evaluated by a flow cytometer.
Caspase 3 activity assay
1 × 106 AML cells were placed in 6-well plates for 48 h-incubation in the respective medium. Then, cells were gathered and dissociated by lysis buffer for 20 min after washing with sterile PBS. Later, the protein supernatant from cell lysates was utilized for the activity determination of caspase 3 via Caspase 3 Activity Assay Kit (Beyotime) referring to the specification. The OD value was monitored at 405 nm by a microplate reader, and the relative activity of caspase-3 was tested through a fold-change versus the control group.
Western blot assay
Total protein isolation from bone marrow specimens and cell lines (HS-5, THP-1, and HL-60) was accomplished utilizing RIPA lysis buffer (Beyotime). 30 μg protein was added onto 10% SDS-PAGE gel (Beyotime), followed by the transference to PVDF membrane (Beyotime). After blockage with 5% fat-free milk for 1 h at indoor temperature, the membrane was reacted with primary antibodies against proliferating cell nuclear antigen (PCNA; 1:3000, ab92552, Abcam), Bcl-2-associated X protein (Bax; 1:1000; ab263897, Abcam), and B-cell lymphoma/leukemia-2 (Bcl-2; 1:1000, ab196495, Abcam), ZBTB20 (1:2000, ab127702, Abcam) and internal protein standard GAPDH (1:2500, ab9485, Abcam) at 4°C for 24 h. Subsequently, the membrane interacted with a secondary antibody (1:25000, ab205718, Abcam) for 1 h. The protein blots were visualized via BeyoECL Star Kit (Beyotime).
Dual-luciferase reporter assay
The fragments of circ_0001602 or ZBTB20 3’UTR harboring the putative miR-192-5p complementary sites or mutant sites were cloned and inserted into pmirGLO luciferase reporter vector (Promega) to establish circ_0001602 wild type (WT), ZBTB20 3’UTR WT, circ_0001602 mutant type (MUT), ZBTB20 3’UTR MUT reporters. Next, these generated plasmids and miR-192-5p mimic or mimic NC and were transduced into HEK293 T cells. The relative luciferase intensity was examined after 48 h by exploiting the Dual-Lucy Assay Kit (Solarbio).
RNA immunoprecipitation (RIP) assay
AML cells were dissociated in RIP lysis buffer from Magna RIP Kit (Millipore, Billerica, MA, USA), and then the lysates interacted with magnetic beads which were combined with Argonaute2 antibody (Anti-Ago2; ab32381; Millipore) or control Immunoglobulin G antibody (Anti-IgG; ab133470; Millipore) for 8 h at 4°C. The RNA was finally separated from the complexes using TRIzol reagent (Takara) and the enrichments of circ_0001602 and miR-192-5p were subjected to qRT-PCR analysis.
Statistical analysis
Data collected from three duplicates were displayed as mean ± standard deviation (SD). Statistical analysis was implemented by SPSS 18.0 software. Student’s t-test and one-way analysis of variance (ANOVA) were used for the different comparisons. The linear relations in AML tissues among circ_0001602, miR-192-5p, and ZBTB20 expression were analyzed via Spearman’s correlation coefficient analysis. P < 0.05 was defined as statistically significant.
Results
Circ_0001602 abundance was enhanced in AML tissues and cell lines
In order to inquire about the function of circ_0001602 in AML progression, the level of circ_0001602 in AML tissues and adjacent non-tumor tissues was determined. The results displayed that circ_0001602 expression was dramatically enhanced in the bone marrow samples from AML patients (AML; n = 52) relative to that in normal subjects (Normal; n = 34) (A). Conformably, circ_0001602 was observably upregulated in AML cell lines (THP-1, HL-60, and NB4) versus that in normal HS-5 cell lines (B). In consideration of the higher expression of circ_0001602 in THP-1 and HL-60 cells, they were chosen for subsequent experiments. Then, the features of circ_0001602 were analyzed using RNase R and transcription inhibitor Actinomycin D. RNase R digestion assay validated that the level of linear CCND3 mRNA was overtly declined after RNase R treatment, while circ_0001602 was hardly affected by RNase R digestion (C,D), implying that circ_0001602 was a highly stable circular RNA molecule in AML cells. Actinomycin D assay exhibited that circ_0001602 level remained stable after Actinomycin D treatment for 24 h, but linear CCND3 mRNA expression was significantly reduced (E,F), further indicating that circ_0001602 was a circular and stable transcript. These data manifested that circ_0001602 was upregulated in AML tumor tissues and cells with a stable circular structure.
Figure 1. Circ_0001602 level was elevated in AML tissues and cells. (A) The expression of circ_0001602 in the bone marrow samples from AML patients (AML; n = 52) and normal subjects (Normal; n = 34) was tested by qRT-PCR. (B) Circ_0001602 abundance was detected in AML cell lines (THP-1, HL-60, and NB4) and normal HS-5 cell line by qRT-PCR. (C and D) The stability of circ_0001602 and linear CCND3 was tested by RNase R assay. (E and F) QRT-PCR assay was performed to examine the levels of circ_0001602 and linear CCND3 mRNA in THP-1 and HL-60 cells at indicated time points after Actinomycin D treatment. **P < 0.01, ***P < 0.001.
Circ_0001602 interference repressed cell proliferation and induced cell cycle arrest and apoptosis in AML cells
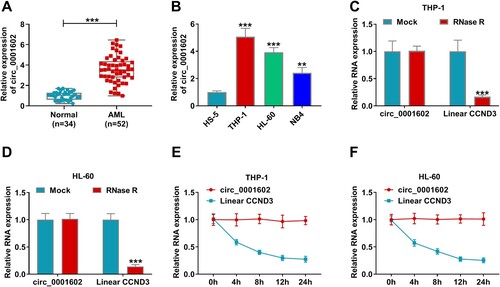
To investigate whether the dysregulation of circ_0001602 was concerned with the development of AML, the effect of circ_0001602 knockdown on AML cells was studied. The loss-of-function experiments were conducted by introducing si-circ_0001602#1, si-circ_0001602#2 or si-NC into THP-1 and HL-60 cells. As depicted in A,B, circ_0001602 expression was conspicuously inhibited by si-circ_0001602#1 or si-circ_0001602#2 transfection, while the expression of CCND3 mRNA was insusceptible in THP-1 and HL-60 cells, indicating that circ_0001602 was successfully downregulated. As demonstrated by the CCK-8 assay, the proliferation capacity of THP-1 and HL-60 cells was distinctly inhibited following the transfection of si-circ_0001602#1 or si-circ_0001602#2 (C,D). Flow cytometry analysis indicated that the number of THP-1 and HL-60 cells was increased at G0/G1 phase and decreased at the S phase by the downregulation of circ_0001602, suggesting that circ_0001602 deficiency promoted cell cycle arrest (E,F). Meanwhile, circ_0001602 silence facilitated the apoptosis of THP-1 and HL-60 cells, reflected by increased apoptotic rate and caspase 3 activity (G,H). Additionally, the protein abundances of proliferation-associated maker (PCNA) and apoptosis-associated makers (Bax and Bcl-2) were inspected. The results revealed that circ_0001602 interference overtly repressed PCNA and Bcl-2 levels and facilitated Bax level (I). These data manifested that circ_0001602 knockdown suppressed the malignant behaviors of AML cells. For the higher transfection efficiency of si-circ_0001602#2, it was selected for subsequent experiments.
Figure 2. Circ_0001602 silence suppressed the development of AML cells. THP-1 and HL-60 cells were transduced with si-circ_0001602#1, si-circ_0001602#2 or si-NC. (A and B) QRT-PCR assay was utilized for circ_0001602 and linear CCND3 mRNA expression in transfected cells. (C and D) Cell viability was tested by CCK-8 assay. (E and F) The percentage of THP-1 and HL-60 cells at G0/G1, S, or G2/M phase was assessed using flow cytometry. (G) Cell apoptosis was monitored using flow cytometry. (H) Caspase 3 activity was explored by corresponding kit. (I) Western blot assay was carried out for the levels of PCNA, Bax and Bcl-2 protein in transfected cells. *P < 0.05, **P < 0.01, ***P < 0.001.
MiR-192-5p was a target of circ_0001602
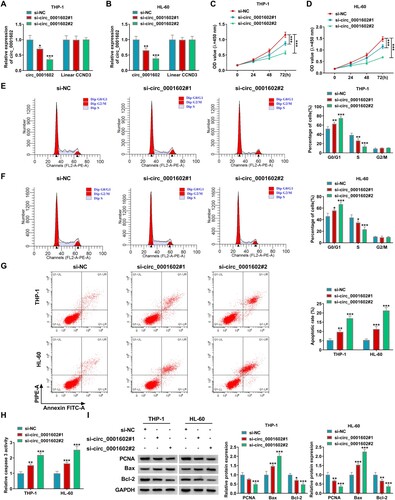
For exploring the molecular mechanism by which circ_0001602 regulated the progression of AML cells, Circular RNA interactome (https://circinteractome.nia.nih.gov) software was utilized to seek the potential target miRNAs of circ_0001602. The prediction results suggested that 3 miRNAs (miR-192-5p, miR-153, miR-215) have complementary sequences with circ_0001602. Through the expression analysis of the above alternative 3 miRNAs in 6 paired bone marrow samples from AML patients and normal subjects, they were all downregulated in AML bone marrow samples (A). Simultaneously, they were all upregulated in THP-1 and HL-60 cells after circ_0001602 deficiency (B,C). Therein, miR-192-5p with the most upregulation was selected as the molecular target in our study. As depicted in D, the putative binding sites between circ_0001602 and miR-192-5p and the mutated binding sites of circ_0001602 on miR-192-5p were presented. Then dual-luciferase reporter assay was conducted to affirm this combination. The results demonstrated that miR-192-5p mimic transfection overtly limited the luciferase activity of circ_0001602 WT reporter, but did not impact the luciferase intensity of circ_0001602 MUT reporter in HEK293 T cells, confirming the combination between circ_0001602 and miR-192-5p (E). RIP assay showed that both circ_0001602 and miR-192-5p were largely enriched in the Anti-Ago2 group but not in the Anti-IgG group, hinting that miR-192-5p was a targeted miRNA of circ_0001602 (F,G). Unlike the expression of circ_0001602, miR-192-5p was overtly downregulated in AML bone marrow samples (n = 52) and cells (THP-1 and HL-60) in comparison to that in normal bone marrow samples (n = 34) and cells (HS-5) (H,I). Overall, this evidence supported that miR-192-5p was a functional target of circ_0001602 in AML cells.
Figure 3. Circ_0001602 directly targeted miR-192-5p in AML cells. (A) The expression of miRNAs (miR-192-5p, miR-153, and miR-215) in AML bone marrow samples and normal bone marrow samples was tested by qRT-PCR. (B and C) The levels of miRNAs in THP-1 and HL-60 cells transduced with si-circ_0001602#2 or si-NC were measured by qRT-PCR assay. (D) The putative binding sites between circ_0001602 and miR-192-5p were presented. (E) The luciferase activity in HEK293 T cells co-transfected with miR-192-5p mimic or mimic NC and circ_0001602 WT or circ_0001602 MUT reporters was detected by dual-luciferase reporter assay. (F and G) The enrichments of circ_0001602 and miR-192-5p in THP-1 and HL-60 cells incubated with Anti-IgG or Anti-Ago2 antibody in RIP assay were examined by qRT-PCR assay. (H) The level of miR-192-5p in AML tissues and normal tissues was tested using qRT-PCR assay. (I) The level of miR-192-5p in HS-5, THP-1, and HL-60 cells was validated by qRT-PCR assay. **P < 0.01, ***P < 0.001.
MiR-192-5p silence partly reversed the inhibitory influences of circ_0001602 knockdown on AML cell progression
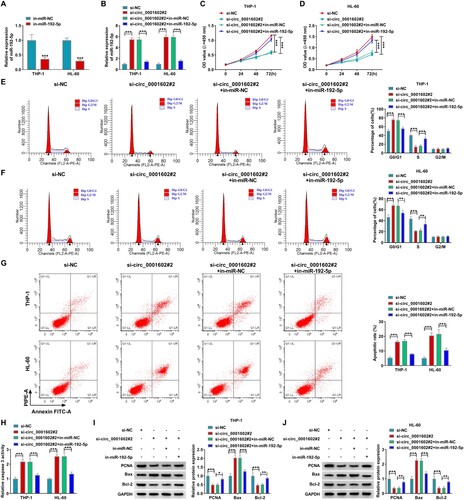
Next, whether circ_0001602 had cross-talk with miR-192-5p in AML cell development was explored through rescue experiments. First of all, the knockdown efficiency of in-miR-192-5p was verified by qRT-PCR analysis in THP-1 and HL-60 cells (A). Rescue experiments were carried out through transfecting si-circ_0001602#2, si-circ_0001602#2 + in-miR-192-5p, or corresponding controls into THP-1 and HL-60 cells. As exhibited in B, circ_0001602 interference apparently elevated the expression of miR-192-5p, while this effect was abrogated by the introduction of in-miR-192-5p in THP-1 and HL-60 cells, confirming that the transfection was successful. In function, transfection of si-circ_0001602#2 alone decreased cell viability (C,D), and facilitated G0/G1 phase arrest (E,F), cell apoptosis (G) and caspase 3 activity in THP-1 and HL-60 cells (H), while these influences were all partially canceled by miR-192-5p silence. Besides, the reduced levels of PCNA and Bcl-2 and the increased expression of Bax in THP-1 and HL-60 cells after circ_0001602 interference were all effectively ameliorated by miR-192-5p inhibition (I,J). To sum up, the above findings evidenced that circ_0001602 deficiency exerted inhibitory impacts on the malignant phenotypes of AML cells partly by targeting miR-192-5p.
Figure 4. MiR-192-5p deficiency reversed the effects of circ_0001602 knockdown on AML cell progression. (A) The efficiency of miR-192-5p inhibitor in THP-1 and HL-60 cells was inspected using qRT-PCR. (B-J) THP-1 and HL-60 cells transfected with si-NC, si-circ_0001602#2, si-circ_0001602#2 + in-miR-NC or si-circ_0001602#2 + in-miR-192-5p. (B) The expression of miR-192-5p in these transfected cells was detected by qRT-PCR. (C and D) CCK-8 assay was carried out to analyze the proliferation capacity of THP-1 and HL-60 cells. (E and F) The cell cycle distribution of lung carcinoma cells in different phases of cell cycle (G0/G1, S and G2/M) were identified by flow cytometry. (G) The apoptotic rate of cells was evaluated by flow cytometry. (H) Caspase 3 activity was tested by the corresponding kit. (I and J) The protein levels of PCNA, Bax, and Bcl-2 in transfected cells were examined by western blot. *P < 0.05, **P < 0.01, ***P < 0.001.
MiR-192-5p directly interacted with ZBTB20 in AML cells
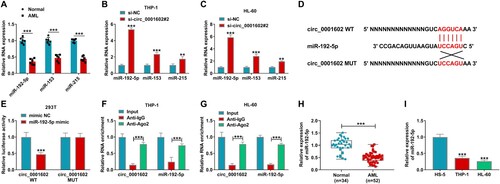
To deeper comprehend the function of miR-192-5p in AML, the molecular targets of miR-192-5p were searched utilizing the online tool Targetscan (http://www.targetscan.org/vert_72/). ZBTB20 was identified to have potential binding position with miR-192-5p (A). Dual-luciferase reporter presented that the co-transfection of miR-192-5p mimic and ZBTB20 3’UTR WT dramatically suppressed the luciferase activity of HEK293 T cells compared to mimic NC and ZBTB20 3’UTR WT co-transfected cells, whereas the luciferase activity was not influenced in ZBTB20 3’UTR MUT group, further affirming the direct interaction between miR-192-5p and ZBTB20 (B). Furthermore, the mRNA and protein abundances of ZBTB20 were upregulated in AML tissues (C,D). Likewise, the mRNA and protein levels of ZBTB20 in THP-1 and HL-60 cells were strikingly increased versus that in normal HS-5 cells (E,F). Collectively, these outcomes illuminated that ZBTB20 was a downstream targeted gene of miR-192-5p.
Figure 5. ZBTB20 was a direct target gene of miR-192-5p. (A) The binding sites between miR-192-5p and 3’UTR of ZBTB20 were exhibited. (B) Dual-luciferase reporter assay was implemented to prove the interaction between miR-192-5p and ZBTB20 in HEK293 T cells. (C and D) The mRNA and protein levels of ZBTB20 in AML tissues and normal tissues were determined by qRT-PCR assay or western blot assay. (E and F) The qRT-PCR and western blot assays were employed to test ZBTB20 abundance in HS-5, THP-1, and HL-60 cells. ***P < 0.001.
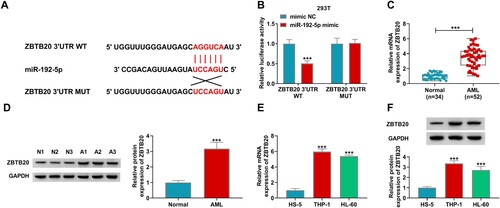
Elevating ZBTB20 partly overturned the inhibitory influences of miR-192-5p overexpression on AML cell development
Next, miR-192-5p mimic and ZBTB20 were respectively successfully transduced into THP-1 and HL-60 cells, and their overexpression efficiencies of them were severally evaluated by qRT-PCR assay and western blot (A,B). In order to determine the regulatory effect of miR-192-5p on ZBTB20 in AML cell progression, THP-1 and HL-60 cells were introduced with miR-192-5p mimic, miR-192-5p mimic + ZBTB20 or corresponding controls. It was discovered that ZBTB20 introduction could conspicuously invert the suppressive influence of miR-192-5p overexpression on ZBTB20 protein expression (C). Functional experiments suggested that miR-192-5p overexpression restrained cell vitality (D,E), cycle progress (F,G), and promoted the apoptosis (H) and caspase 3 activity (I) in AML cells, whereas these impacts were all effectively attenuated by the addition of ZBTB20. In addition, it was found that miR-192-5p overexpression resulted in an apparent reduction in PCNA and Bcl-2 levels and an obvious elevation in Bax level in both THP-1 and HL-60 cells, whereas ZBTB20 elevation partly mitigated the effects (J,K). In summary, it was concluded that miR-192-5p could hamper the malignant progression of AML cells through downregulating ZBTB20.
Figure 6. MiR-192-5p overexpression restrained the progression of AML cells by sponging ZBTB20. (A) The expression of miR-192-5p in THP-1 and HL-60 cells transfected with mimic NC or miR-192-5p mimic was detected by qRT-PCR. (B) The efficiency of ZBTB20 overexpression was measured by western blot. (C-K) THP-1 and HL-60 cells were transfected with mimic NC, miR-192-5p mimic, miR-192-5p mimic + pcDNA or miR-192-5p mimic + ZBTB20. (C) The protein level of ZBTB20 in THP-1 and HL-60 cells was detected by western blot assay. (D-K) CCK-8 assay for cell viability (D and E), flow cytometry analysis for cell cycle distribution (F and G) and apoptosis (H), Caspase 3 Activity Assay Kit for caspase 3 activity (I), as well as western blot assay for protein levels of PCNA, Bax, and Bcl-2 (J and K) in transfected cells were respectively implemented. *P < 0.05, **P < 0.01, ***P < 0.001.
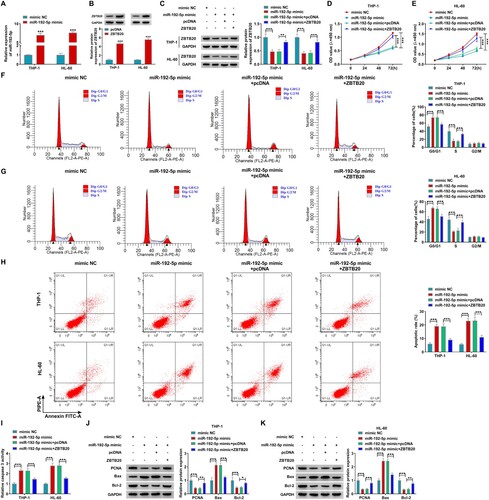
Circ_0001602 modulated ZBTB20 expression via sponging miR-532-5p
Last, the relationships among circ_0001602, miR-192-5p, and ZBTB20 in AML cells were explored. Meanwhile, it was confirmed that miR-192-5p level was inversely correlated with circ_0001602 level (r = −0.6528, P < 0.0001) and ZBTB20 mRNA level (r = −0.6118, P < 0.0001) in AML tissues (A,B). Furthermore, ZBTB20 mRNA expression was positively associated with circ_0001602 expression (r = 0.5399, P < 0.0001) in AML tissues (C). Moreover, circ_0001602 interference obviously inhibited the protein abundance of ZBTB20 in THP-1 and HL-60 cells, while miR-192-5p inhibition partly restored the effect (D,E). These results clarified that circ_0001602 could positively regulate ZBTB20 expression through sponging miR-192-5p in AML cells. The interaction mode of circ_0001602/miR-192-5p/ZBTB20 regulatory network in AML cells was exhibited in .
Figure 7. Circ_0001602 modulated ZBTB20 expression via targeting miR-192-5p. (A-C) The correlations among the levels of circ_0001602, miR-192-5p, and ZBTB20 in AML tissues were analyzed by spearman’s correlation coefficient analysis. (D and E) The level of ZBTB20 in THP-1 and HL-60 cells transfected with si-NC, si-circ_0001602#2, si-circ_0001602#2 + in-miR-NC or si-circ_0001602#2 + in-miR-192-5p was examined by western blot. *P < 0.05, **P < 0.01, ***P < 0.001.
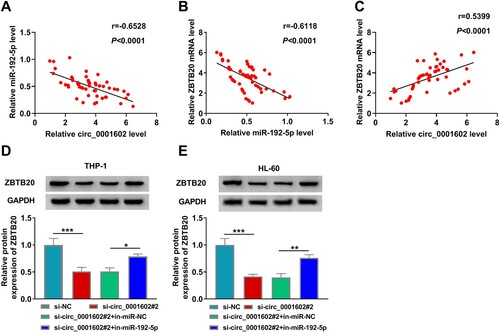
Figure 8. The interaction mode of circ_0001602/miR-192-5p/ZBTB20 regulatory network in AML cells. Circ_0001602 was upregulated in AML cells, and it could suppress miR-192-5p activity by competitively sponging miR-192-5p from ZBTB20. Thus, the expression of ZBTB20 was increased in AML cells, which promoted cell proliferation and inhibited cell apoptosis.
Discussion
Presently, growing research articles have expounded that circRNAs exert pivotal roles in the biological processes of multifarious cancers [Citation28,Citation29]. Nevertheless, the effect and precise mechanism of circ_0001602 in AML evolution has not been explored. Here, the function of circ_0001602 and the mechanism of novel circ_0001602/miR-192-5p/ZBTB20 pathway based on circ_0001602 were first certified in AML etiology.
Emerging studies have discovered that circRNAs participate in the biological courses of cancer through regulating diverse pathways [Citation28,Citation30]. Likewise, the dysregulation of several circRNAs could accelerate or restrain tumor development in blood malignancies including leukemias, and they might serve as the biological markers for diagnosis or prognosis and new curative targets [Citation11,Citation31]. For example, circ_0002483 silence suppressed the vicious phenotypes of AML cells through modulation of the miR-758-3p/MYC axis [Citation32]. And circCRKL impeded AML cell advancement via upregulating p27 expression through sponging miR-196a-5p and miR-196b-5p [Citation33]. However, the effect of circ_0001602 in AML development has not been explored. Hu et al. illuminated that circ_0001602 had high expression in the bone marrow specimens of AML patients versus that of healthy individuals [Citation13]. In keeping with this research, we also observed that circ_0001602 had enhanced levels in AML tissues and cells. Furthermore, circ_0001602 deficiency restrained the proliferation, and cell cycle process and facilitated the apoptosis in AML cells. Hence, we speculated that circ_0001602 exerted carcinogenic function in AML progression.
CircRNAs have been reported to participate in AML pathogenesis by serving as miRNA sponges [Citation26,Citation34]. In AML, miRNAs might serve as important regulators through acting as oncogenes or tumor suppressors [Citation17,Citation35]. Later, the latent theory of circ_0001602 in regulating AML development was further probed. By screening and verifying, miR-192-5p was suggested to be targeted by circ_0001602. As reported, miR-192-5p exerts a tumor-suppressing function in diverse malignancies, such as papillary thyroid carcinoma [Citation36], glioblastoma [Citation37], and cervical cancer [Citation38]. Similarly, Li et al. indicated that miR-192-5p was concerned with the vicious phenotypes of AML through targeting ULK1 and serving as a target of circ_0005774 [Citation22]. Besides, it was also verified that the low level of miR-192 predicted poor prognosis in pediatric AML [Citation20], and miR-192 could inhibit AML cell proliferation by regulating the expression of CCNT2 [Citation21]. Herein, we confirmed that miR-192-5p was lowly expressed in AML tissues and cells. Meanwhile, miR-192-5p overexpression inhibited AML cell proliferation, cell cycle progress, and facilitated cell apoptosis. Moreover, the miR-192-5p level was reversely correlated to circ_0001602 expression in AML tissues, and miR-192-5p deficiency partly neutralized the malignancy of AML cells mediated by circ_0001602 interference. Thus, we deduced that circ_0001602 knockdown suppressed AML cell development through interacting with miR-192-5p.
The circRNA/miRNA/mRNA cascade has been identified to be a significant modulatory pattern in AML pathogenesis, and it might also provide novel prognostic biomarkers for AML [Citation39]. Therefore, whether there were miRNAs that were involved in AML progression under the downstream circ_0001602/miR-192-5p pathways was further probed. This study indicated that ZBTB20 was a molecular target of miR-192-5p. Previous studies have given evidence that ZBTB20 is highly expressed and served a crucial part in the development of multifarious malignancies, including glioblastoma [Citation40], gastric cancer [Citation41], and breast cancer [Citation42]. Likewise, ZBTB20 was also certified to exert a tumor-promoting function in AML through LINC00641/miR-378a/ZBTB20 axis [Citation23]. Consistent with this, this work observed that ZBTB20 abundance was distinctly elevated in AML tissues and cells. Furthermore, rescue experiments manifested that the influences of miR-192-5p overexpression on the malignant characteristics of AML cells were partly attenuated by increasing ZBTB20 expression, suggesting that miR-192-5p repressed AML cell development by targeting ZBTB20. In addition, the correlations among circ_0001602, miR-192-5p, and ZBTB20 were explored. The results revealed that the ZBTB20 level was reversely relevant to the miR-192-5p level and positively relevant to circ_0001602 expression. Besides, circ_0001602 could positively modulate ZBTB20 expression by absorbing miR-192-5p. Therefore, we summarized that circ_0001602 could aggravate the vicious properties of AML cells by competitively sequestering miR-192-5p to elevate ZBTB20 expression in vitro.
In conclusion, our results disclosed that circ_0001602 level was remarkably elevated in AML, and circ_0001602 could aggravate the development of AML cells through modulating the miR-192-5p/ZBTB20 axis. Our stupdy manifested that circ_0001602 might serve as a hopeful target for AML therapy.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Rubnitz JE, Gibson B, Smith FO. Acute myeloid leukemia. Hematol Oncol Clin North Am. 2010;24(1):35–63. doi:10.1016/j.hoc.2009.11.008
- Saleh K, Khalifeh-Saleh N, Kourie HR. Acute myeloid leukemia transformed to a targetable disease. Future Oncol. 2020;16(14):961–972. doi:10.2217/fon-2019-0670
- Appelbaum FR, Gundacker H, Head DR, et al. Age and acute myeloid leukemia. Blood. 2006;107(9):3481–3485. doi:10.1182/blood-2005-09-3724
- Stolzel F, Stölzel F, Mohr B, et al. Karyotype complexity and prognosis in acute myeloid leukemia. Blood Cancer J. 2016;6:e386), doi:10.1038/bcj.2015.114
- Stanchina M, Soong D, Zheng-Lin B, et al. Advances in acute myeloid leukemia: recently approved therapies and drugs in development. Cancers (Basel). 2020;12(11).
- Mukherjee S, Sekeres MA. Novel therapies in acute myeloid leukemia. Semin Oncol Nurs. 2019;35(6):150955), doi:10.1016/j.soncn.2019.150955
- Qu S, Zhong Y, Shang R, et al. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14(8):992–999. doi:10.1080/15476286.2016.1220473
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi:10.1038/s41576-019-0158-7
- Shang Q, Yang Z, Jia R, et al. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18(1):6), doi:10.1186/s12943-018-0934-6
- Lei M, Zheng G, Ning Q, et al. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19(1):30), doi:10.1186/s12943-020-1135-7
- Liu Y, Cheng Z, Pang Y, et al. Role of microRNAs, circRNAs and long noncoding RNAs in acute myeloid leukemia. J Hematol Oncol. 2019;12(1):51), doi:10.1186/s13045-019-0734-5
- Xue F, Li M, Liu Y, et al. Circ_0035381 regulates acute myeloid leukemia development by modulating YWHAZ expression via adsorbing miR-582-3p. Biochem Genet. 2023;61(1):354–371. doi:10.1007/s10528-022-10244-1
- Hu Q, Gu Y, Chen S, et al. Hsa_circ_0079480 promotes tumor progression in acute myeloid leukemia via miR-654-3p/HDGF axis. Aging (Albany NY). 2020;13(1):1120–1131.
- Peter ME. Targeting of mRNAs by multiple miRNAs: the next step. Oncogene. 2010;29(15):2161–2164. doi:10.1038/onc.2010.59
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi:10.1146/annurev-biochem-060308-103103
- Mo YY. MicroRNA regulatory networks and human disease. Cell Mol Life Sci. 2012;69(21):3529–3531. doi:10.1007/s00018-012-1123-1
- Wang X, Chen H, Bai J, et al. MicroRNA: an important regulator in acute myeloid leukemia. Cell Biol Int. 2017;41(9):936–945. doi:10.1002/cbin.10770
- Wang G, Yu X, Xia J, et al. MicroRNA-9 restrains the sharp increase and boost apoptosis of human acute myeloid leukemia cells by adjusting the Hippo/YAP signaling pathway. Bioengineered. 2021;12(1):2906–2914. doi:10.1080/21655979.2021.1915727
- Shang Z, Ming X, Wu J, et al. Downregulation of circ_0012152 inhibits proliferation and induces apoptosis in acute myeloid leukemia cells through the miR-625-5p/SOX12 axis. Hematol Oncol. 2021;39(4):539–548. doi:10.1002/hon.2895
- Tian C, Zhang L, Li X, et al. Low miR-192 expression predicts poor prognosis in pediatric acute myeloid leukemia. Cancer Biomark. 2018;22(2):209–215. doi:10.3233/CBM-170657
- Ke S, Li R-c, Lu J, et al. MicroRNA-192 regulates cell proliferation and cell cycle transition in acute myeloid leukemia via interaction with CCNT2. Int J Hematol. 2017;106(2):258–265. doi:10.1007/s12185-017-2232-2
- Li Q, Luan Q, Zhu H, et al. Circular RNA circ_0005774 contributes to proliferation and suppresses apoptosis of acute myeloid leukemia cells via circ_0005774/miR-192-5p/ULK1 ceRNA pathway. Biochem Biophys Res Commun. 2021;551:78–85. doi:10.1016/j.bbrc.2021.02.058
- Wang J, Liu ZH, Yu LJ. Long non-coding RNA LINC00641 promotes cell growth and migration through modulating miR-378a/ZBTB20 axis in acute myeloid leukemia. Eur Rev Med Pharmacol Sci. 2019;23(17):7498–7509.
- Chang W, et al. Circ-SFMBT2 facilitates the malignant growth of acute myeloid leukemia cells by modulating miR-582-3p/ZBTB20 pathway. Histol Histopathol. 2022;37(2):137–149.
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
- Panda AC. Circular RNAs Act as miRNA Sponges. Adv Exp Med Biol. 2018;1087:67–79. doi:10.1007/978-981-13-1426-1_6
- Polyatskin IL, Artemyeva AS, Krivolapov YA. Revised WHO classification of tumors of hematopoietic and lymphoid tissues, 2017 (4th edition): lymphoid tumors. Arkh Patol. 2019;81(3):59–65. doi:10.17116/patol20198103159
- Patop IL, Kadener S. circRNAs in Cancer. Curr Opin Genet Dev. 2018;48:121–127.
- Zhu LP, He YJ, Hou JC, et al. The role of circRNAs in cancers. Biosci Rep. 2017;37(5). doi:10.1042/BSR20170750
- Arnaiz E, Sole C, Manterola L, et al. CircRNAs and cancer: Biomarkers and master regulators. Semin Cancer Biol. 2019;58:90–99. doi:10.1016/j.semcancer.2018.12.002
- Perez de Acha O, Rossi M, Gorospe M. Circular RNAs in blood malignancies. Front Mol Biosci. 2020;7:109, doi:10.3389/fmolb.2020.00109
- Xiao Y, Ming X, Wu J. Hsa_circ_0002483 regulates miR-758-3p/MYC axis to promote acute myeloid leukemia progression. Hematol Oncol. 2021;39(2):243–253. doi:10.1002/hon.2829
- Liu W, Cheng F. Circular RNA circCRKL inhibits the proliferation of acute myeloid leukemia cells via the miR-196a-5p/miR-196b-5p/p27 axis. Bioengineered. 2021;12(1):7704–7713. doi:10.1080/21655979.2021.1982310
- Zhang S. The characteristics of circRNA as competing endogenous RNA in pathogenesis of acute myeloid leukemia. BMC Cancer. 2021;21(1):277), doi:10.1186/s12885-021-08029-7
- He C, Luo B, Jiang N, et al. Oncomir or antioncomiR: role of miRNAs in acute myeloid leukemia. Leuk Lymphoma. 2019;60(2):284–294. doi:10.1080/10428194.2018.1480769
- Fu S, et al. MiR-192-5p inhibits proliferation, migration, and invasion in papillary thyroid carcinoma cells by regulation of SH3RF3. Biosci Rep. 2021;41(9).
- Wang H, et al. LncRNA SOX2-OT regulates miR-192-5p/RAB2A axis and ERK pathway to promote glioblastoma cell growth. Cell Cycle. 2021: 1–11.
- Dong R-F, Zhuang Y-J, Wang Y, et al. Tumor suppressor miR-192-5p targets TRPM7 and inhibits proliferation and invasion in cervical cancer. Kaohsiung J Med Sci. 2021;37(8):699–708. doi:10.1002/kjm2.12398
- Cheng Y, Su Y, Wang S, et al. Identification of circRNA-lncRNA-miRNA-mRNA competitive endogenous RNA network as novel prognostic markers for acute myeloid leukemia. Genes (Basel). 2020;11(8).
- Liu J, Jiang J, Hui X, et al. Mir-758-5p suppresses glioblastoma proliferation, migration and invasion by targeting ZBTB20. Cell Physiol Biochem. 2018;48(5):2074–2083. doi:10.1159/000492545
- Zhang Y, Zhou X, Cheng L, et al. ZBTB20 promotes cell migration and invasion of gastric cancer by inhibiting IκBα to induce NF-κB activation. Artif Cells Nanomed Biotechnol. 2019;47(1):3862–3872. doi:10.1080/21691401.2019.1670188
- Xu X, Xie Q, Xie M, et al. Lncrna SNHG8 serves as an oncogene in breast cancer through miR-634/ZBTB20 axis. Cancer Manag Res. 2021;13:3017–3028. doi:10.2147/CMAR.S270128
