Abstract
Objectives: This study aims to identify the skin microbiota of a Saudi female population and investigate topically applying a specific bacterial strain isolated from the skin to understand its effects on skin cells’ basic functions, including cell viability, using a primary human keratinocyte cell culture system.
Methods: Microbiota samples were collected from four body parts of 200 females from AL-Madinah, Saudi Arabia, who were clinically screened for the absence of any disease. The four skin samples were collected by swabbing from the forehead, forearms, underarms, and behind both ears with no prior preparation or clearing of the skin surface. Samples were analyzed with the VITEK®2 Compact system after preparations according to the manufacturer's instructions. Staphylococcus aureus and commensal Staphylococcus epidermidis were isolated from the study participants and grown to investigate their effects on normal human epidermal keratinocyte (NHEK) basic functions, including cell viability in vitro.
Results: Three bacterial phyla (Firmicutes, Actinobacteria, and Proteobacteria) were most abundant in Saudi female populations, while Bacteroidetes was not observed in these individuals. The most abundant phylum in the underarm area was Firmicutes (91.01%), whereas Proteobacteria was abundant in the forearm area (76.92%), and Actinobacteria showed up in the behind-ear area (24.28%). Staphylococcus (S. aureus and S. epidermidis) was the most abundant genus, on average making up >80% of the microbial community within every sample. The effect of S. aureus and S. epidermidis on NHEK viability was investigated. Results indicated that S. aureus reduced the viability of NHEKs, while S. epidermidis strains did not. Next, the effects of co-incubating NHEKs with S. epidermidis and S. aureus on NHEK viability were investigated to determine if S. epidermidis could inhibit the effect of S. aureus. Monolayers incubated with S. aureus and S. epidermidis had higher percentage viability than monolayers incubated with S. aureus alone (p = 0.04, n = 3). NHEK monolayers treated with S. epidermidis two hours before adding S. aureus had higher percentage viability than monolayers exposed to S. aureus alone (p = 0.03, n = 3).
Conclusions: These data demonstrate that the most abundant bacterial phyla in a Saudi female population are Firmicutes, Actinobacteria, and Proteobacteria, and the most abundant genus is Staphylococcus (S. aureus and S. epidermidis). The data also reveal that S. epidermidis can protect human keratinocytes from the effects of the skin pathogen S. aureus in vitro. Further analysis of the mechanisms behind this is necessary.
1. Introduction
For a long time, the skin microbiota has been described by taking swabs from the skin surface and cultivating the samples. By adopting this approach, small subsets of microbiota generally present as skin bacteria, otherwise unculturable, could be specifically cultured. Several studies have characterized and detected skin microbiota by adopting molecular analysis and gene sequencing analysis [Citation1–3]. In studies resembling the work done by Segre et al., an analysis using 16S rRNA sequencing was done to show bacterial diversity at 20 diverse skin sites in 10 healthy humans. Nineteen bacterial phyla were identified, but skin bacteria were mainly classified into four phyla: Bacteroidetes (6.3%), Proteobacteria (16.5%), Firmicutes (e.g. Staphylococci; 24.4%) and Actinobacteria (e.g. Corynebacteria and Propionibacteria; 51.8%) [Citation1,Citation2]. These four phyla mainly existed in the oral cavity and the gut, but the proportions varied on the skin. Whereas Bacteroidetes and Firmicutes were abundant in the gut, Actinobacteria were found on the skin in large numbers.
The nature of skin differs depending on the anatomical locations and can be represented as either sebaceous, dry, or moist [Citation4–6]. Bacteria occupying these sites are as varied as each of these distinct ecological niches, having characteristic nutrients available. The sebaceous areas, such as the face, back, and back of the ears, have high proportions of “lipophilic” bacteria called Propionibacterium. Moist areas, such as the axilla, are rich in bacteria like Corynebacterium and Staphylococcus that prefer environments high in humidity. Dry skin, like the forearms, shows great bacterial variety and houses microbes such as Bacteroidetes and Proteobacteria, but these are in less proportion compared to moist and sebaceous areas [Citation1–4].
Human skin is a dynamic and complex ecosystem occupied by a large number of microorganisms. Moreover, variability in the skin microbiota is due to regional and local environmental conditions of host behaviour, genetics, and demographics. These differences have significant impact on individual health and morbidity. It has been reported that specific patterns of microbial diversity in individuals may help predict diseases and, therefore, help in accurate and speedy diagnosis [Citation7]. As normal microbiota of human skin has an impact on human health, studying their effects on the healthy skin of Saudi females may significantly add to the knowledge of the normal microbiota on healthy human skin in an environment different from those of European countries. New and diverse microbial species may be identified in environmental conditions that are ethnically varied. Environmental influences on the skin microbiota have been studied earlier. The factors that significantly influence microbial colonization on the skin include variations in temperature and moisture [Citation8]. To our knowledge, no study that reports the identification and investigation of the skin microbiota of the Arabic ethnic population has been done to date. Additionally, no research has been conducted to compare the skin microbiota of different racial groups. Hence the present study was designed to study and describe the skin microbiota of Saudi women.
In addition, many studies have demonstrated the importance of skin microbiota and its potential effects on the skin [Citation9], but comparatively few studies have analyzed the topical use of specific bacterial strains from skin. According to Prescott et al. [Citation9], the skin acts as a barrier, and its structure and functions are fundamental for human health. The skin plays a significant role in adapting the body's physiology to the altering environments. Skin microbiota plays a vital role in the maturation of keratinocytes and the host immune network with systemic effects and symmetric regulation. Evidence suggests that the skin microbiota could have important therapeutic value in preventing infection and promoting skin health. However, to date, comparatively few strains have been investigated. Therefore, this study also aimed to investigate the potential of topically applied specific strains isolated from participants’ skin to examine their effects on keratinocyte basic functions, including cell viability and cell proliferation in vitro.
2. Materials and methods
2.1. Identification of skin microbiota
Microbiome samples were collected by swabbing from four body parts of 200 individual females from AL-Madinah AL-Munawwarah, Saudi Arabia, who were clinically screened for the absence of any disease. Four skin samples were collected from the forehead, forearms, underarms, and behind both ears with no prior cleaning of the skin surface. Samples were analyzed using the VITEK® 2 Compact, which is a fully automated system that performs bacterial identification by biochemical analysis using colorimetry (VITEK® 2 Compact System, BioMerieux, US). The samples were prepared according to the manufacturer's instructions.
Staphylococcus aureus and commensal Staphylococcus epidermidis were isolated from our study participants and grown in aerobic conditions at 37°C in Nutrient Broth (Oxoid) or on Nutrient Agar (Oxoid). Cultures were adjusted spectrophotometrically with medium to contain approximately 106 cfu/ml based on the OD600 for each organism. For experiments, bacteria were washed twice in 0.8% NaCl solution and reconstituted in keratinocyte cell media, ready to infect cell cultures.
2.2. Ethics approval
This study was approved by the Medical Ethical Committee of the Faculty of Applied Medical Sciences at Taibah University (MLT 201622).
2.3. Keratinocyte viability assay
Normal human epidermal keratinocytes (NHEKs) were obtained from Promocell (Heidelberg, Germany). NHEKs in culture were used as a model system. These were cultured usually as described in Prince et al. (Citation2012) [Citation10]. The viability of confluent NHEK monolayers was determined by trypan blue exclusion assay following infection for 24 hours with either S. aureus or commensal S. epidermidis at concentrations of approximately 106 cfu/ml.
In some experiments, NHEK monolayers were exposed to S. aureus and commensal S. epidermidis at the same time. Uninfected NHEKs (control group), Infected NHEKs (NHEKs with bacteria).
In separate experiments, cells were exposed to S. epidermidis, and after two hours S. aureus was added. Cell viability was defined using trypan blue exclusion assays as described in Prince et al. (Citation2012) [Citation10].
2.4. Statistical analysis
All data were represented as percentage values. Experiments were performed three times, and one-way analysis of variance (ANOVA) in Graph Pad Prism software was used to compare the viability of NHEKs treated with bacteria (experimental group) and untreated cells (control group). Results were considered to be statistically significant if p < 0.05.
3. Results
3.1. Demographic characteristics of participant women (n = 200)
The study screened 200 women; 60% were aged 20–30 years, 55% were single, and 70% had university or college education (Table ). Participants had no skin diseases, and 89% of the women had no history of chronic diseases (Table ).
Table 1. Demographic and clinical characteristic of women (n = 200).
3.2. Overview of Saudi female skin microbiota
The skin microbiota was distinguished by cultivating from swabs of the skin surface. This approach gave only a small subset of consistent microbiota present, as most skin bacteria were unculturable (data not shown). However, using the VITEK® 2 Compact system for samples from four different skin sites in 200 healthy females, 33 bacterial phyla were defined. Most skin bacteria were classified into three phyla: Actinobacteria (e.g. Corynebacteria and Propionibacteria; 17%), Firmicutes (e.g. Leuconostoc and Staphylococci; 79.5%), and Proteobacteria (3.5%) (Figure and Table ). At the genus level, the well-documented skin colonizer Staphylococcus was the most abundant genus, on average making up >80% of the microbial community in every sample (Table ).
Figure 1. A healthy skin Phyla that was identify by Vitek 2. Actinobacteria (17%), Firmicutes (79.5%) and Proteobacteria (3.5%).
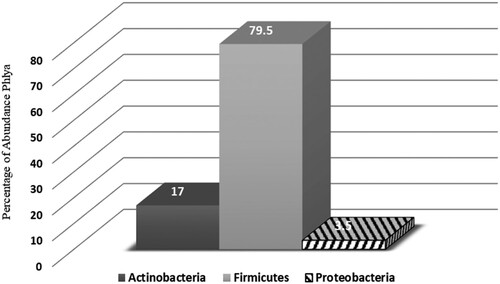
Table 2. Taxonomic identification of skin microbiota of Saudi Females. Relative abundances of the top three phyla across 200 samples included in this study with relative abundance of the top genera within sample.
3.3. Colonization prevalence by body part in study samples
Skin differs depending upon its location of anatomy and can be presented as dry (forearm), moist (underarm), and sebaceous (forehead and behind the ears) (Table ). The bacteria occupying these sites are varied, as each site shows a distinct ecological niche with its special nutrient availability. The most abundant phylum in the underarm area is Firmicutes (91.01%), whereas Proteobacteria is abundant in the forearm area (76.92%), and Actinobacteria was present in the behind-ear area (24.28%).
Table 3. Classification and distributing skin microbiota and its connected microenvironments; moist (Underarm), sebaceous (forehead and behind- ears) and dry (forearm).
3.4. Occurrence of potential opportunistic pathogens within Staphylococcus
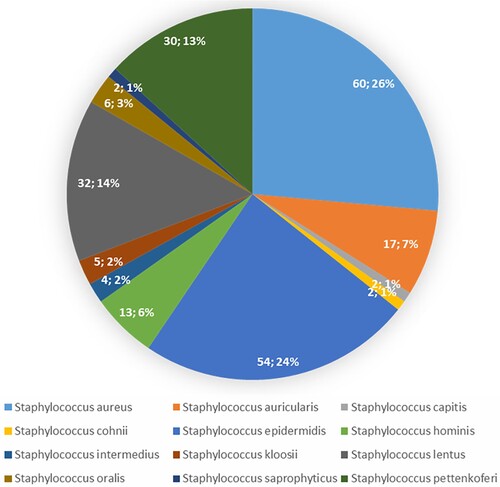
Staphylococcus was chosen for analysis as this genus is clinically important (Figure ). The potential human pathogen S. aureus overwhelmingly predominated the staphylococcal population across samples, making up an average of 60.26% of the staphylococcal population, while S. epidermidis made up 54.24%. S. aureus and S. epidermidis abundance were significantly different between various body sites. Forehead samples had greater abundance of S. epidermidis compared to other sites (p = 0.02), whereas S. aureus was more abundant in the underarm area than other sites (p = 0.01; Figure ).
3.5. Effects of S. aureus and S. epidermidis on keratinocyte viability using trypan blue assay
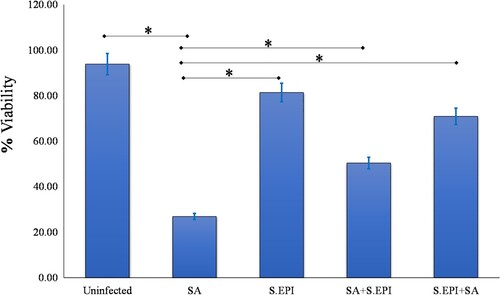
An experiment was performed to determine the effect of the common S. aureus and S. epidermidis on normal human epidermal keratinocyte (NHEK) viability. NHEKs were exposed to 106 cfu/ml bacterial cells for 24 hours to determine any toxic effects of the organisms. Viability was monitored using a trypan blue exclusion assay. After uninfected NHEKs (control group) were incubated for 24 hours, 93.83% of the cells were viable. By contrast, the percentage of NHEKs that were viable 24 hours after infection with S. aureus was significantly lower (26.87%; p = 0.002). After 24 hours of exposure to a concentration of 106 S. epidermidis/ml, 81.38% of the NHEKs remained viable (Figure ). S. aureus decreased viability of NHEKs, while S. epidermidis strains did not.
Figure 2. Relative abundance of staphylococcal species in current samples. Pie chart performs the abundance of species as a percentage of the entire microbial community.
The next experiments investigated the combination of S. aureus and S. epidermidis on NHEK viability to determine if S. epidermidis could reduce the effect of S. aureus. NHEKs were incubated with a combination of S. epidermidis and S. aureus at a concentration of 106 cells/ml. Cells were incubated overnight and the mean percentage viability of NHEKs was established using trypan blue assay. When S. aureus and S. epidermidis were added to NHEKs at the same time, only 50.38% of NHEKs remained viable after 24 hours, whereas 70.91% of NHEKs were viable when treated with S. epidermidis two hours before adding S. aureus (p = 0.03, Figure ).
4. Discussion
The Human Microbiome Project presents a large combination of data by employing high-throughput sequencing (The Human Microbiome Project Consortium). It has generated some useful reports on the skin microbiota, but most of these studies include a majority of western subjects. Several studies have investigated the distinct microbiota of European, Chinese, and American populations [Citation2,Citation11,Citation12], while studies including populations of Arabic ethnicity are rare. Therefore, studying the normal microbiota residing on the skin of healthy Saudi females may significantly add to the knowledge of normal microbiota of healthy human skin in an environment very different from the European milieu.
The current study characterizes the skin microbiota of Saudi Arabian females. The microbes that were found to be prevalent on their skin belonged to the genera that include Propionibacterium, Corynebacterium, and Staphylococcus. In addition, the subset data of this study contained three phyla and 400 genera (Figure ). Three phyla made up over 98% of all the skin microbiota: Firmicutes (79.5%), Actinobacteria (17%), and Proteobacteria (3.5%). At the genus level, Staphylococcus was the most abundant genus, on average making up >80% of the microbial community within each sample. In contrast, Bacteroidetes has been reported in American individuals [Citation1–3], but not in Saudi individuals. Significant differences have also been observed between Saudi females and Chinese females. The phylum Firmicutes was seen to be most abundant in Saudi individuals, while Marcus [Citation13] found that Proteobacteria and Actinobacteria were significantly more abundant in Chinese females.
Grice and colleagues studied the microbiota from various skin sites and found that the dominant microbiota was heavily influenced by location [Citation1–4]. This is in accordance with our results, which showed that the most abundant phylum in the underarm area was Firmicutes, whereas Proteobacteria was abundant in the forearm area (76.92%), and Actinobacteria was present in the behind-ear area in Saudi females (Table ). However, another set of studies found that Staphylococci and Propionibacteria dominated in sebaceous sites, while Corynebacteria dominated in moist sites [Citation2]. Gao et al. [Citation5] showed that the primary phyla found on the skin of the forearm were Actinobacteria, Firmicutes, and Proteobacteria. Other studies have also concluded that anatomically distinct skin sites such as the ear [Citation11], hands [Citation14], and forehead [Citation12] include different bacterial species.
Our studies also confirm that skin microbial communities differ based on varying racial backgrounds and cannot be purified by methodological variations alone. Both the community structure and membership are responsible for microbial differences between racial groups, essentially leading to the growth of the global pan-microbiome. Moreover, some variations can be explained by the absence or presence of specific phyla that may be low in abundance. Some evidence confirms that changes in the overall balance of organisms may be due to a variety of internal and external factors, such as nutrient availability, water, oxygen, temperature, and pH, along with the individual's gender and ethnicity.
Figure 3. The percentage of Staph. Abundance in different body parts. A significant difference in S. aureus and S. epidermidis distributed between various body parts.
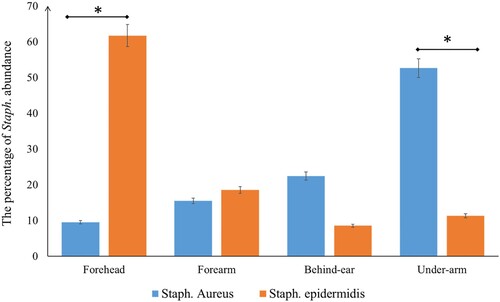
Figure 4. S. epidermidis protects NHEK from the effects of S. aureus. NHEK viability was unaffected by S. epidermidis (S.EPI). S. epidermidis protected the NHEK from the effects of S. aureus (SA) and the viability of NHEK treated with combination of two bacteria was 50.38%. The viability of NHEK treated with S. epidermidis 2 hours before adding the S. aureus was 70.91% compared to 26.87% in NHEK infected with S. aureus alone (P = 0.03, n = 3).
In general, bacteria on the skin can be classified as either “transient” or “resident”. For reasons that seem obvious, environments that are exposed to the outside generally have a higher population of transient species than the deeper unexposed areas [Citation15]. According to the present study, coagulase-negative Staphylococci (CoNS) are the most prominent gram-positive cocci isolated from the skin of the Saudi population (Figure ), mainly S. epidermidis and S. aureus. Altogether, at least 12 species of CoNS have been isolated from the skin and are found over all parts of the body, which partially agrees with some studies, such as those reported by Roth and James [Citation15] and Hamory et al. [Citation16].
Normal microbiota on healthy human skin is also a major component of the barrier against colonization of potentially pathogenic microbes or excessive growth of “opportunistic” pathogens [Citation17]. For example, S. epidermidis can produce antimicrobial factors, such as phenol, that affect the colonization of pathogens [Citation17,Citation18,Citation22,Citation23]. In addition, S. epidermidis has been revealed to produce antimicrobial peptides that kill S. aureus, and transferring this species onto the skin of patients with atopic dermatitis decreased S. aureus colonization [Citation18]. Based on the positive influence of skin commensals on skin health and their association with skin diseases, we hypothesize that skin commensal bacteria may inhibit the toxic effects of S. aureus on keratinocytes and improve cell viability. This preliminary study was undertaken to begin to characterize the effects of S. epidermidis strains on the functions of keratinocytes, in particular, the possibility that S. epidermidis could protect keratinocytes from the common pathogen S. aureus. To begin, the effect of S. epidermidis on keratinocyte viability was tested. The common skin pathogen S. aureus killed significant numbers of NHEKs when incubated with cells for 24 hours. By contrast, the same concentration (106 cfu/ml) of commensal S. epidermidis did not affect the viability of NHEKs, suggesting that S. epidermidis has no adverse effects on NHEKs. Therefore, in the next part of the study, the possibility that S. epidermidis can protect cells from the effects of S. aureus was investigated. Co-incubation of NHEKs with approximately 106 cfu/ml of the pathogen S. aureus and 106 cfu/ml of S. epidermidis increased the viability of cells when compared to cells infected with S. aureus alone (Figure ).
The protective effects of probiotics could be due to several different mechanisms. One likely mechanism is competitive inhibition between the pathogen and commensal for nutrients and receptor binding sites [Citation19,Citation20,Citation21] although this was not investigated in this study. Furthermore, the timing of adding S. epidermidis compared to the pathogen is important; S. epidermidis protected NHEKs if it was added two hours before or at the same time as the pathogen, but not after the infection had begun (data did not show), suggesting a mechanism of competition with the pathogen for NHEK binding sites. The present study did not explore the possible mechanisms involved in the protective effects of the S. epidermidis, but this will be done in future work.
5. Conclusion
The last few years have seen the effective application to study the microbiota of all major colonized body sites. To the best of our knowledge, this study is the first to identify and characterize the microbiota of human skin of Saudi Arabian females. The current study advocates that the normal microbiota on healthy human skin of Saudi females is significantly different from other populations. We identified three phyla (Firmicutes, Actinobacteria, and Proteobacteria) that made up over 98% of all the skin microbiome. Staphylococcus was the most abundant genus of the microbial community within each sample. These findings are also the first to demonstrate S. epidermidis and S. aureus interactions on human keratinocytes and suggest that isolated commensal S. epidermidis strains may be a useful means of counteracting S. aureus infection and reducing the toxicity of pathogens. The use of commensal bacteria thus offers options to develop a new therapy that could reduce skin infection. However, more investigation is required to improve our understanding of S. epidermidis functions.
Statement of ethics
The volunteers have given their written or verbal informed consent prior to the start of the study. They were assured that the information obtained will only be shared between the researchers and that they were free to withdraw from study at any time without any pressure on them. Participants were identified by codes and never by their real names.
The study protocol has been approved by the Faculty of Applied Medical Sciences’ ethical committee on human research (MLT 201622).
Acknowledgement
The Author extend her appreciation to the Deanship of Scientific Research at Taibah University for funding the work through the research group project (8016/5/1437). I acknowledge Maternity and Children Hospital, Madinah especially the Microbiology Lab (Dr. Sahar Amin, Consultant of Microbiology) for resources and supporting us to do the experiments. The author would also like to thank Miss Rania Osta and Miss Enass Naeem, the lab technicians who assisted us in the current study for their cooperation, kindness, and time.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Grice EA, Kong HH, Renaud G, et al. A diversity profile of the human skin microbiota. Genome Res. 2008a;18:1043–1050.
- Grice EA, Kong HH, Conlan S, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009a;324:1190–1192.
- Grice EA, Segre JA. The skin microbiome. Nature Reviews in Microbiology. 2011;9:244–253.
- Grice EA. The skin microbiome: potential for novel diagnostic and therapeutic approaches to cutaneous disease. Medical Surgery. 2014;33:98–103.
- Gao Z, Perez-Perez GI, Chen Y, et al. Quantitation of major human cutaneous bacterial and fungal populations. Journal of Clinical Microbiology. 2010;48:3575–3581.
- Gao Z, Tseng CH, Pei Z, et al. Molecular analysis of human forearm superficial skin bacterial biota. The National Academy of Sciences of the USA. 2007;104:2927–2932.
- Wilson M. Microbial inhabitants of humans: their ecology and role in health and disease. Cambridge: Cambridge University Press; 2010.
- McBride ME. Physical factors affecting skin flora and disease. In: The skin microflora and microbial skin disease. Cambridge: Cambridge University Press; 1993. p. 73–101.
- Prescott SL, Larcombe D-L, Logan AC, et al. The skin microbiome: impact of modern environments on skin ecology, barrier integrity, and systemic immune programming. World Allergy Organization Journal. 2017;10:29.
- Prince T, Mcbain AJ, O’Neill CA. Lactobacillus reuteri protects epidermal keratinocytes from Staphylococcus aureus-induced cell death by competitive exclusion. Applied and Environmental Microbiology. 2012;78:5119–5126.
- Leung MHY, Wilkins D, Lee PKH. Insights into the pan-microbiome: skin microbial communities of Chinese individuals differ from other racial groups. Scientific Reports. 2015;5:11845.
- Fierer N, Hamady M, Lauber CL, et al. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. National Academy of Sciences of the USA. 2008;105:17994–17999.
- Park YJ, Lee HK. The role of skin and orogenital microbiota in protective immunity and chronic immune-mediated inflammatory disease. Front Immunol. 2018;8:1955.
- Frank DN, Spiegelman GB, Davis W, et al. Culture-independent molecular analysis of microbial constituents of the healthy human outer ear. Journal of Clinical Microbiology. 2003;41:295–303.
- Dekio I, Hayashi H, Sakamoto M, et al. Detection of potentially novel bacterial components of the human skin microbiota using culture-independent molecular profiling. Journal of Medical Microbiology. 2005;54:1231–1238.
- Roth RR, James WD. Microbial ecology of the skin. Annual Reviews in Microbiology. 1988;42:441–464.
- Hamory BH, Parisi JT, Hutton JP. Staphylococcus epidermidis: a significant nosocomial pathogen. American Journal of Infection Control. 1987;15:59–74.
- Byrd AL, Deming C, Cassidy SKB, et al. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci Transl Med. 2017;9:eaal4651.
- Nakatsuji T, Chen TH, Narala S, et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med. 2017;9:eaah4680.
- Ghoul M, Mitri S. The ecology and evolution of microbial competition. Trends Microbiol. 2016;24:833–845.
- Jenni L, Karkman A, Laatikainen T, et al. Patterns in the skin microbiota differ in children and teenagers between rural and urban environments. Scientific Reports. 2017;7:45651.
- Chetan S, Rokana N, Chandra M, et al. Antimicrobial resistance: its surveillance, impact, and alternative management strategies in dairy animals. Frontiers in Veterinary Science. 2017;4:237.
- O’Sullivan JN, Rea MC, O’Connor PM, et al. Human skin microbiota is a rich source of bacteriocin-producing staphylococci that kill human pathogens. FEMS Microbiology Ecology. 2019;95:fiy241.
