ABSTRACT
The phenotypic, distributional, and genetic distinctiveness of the bandtail goatfish Upeneus taeniopterus within the genus Upeneus (Mullidae) is elaborated using a comprehensive alpha-taxonomic and barcoding approach. Based on a large number of morphometric, meristic and colour characters obtained from 71 preserved or freshly photographed specimens, an updated diagnosis, a redescription, and detailed inter- and intraspecific comparisons are provided. The distribution information is revised with strong emphasis on ensuring correct species identification. Upeneus taeniopterus shows intraspecific variation in morphology and number of oblique bars on the caudal fin related to two size classes, ‘subadults’ (< 12 cm SL) and ‘adults’ (12 cm SL or larger). Indications for population differences were only detected for the smaller size class, possibly reflecting geographic developmental differences. This species is widely distributed in the Indo-Pacific from Mozambique to the Tuamoto Archipelago and from the Ryukyu Islands to Tonga and occurs mostly in very shallow subtidal sandy beach or lagoon habitats of oceanic islands and atolls. Four new records of the species for Palau, Papua (Indonesia), Tonga and Vanuatu are reported. Comparisons with all other 36 congeners revealed clear differences from U. taeniopterus in the combination of maximum size, eight meristic and colour characters, distributional range and habitat selection. The only congeneric species with similarly large maximum size and wide distributional range is Upeneus vittatus, which differs however in morphology, colour and habitat. The congeneric species differ from U. taeniopterus with sequence divergences which are comparable to those observed among genera. More genetic tissue samples are needed to further investigate the relatedness among Upeneus species and to search for phylogeographic patterns in U. taeniopterus. The need to thoroughly study the insufficiently explored subtidal sandy habitats of oceanic islands and atolls is emphasized.
RESPONSIBLE EDITOR:
Introduction
The bandtail goatfish Upeneus taeniopterus Cuvier, 1829 (Mullidae), originally described from Trincomalee, Sri Lanka, occurs mostly in shallow coastal waters of oceanic islands and atolls within a vast area extending from Mozambique to Hawaii (Uiblein & Heemstra Citation2010). In a regional taxonomic review of the genus Upeneus based on a large number of morphometric, meristic and colour characters from 24 species, Uiblein & Heemstra (Citation2010) established the senior synonymy of U. taeniopterus with U. arge Jordan & Evermann, 1903, described from Hawaii. They included U. taeniopterus in the so-called tragula species group due to low gill raker and pectoral fin-ray counts and the presence of dark oblique bars on both caudal-fin lobes. This assignment facilitated the preparation of identification keys and was followed in subsequent studies that added additional species to the tragula group (Uiblein & Heemstra Citation2011a; Uiblein & Gouws Citation2014).
In earlier taxonomic accounts featuring species of the tragula group (Uiblein & Gouws Citation2014, and citations therein), considerable differences from U. taeniopterus were encountered, but not elaborated further. Regarding the currently available distribution information, none of the other nine tragula-group species matches the wide range of U. taeniopterus (cf. Uiblein et al. Citation1998; Randall & Kulbicki Citation2006; Uiblein & Heemstra Citation2010, Citation2011a; Uiblein & Gouws Citation2014): U. heemstra Uiblein & Gouws, Citation2014 occurs in the Western Indian Ocean and off SE India; U. luzonius Jordan & Seale, 1907 is currently known only from the Philippines (reported occurrences from other areas have not been verified so far); U. margarethae Uiblein & Heemstra, Citation2010 occurs in the Indian Ocean and the Arafura Sea; U. mouthami Randall & Kulbicki, Citation2006 has only been recorded from the Chesterfield Islands, New Caledonia; U. niebuhri Guézé, 1976 is restricted to the Gulf of Suez, Red Sea; U. oligospilus Lachner, Citation1954 and U. randalli Uiblein & Heemstra, Citation2011 occur in the Persian Gulf; U. sundaicus (Bleeker, 1855) is known from the Persian Gulf and the Eastern Indian Ocean to the South China Sea and Australia; and U. tragula Richardson, 1846 occurs from off the Andaman Islands to New Caledonia and Japan. With the exception for U. mouthami, all these species occur mostly on continental shelves and much less around islands or atolls than does U. taeniopterus.
Regarding phenotypic characters (cf. Uiblein & Heemstra Citation2010; Uiblein & Gouws Citation2014), U. taeniopterus differs from the other tragula-group species in a higher lateral-line scale count and in having at least two body stripes vs. only a single or no body stripe. With a reported maximum size of 27 cm SL (Uiblein & Heemstra Citation2010) it appears to attain much larger size than the other nine species, which range from 9.4 (U. mouthami) to 19 cm SL (U. tragula) (Uiblein & Gouws Citation2014). These characteristics combined with the differences between the tragula group and the other species and species groups of the genus Upeneus (e.g., Uiblein & Heemstra Citation2011b; Uiblein & Gouws Citation2015) indicate the rather distinctive status of U. taeniopterus. However, detailed alpha-taxonomic comparisons with all other 36 currently known Upeneus species (Uiblein & White Citation2015) have not been conducted yet. Also, no genetic studies have been made so far to elaborate the relatedness of U. taeniopterus with congeneric species.
This paper investigates the phenotypic, distributional and genetic distinctiveness of U. taeniopterus within the genus Upeneus in a comprehensive alpha-taxonomic and barcoding approach. A large amount of phenotypic and occurrence data were collected using museum specimens, photographs and/or in situ observations of fresh fish, and relevant literature. An updated taxonomic account for U. taeniopterus was prepared featuring a detailed diagnosis and a redescription. Growth-related allometric changes and population differences were also considered and maximum size and distribution information were revised. These data were compared with partly updated information from all other currently known congeneric species (n = 36). Finally, all barcoding data of U. taeniopterus available to the authors, from two specimens from the Seychelles and Hawaii, respectively, were analysed with comparative data from 12 congeneric species. These findings and the need for further research to better understand the interspecific relationships among Upeneus species and the biology and ecology of U. taeniopterus are discussed.
Materials and methods
Taxonomy
In total 71 specimens of Upeneus taeniopterus were examined. From 53 scientific collection specimens standard length (SL), 40 measurements, 10 counts, and colour characters were gathered ( and ). Through communication with colleagues from scientific fish collections and/or examination of photographs, size information of six additional preserved specimens were obtained, with counts of oblique bars on the caudal fin available from five of these and the lateral-line scale count from a single specimen (). Additional data were obtained using photographs of one preserved and 14 freshly caught or live U. taeniopterus, the latter including three of the 53 above-mentioned museum specimens.
Table I. Morphometric, meristic, and colour characters for Upeneus taeniopterus along its distributional range.
Table II. Taxonomic group assignment, number of specimens examined, and maximum size, morphological and colour characters for 37 species of Upeneus.
Methods for obtaining morphometric, meristic and colour data follow Uiblein & Heemstra (Citation2010) and Uiblein & Gouws (Citation2015). Goatfishes may show considerable allometric changes during ontogeny that relate to changes in lifestyle and the possible onset of sexual maturity (Uiblein & Gledhill Citation2015). To account for allometric changes in U. taeniopterus, plots of SL and morphometric, meristic and quantitative colour characters were examined and two distinct size groups established, with fish smaller than 12 cm SL assigned to ‘subadults’ and individuals of 12 cm SL or larger to ‘adults’.
Regarding distribution information, a large number of literature sources including taxonomic accounts, annotated species lists, books and fisheries reports were screened to get an overview of previously published records. For the detailed elaboration and mapping of the distribution, priority was however given to information available from scientific collection specimens and photographs of well identifiable live, freshly collected, or preserved specimens that were examined by us. If literature was used to refer to records that were not directly verified, attention was paid to account for the correctness of species identification by seeking additional support and/or documentation through contact with local experts. Archipelagos or atolls were treated as single geographic entities for mapping the distribution. For the verified records all available occurrence details were included in the material list.
For the interspecific alpha-taxonomic comparisons, a comprehensive data set obtained from 810 specimens of all other 36 congeneric species was used, focusing primarily on maximum size (SL in mm) and eight diagnostically important meristic and colour characters. New comparative data were obtained for the recently described U. nigromarginatus and four other species (see material list below). In addition, complementary information from four published accounts (Fischer & Bianchi Citation1984; Randall Citation1996; Randall & Kulbicki Citation2006; Yamashita et al. Citation2011) was referred to when published values exceeded the ranges obtained by us. For the distributional comparisons, 10 recently published accounts (Uiblein & Heemstra Citation2010, Citation2011a, Citation2011b; Uiblein & McGrouther Citation2012; Uiblein & Causse Citation2013; Uiblein & Lisher Citation2013; Uiblein & Gouws Citation2014, Citation2015; Uiblein & Gledhill Citation2015; Uiblein & White Citation2015) and new occurrence data from the material examined in the present study were used.
Institutional abbreviations follow Fricke & Eschmeyer (Citation2016). Other abbreviations are: HIFIRE = Fish collection of the Institute of Marine Research, Bergen, Norway; HT = holotype; LL = lateral-line scales; PT = paratype(s).
Comparative material examined
Only the five species for which formerly unpublished comparative data were generated for the present study are listed below. For all other 31 Upeneus species complete material lists have been provided in earlier publications which are referred to accordingly in .
Upeneus filifer (7 specimens, 68–106 mm SL): eastern Australia: AMS 12541, PT, 132 mm, Queensland, Cape Gloucester, 20°04′S, 148°27′E, 106 m; CSIRO H 6758-02, 99 mm, Queensland, NE of Shoalwater Bay, 21°47.08′S, 151°21.95′E; AMS 32120-003, 83 mm, New South Wales, off Clarence River, 29°20′S, 153°34′E, 67–73 m; AMS 32196-001, 4 specimens, 68–82 mm, New South Wales, off Clarence River, 29°25′S, 153°34′E, 65–70 m.
Upeneus mouthami (8 specimens, 47–94 mm SL): Coral Sea, New Caledonia, Chesterfield Islands: BPBM 33855, PT, 94 mm, Chesterfield Bank, 20°51′0″S, 158°45′00″E, 71 m; MNHN 2004-1571, PT, 73 mm, Chesterfield Bank; USNM 378143, PT, 81 mm, 19°12′23″S, 158°42′02″E; BPBM 39467, PT, 88 mm, Bellona Reefs, 21°24′54″S, 159°09′18″E, 60 m; Vanuatu (new record): MNHN 2002-0070, 1 of 3, 55 mm, 15°37.98′S, 167°03′E, 140–175 m; MNHN 2008-1433, 1 of 5, Malo Island, Bruat Channel, 15°37.32′S, 167°09.60′E, 52–66 m; MNHN 2008-1459, 47 mm, Espirito Santo Island, 15°31.68′S, 167°10.80′E, 36–43 m; MNHN 2010-0616, 51 mm, NW Malo Island, 15°39.90′S, 167°03.78′E, 114–132 m.
Upeneus nigromarginatus: (5 specimens, 154–201 mm SL): Philippines, Panabo City, fish market, 7°18′23″N, 125°41′1″E: RMNH.PISC.37991, HT, 201 mm; RMNH.PISC.36422, PT, 154 mm; RMNH.PISC. 36423, PT, 156 mm; RMNH.PISC.36424, PT, 162 mm; RMNH.PISC.37992, PT, 154 mm.
Upeneus randalli (9 specimens, 60–106 mm SL): Persian Gulf: BPBM 33180, HT, 101 mm, off southern Kuwait, 29˚00′N, 48˚25′E, 15–20 m; BPBM 21201, 6 PT, 66–88 mm, Bahrain, fish market; BPBM 29498, 60 mm, Bahrain; Iran, Gulf of Oman (new record): ZMUC P49161, PT, 106 mm, Chahabar (erroneously reported as from Persian Gulf in Uiblein & Heemstra Citation2011a).
Upeneus quadrilineatus (10 specimens, 62–123 mm SL): Indonesia, Java: CSIRO H 7696-01, 81 mm, East Java, Pacitan, 08°13′S, 111°04′E; Central Java, Cilacap, fish market: CSIRO H 7697-01, 102 mm; CSIRO H 7469-02, 133 mm; CSIRO H 7697-02, 2, 112–117 mm; CSIRO H 7469-03, 123 mm; NCIP 3495, 90 mm, West Java, Tanjung Pasir; Lombok, Tanjung Luar, fish market: MZB 22936, 62 mm; MZB 22937, 63 mm; CSIRO H 7217-04, 66 mm.
Genetic studies
Phylogenetic and genetic distance analyses, using the barcoding cytochrome c oxidase subunit I (COI) gene fragment (Hebert et al. Citation2003), were conducted to investigate and confirm the placement of Upeneus taeniopterus within the tragula group or, failing that, establish the likely group affinity of this species. Data used in previous species delineation studies (Uiblein & Gouws Citation2014, Citation2015) of Upeneus were secured from GenBank (Citation2015) or obtained from the sources documented previously (Uiblein & Gouws Citation2014, Citation2015). At least one representative of each species examined in these studies was included in the present analysis, with multiple specimens/sequences included from geographically separated localities and paragenetypes of particular species, where possible. Sequences of U. taeniopterus were obtained from Barcode of Life Data Systems v3 (BOLD; Boldsystems Citation2015), from barcoding projects managed by SAIAB and USNM. Collectively, the included data () represented all six described species groups within Upeneus (with U. filifer excluded): the japonicus, moluccensis, suahelicus, stenopsis, tragula and vittatus groups. Sequences of Mulloidichthys vanicolensis and Parupeneus barberinus, used previously as outgroups (Uiblein & Gouws Citation2015), were again used as outgroups in the present analysis.
Table III. Sources and accession details (GenBank accession numbers and/or BOLD process IDs) of the cytochrome c oxidase subunit I (COI) sequence data for the Upeneus representatives and the two outgroup (Mulloidichthys vanicolensis (Valenciennes, 1831) and Parupeneus barberinus (Lacepède, 1801)) specimens included in the current genetic study.
Final sequence alignment and phylogenetic analysis proceeded as documented by Uiblein & Gouws (Citation2014), with trees being constructed under maximum likelihood (ML) and unweighted parsimony (UP) frameworks in PAUP* 4b10 (Swofford Citation2002). The use of jModelTest 4.1.2 (Darriba et al. Citation2012) to determine the most suitable model of nucleotide evolution prior to the ML analysis, and the use of RAxML 8.2.4 (Stamatakis Citation2014) to conduct the bootstrapping (with 1000 replicates) to determine nodal support (Felsenstein Citation1985) under a ML framework were the only alterations to the previously published (Uiblein & Gouws Citation2014) methodology. The likelihoods of alternative topologies to those obtained through the above analyses, constrained to enforce certain species relationships or species group membership, were evaluated by means of Shimodaira & Hasegawa (Citation1999: SH) tests in PAUP*, with full optimization and 1000 bootstrap replicates. Kimura’s (Citation1980) 2-parameter (K2P) distances were calculated among included specimens, species and species groups, as described previously (Uiblein & Gouws Citation2014, Citation2015).
Results
Taxonomy
Upeneus taeniopterus Cuvier, 1829
Bandtail goatfish
( and ; –)
Figure 1. Upeneus taeniopterus. (A) Seychelles, Mahé Island (Baie Ternay), SAIAB 76409, 93 mm (P.C. Heemstra); (B) Mascarenes, Mauritius, Rodrigues (Antonio’s Finger), SAIAB 69803, 134 mm (P.C. Heemstra); (C) Cook Islands, Aitutaki Island, 255 mm (G. McCormick); (D) Solomon Islands, Fenualoa Island, USNM 389116, 189 mm SL (J.T. Williams); (E) Indonesia, Papua, Biak (Biak city fish market), 235 mm (F. Uiblein); (F) Hawaii, Molokai Island, adult (Hallelujah Hou Fishing; www.hallelujahhoufishing.com); (G–H) Tonga, Vava’u Archipelago: (G) Vava’u Island (W of Neiafu), adult (E. Clua); (H) Kapa Island, 240 mm (F. Uiblein).
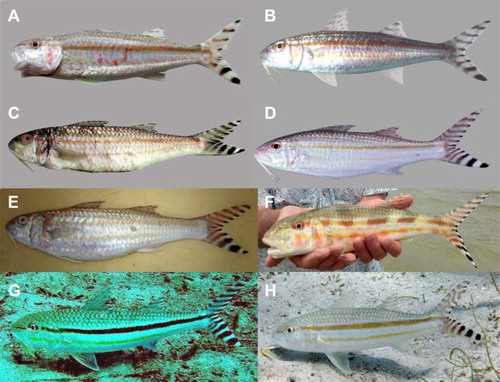
Figure 2. Scatter plots of morphological characters and number of oblique bars on caudal fin for Upeneus taeniopterus, the dotted line indicating the separation of the two size classes (adults and subadults).
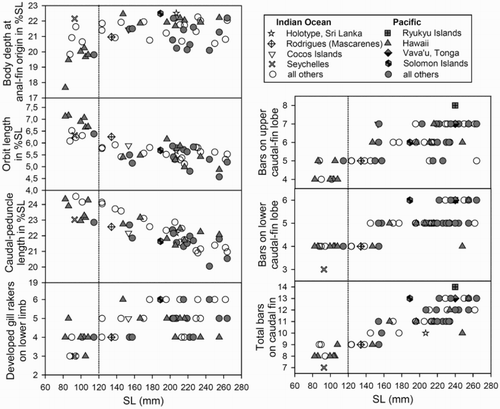
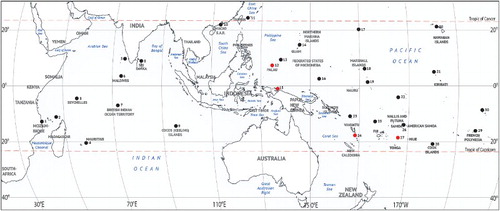
Figure 3. Geographic distribution of Upeneus taeniopterus. (1) Mozambique: Pinda Bank; (2) Seychelles: Aldabra Islands; (3) Seychelles: Mahé Island; (4) Mascarenes, Mauritius: Rodrigues; (5) India: Laccadives; (6) Maldives: Villingili Island; (7) British Indian Ocean Territory (BIOT), Chagos: Diego Garcia and Peros Banhos Atolls; (8) Sri Lanka: Trincomalee; (9) Australia, Cocos (Keeling) Islands; (10) People's Republic of China, Macau; (11) Japan, Ryukyu Islands: Ishigakijima; (12) Palau: Arakabesan Island; (13) Micronesia, Caroline Islands: Yap Island; (14) Mariana Islands: Saipan and Guam Islands; (15) Indonesia, Papua: Biak Island; (16) Micronesia, Caroline Islands: Pohnpei Island; (17) Wake Atoll (US territory); (18) Marshall Islands: Arno Atoll; (19) Kiribati, Gilbert Islands: Onotoa and Tarawa Atolls; (20) Hawaii (USA): Hilo, Molokai and Oahu Islands; (21) Line Islands (US territory): Palmyra Atoll; (22) Kiribati, Phoenix Islands: Canton Island; (23) Solomon Islands, Santa Cruz Islands: Fenualoa Island; (24) Vanuatu: Efate Island; (25) Fiji: Rotuma Island; (26) Samoa Islands; (27) Tonga: Vava'u Archipelago, Vava’u and Kapa Islands; (28) Cook Islands, Southern Cook Islands: Aitutaki Island; (29) French Polynesia, Society Islands: Tahiti Island; (30) French Polynesia, Tuamotu Archipelago: Rangiroa Island. New records are plotted in red.
Upeneus taeniopterus Cuvier in Cuvier & Valenciennes, 1829: 451 (type locality: Sri Lanka, Trincomalee); Fischer & Whitehead Citation1974; Wass Citation1984; Winterbottom et al. Citation1989; Zug et al. Citation1989; Seeto & Baldwin Citation2010; Uiblein & Heemstra Citation2010; Uiblein & Gouws Citation2014.
Upeneus arge Jordan & Evermann, 1903: 187 (type locality: Hawaii); Fowler Citation1928; Schultz Citation1943; Lachner Citation1954; Jones & Kumaran Citation1980; Fisheries Division Citation1983; Masuda et al. Citation1984; Myers & Donaldson Citation2003; Lobel & Lobel Citation2004; McCormack Citation2007; Randall Citation2007; Mundy et al. Citation2010; Allen & Bailey Citation2011.
Material examined
Preserved specimens (59 specimens, 82–290 mm SL, fresh photograph information in parentheses): HT: MNHN 0000-9568, 207 mm, Sri Lanka, Trincomalee, 8°34′0″S, 81°13′00″E; Mozambique: SAIAB 13915, 101 mm, Pinda Bank, 14°13′S, 40°46′E; Seychelles, Aldabra Islands: USNM 267590, 10 specimens, 170–264 mm, Aldabra Atoll, Ile Picard, lagoon inside SE portion of island, 9°22′40″S, 46°14′40″E, 1 m; Seychelles, Mahé: SAIAB 76409, 93 mm, Baie Ternay, 4°38′31″S, 55°22′49″E (Phil C. Heemstra, colour photo of freshly collected fish (A); genetic tissue sample); Mascarenes, Mauritius: SAIAB 69803, 134 mm, Rodrigues, Antonio's Finger, 19°39′41″S, 63°28′1″E, 25–50 m (Phil C. Heemstra, colour photo of freshly collected fish (B)); Maldives, Villingili Island: BPBM 18863, 214 mm, E side of island, lagoon; Chagos (British Indian Ocean Territory): USNM 396089, 8 specimens, 88–177 mm, Diego Garcia Atoll, 7°25′56″S, 72°25′43″E, 1 m; USNM 396090, 2 specimens, 211–250 mm, Diego Garcia Atoll, 7°15′33″S, 72°22′40″E, 3 m; Australia, Cocos (Keeling) Islands: BMNH 1949.11.29.220, 153 mm, Horsburgh Island; China, Macau: MNHN A-3500, 220 mm, 22°3′N, 116°36′E; Palau: CAS 206472, 114 mm, SE Arakabesan Island, 7°20′30″N, 134°27′26″E; Micronesia, Yap Island: CAS 232880, 2 specimens, 150–154 mm, NE Colonia, reef flat, 9°32′03″N, 138°07′59″E; Mariana Islands, Saipan Island: ANSP 114755, 2 specimens, 207–219 mm, lagoon between Managaha Island and Tanapag; Kiribati, Phoenix Islands: USNM 115685, 2 specimens, 222–255 mm, Canton Island, lagoon; Kiribati, Gilbert Islands: AMS IB-5538, 226 mm, Tarawa Atoll; USNM 167479, 264 mm, Onotoa Atoll, N island, lagoon; Hawaii, Oahu, Honolulu: USNM 50667, HT of Upeneus arge, 166 mm; ANSP 24227, PT of U. arge, 206 mm; ANSP 89186, 234 mm; ANSP 179672, 2 specimens, 216–255 mm; USNM 55100, 196 mm; ZMUC P49132-38, 7 specimens, 82–147 mm; Hawaii, Hawaii Island, Hilo: USNM 83449, 211 mm; Line Islands (US territory), Palmyra Atoll: USNM 429177, 244 mm, Palmyra Island, inner lagoon; ANSP 77564, 215 mm; Solomon Islands, Santa Cruz Islands: USNM 389116, 189 mm, Fenualoa Island, SW side, 10°15′S, 166°17′E, 0–15 m (Jeff T. Williams, colour photo of freshly collected fish (D)); Vanuatu, Efate Island, Port Vila: AMS I-11299, 158 mm; AMS I-11300, 154 mm, 17°45′S 168°18′E; French Polynesia, Society Islands, Tahiti: ANSP 47550, 2 specimens, 203–204 mm; BMNH 1873.8.1.5, 233 mm; French Polynesia, Tuamotu Archipelago: MNHN 1980-0093, 290 mm, Rangiroa Island, 20°0′N, 152°30′E.
Photographs (12 specimens, all >120 mm SL): Chagos (British Indian Ocean Territory): b/w photo of a preserved fish by Arthur Strange in Winterbottom et al. (Citation1989), 196 mm, Peros Banhos Atoll; Indonesia, Papua, Biak Island: colour photo of a freshly collected fish by Franz Uiblein (2016; previously unpublished (E)), 235 mm, Biak city fish market, 1°11′08″S, 136°04′54″E; Japan, Ryukyu Islands: colour photo of a freshly collected fish by Tetsuo Yoshino in Masuda et al. (Citation1984), 220 mm, Ishigakijima; Mariana Islands, Guam Island: two colour photos of at least two live adults (no size information) by Robert F. Myers (unpublished photographs taken in 2011; Robert F. Myers, personal communication, 2015), Piti, 2 m; Micronesia, Caroline Islands, colour photo of a live fish by Yasumasa Kobayashi in Okamura & Amaoka (Citation1997), ca. 240 mm, Pohnpei Island; Marshall Islands: colour photo of a freshly collected fish by Michael Trevor in Froese & Pauly (Citation2016), 130 mm, Arno Atoll; Hawaii (USA): colour photo of freshly collected fish by John E. Randall in Froese & Pauly (Citation2016), 248 mm, Oahu Island; colour photo of freshly collected adult by Hallelujah Hou Fishing (Clay Ching, personal communication, 2016 (F)), Molokai Island; Tonga, Vava’u Archipelago: colour photo of live adult by Eric Clua (2002; previously unpublished (G)), Vava'u Island, W of Neiafu, 18°39′40″S, 174°01′38″W, ca. 10 m; colour photo of a live fish by Franz Uiblein (2014; previously unpublished (H)), 240 mm, Kapa Island, 18°42′43″S, 174°02′13″E, 1–2 m; Cook Islands, Southern Cook Islands: colour photo of freshly collected fish by Gerald McCormack in McCormack (Citation2007) (), 255 mm, Aitutaki Island.
Diagnosis of adults
Dorsal fin VIII spines, 9 rays; pectoral fins 13–15 rays; gill rakers 5–7 + 15–18 = 21–24; lateral-line scales 36–38; maximum verified size 290 mm SL; adults, measurements in % of SL: body depth at first dorsal-fin origin 22–26, at anus 20–23; caudal-peduncle depth 9.2–11; maximum head depth 19–22; head depth through eye 14–18; head length 25–29; orbit length 4.6–6.3; upper jaw length 11–13; barbel length 17–22; caudal-fin length 28–32; anal-fin height 14–17; pelvic-fin length 17–20; pectoral-fin length 17–20; first dorsal-fin height 20–23; second dorsal-fin height 15–17; total oblique bars on caudal fin 9–14, upper caudal-fin lobe with 5–8 black bars, the proximal bars slightly curved; 4–6 bars on lower lobe; bars on or close to both lobe tips and penultimate bar on lower lobe mostly black, other bars mostly red or brown, becoming black at distal inner margin of lobes; at least the black parts of bars retained on preserved fish; two lateral body stripes, one pale brown or reddish brown at mid-body from snout or eye to caudal-fin base, the other fainter, mostly shorter, and more yellowish below; first dorsal-fin tip pale; barbels white at bases and yellow distally in fresh fish, sometimes entirely white; body uniformly pale brown in preserved fish.
Diagnosis of subadults
Measurements in % of SL: body depth at first dorsal-fin origin 22–25, at anus 18–22; caudal-peduncle depth 9.1–10; maximum head depth 18–21; head depth through eye 15–17; head length 27–30; orbit length 6.1–7.2; upper jaw length 11–13; barbel length 17–20; caudal-fin length 30–31; anal-fin height 15–17; pelvic-fin length 18–20; pectoral-fin length 18–20; first dorsal-fin height 20–22; second dorsal-fin height 15–17; total oblique bars on caudal fin 7–9, upper caudal-fin lobe with 4–5 black bars, the proximal bars slightly curved; 3–4 bars on lower lobe; see diagnosis for adults for meristic characters and other colour patterns.
Description
For a complete list of holotype measurements (in % SL) and counts see Uiblein & Heemstra (Citation2010; table 6). Morphometric data as ratios of SL for holotype, followed by data for adults and subadults (both in brackets with subadults in parentheses): body moderately deep, its depth at first dorsal-fin origin 4.6 [3.9–4.6 (4.0–4.6)]; body depth at anal-fin origin 4.4 [4.4–5.0 (4.5–5.7)]; maximum head depth 5.3 [4.5–5.4 (4.8–5.4)]; head depth through eye 7.0 [5.6–6.8 (5.7–6.7)]; head length 4.0 [3.4–3.9 (3.3–3.7)], larger than maximum depth of body and subequal to or slightly shorter than caudal-fin length (3.5 [3.1–3.6 (3.2–3.4)]); snout length 9.9 [8.4–9.7 (8.7–10)], subequal to postorbital length (10.1 [8.1–9.6 (7.9–9.6)]); orbit length 18 [16–22 (14–16)], smaller than interorbital width (12.7 [11–13 (12–13)]) and caudal-peduncle depth (10.1 [8.7–11 (9.8–11)]); barbel length 5.5 [4.5–5.8 (4.9–6.0)]; anal-fin height 6.7 [5.8–6.9 (5.8–6.7)], pelvic-fin length (5.7 [5.1–6.0 (5.0–5.7)]) subequal to interdorsal distance (5.7 [5.2–6.8 (5.4–6.2)]) and shorter than caudal peduncle length (4.5 [4.1–5.0 (3.9–4.4)]).
Fresh colour (Figure 1)
Body and head silvery white to pale grey, dorsally and anteriorly of eye darker, with red or light-brown scale markings or skin pigmentation; at least two lateral body stripes, one pale brown or reddish brown at mid-body from snout or eye to caudal-fin base, another fainter and more yellowish stripe right below pectoral-fin base from level of eye or behind operculum to caudal-fin base or to caudal peduncle; rarely, a very weak third yellow stripe further below, from level of pectoral-fin base to about anal-fin origin; eyes with black pupils and orbit forming partly or entirely a reddish-brown ring that connects to the lateral body stripe; in one live specimen collected by flyfishing off Molokai Island, Hawaii (F), five broad, dark vertical bands appear on the body which extend from dorsal margin down to the lateral body stripes; in addition in this specimen, as well as in the subadult and small adult (A, B), weak red patches laterally on ventral side of head and body; barbels generally white at bases and yellow distally, or entirely white; total oblique bars on caudal fin 9–14 (7 bars in freshly photographed subadult; A), upper caudal-fin lobe with 5–8 black bars (4 bars in single subadult), the proximal bars slightly curved; 4–6 bars on lower lobe (3 bars in single subadult); bars on or close to both lobe tips and penultimate bar on lower lobe mostly black, other bars mostly red-brown or brown becoming black at distal inner margin of lobes; caudal-fin bars subequal (slightly wider in adults, slightly narrower in subadults) to hyaline or white interspaces between bars in larger fish (C–H); bars narrower than interspaces in subadults (A; also evident from preserved material, see also below); the 2 to 3 most proximal bars on lower caudal-fin lobe wider than the other bars and as wide or wider than orbit length; in one instance of a 240 mm SL specimen encountered by the first author off Kapa Island, Vava’u Archipelago, Tonga, the bars on the upper caudal-fin lobe were fused, resulting in a marbled colour pattern (H); dorsal fins hyaline with 3 to 5 grey stripes, one at or close to fin bases; first dorsal-fin tip pale; anal and paired fins hyaline and unpigmented.
Preserved colour
Body uniformly pale or pale brown in preserved fish, including holotype (photograph in Uiblein & Heemstra Citation2010); caudal-fin bars pale-brown to dark brown, usually well retained; other fins and barbels uniformly pale.
Distribution
Among the geographic areas of the Indo-Pacific that were screened for the current study, Upeneus taeniopterus has been found to occur in 30 areas between Pinda Bank, Mozambique, in the west and Rangiroa Island, Tuamoto Archipelago (French Polynesia), in the east, and between Ishigakijima, Ryukyu Islands (Japan), in the north and Aitutaki Island, Cook Islands, in the south (). Twenty of these area records are primarily based on preserved fish from scientific collections, six are based on photographs of live or freshly collected fish, and four are based on literature. Two of the four literature records rely on taxonomic examination: the record from the Laccadive Islands (India) is based on alpha-taxonomic studies of material collected from four islands and documented with a good-quality drawing of a 139 mm SL specimen collected most certainly off Karavathi Island (Jones & Kumaran Citation1980). The record from Wake Atoll by Lobel & Lobel (Citation2004) refers to Myers (Citation1999) and is based on photographic documentation and identification by a fish taxonomist (Robert Myers, personal communication, 2015). Of the other two literature records, the one from Fiji is based on a fisheries survey (Fisheries Division Citation1983), cited by Zug et al. (Citation1989) and Seeto & Baldwin (Citation2010), and the record from Samoa is based on a comprehensive annotated checklist by Wass (Citation1984).
Most of the areas recorded (26 of 30) are oceanic islands or atolls. Four well-documented new area records are reported here: the records for Palau and Vanuatu are both based on preserved scientific collection specimens and the records for Papua (Indonesia) and Tonga are based on photographic documentation, fish market visits, or direct in situ observations by the first author. According to the current knowledge, U. taeniopterus does not occur in the large area between the Andaman Sea, Malaysia, central and western Indonesia, Thailand, Cambodia, Vietnam, Taiwan, Philippines, most of Papua New Guinea, New Caledonia and off mainland Australia.
Habitat
Upeneus taeniopterus occurs in shallow sandy areas, mostly in lagoons and subtidal beaches of oceanic islands and atolls from one to 10 m. Only two of 12 verified depth records available for this species are deeper than 10 m: 0–15 m off Fenualoa Island, Santa Cruz Islands (Solomon Islands) and 25–50 m off Rodrigues (Mauritius).
Comparisons
Upeneus taeniopterus clearly differs from all 36 congeneric species in the combination of maximum size and eight meristic and colour characters (see for a detailed list of distinguishing characters and their value ranges). From 34 Upeneus species U. taeniopterus differs in maximum size and at least in one meristic and one colour character. From U. luzonius (for which the colour in live or fresh conditions is not yet known), it differs in larger maximum size and higher number of lateral-line scales. From U. vittatus, which reaches a similar maximum size, it differs in a lower number of gill rakers, a smaller number of lateral body stripes (2–3 vs. 4 stripes), smaller eyes in adults (orbit length 4.6–6.3 vs. 6.6–8.8%SL), and the absence of dark pigmentation on the first dorsal-fin tip (see Uiblein & Gouws Citation2015, for additional comparative data). Moreover, adult U. vittatus differ in the number of oblique bars on the lower caudal fin lobe, having 3 (rarely 4) oblique bars (Uiblein & Gouws Citation2015) vs. 4–6 bars in U. taeniopterus.
Regarding distribution patterns, U. taeniopterus differs from all other congeneric species in the combination of a wide distributional range and the primary occurrence off oceanic islands and atolls. Only U. vittatus has a similarly wide distribution range (South Africa and Red Sea to French Polynesia), but this species occurs frequently on continental shelves throughout the Indo-Pacific (Randall & Kulbicki Citation2006; Uiblein & Gouws Citation2015) where U. taeniopterus is mostly absent. Also, while these two species frequently co-occur around oceanic islands and atolls, they may separate in habitat, as U. vittatus ranges to greater depths (at least 100 m) and can be found on mud or silty sand bottoms of estuarine areas (Randall & Kulbicki Citation2006; Uiblein & Gouws Citation2015). Three other species with relatively wide ranges, U. guttatus (South Africa and Red Sea to Japan and New Caledonia), U. moluccensis (Mediterranean to Japan) and U. sulphureus (Mozambique to Japan and Fiji), mostly occur in the coastal waters of continents and large islands (Randall & Kulbcki 2006; Uiblein & Heemstra Citation2010; Uiblein & Gledhill Citation2015). The frequent association with continents and large islands applies also to the distribution of all other Upeneus species, except for the three very narrowly distributed U. mouthami, U. seychellensis and U. vanuatu. Upeneus mouthami occurs exclusively in two Melanesian island archipelagos (W Central Pacific), the Chesterfield Islands (New Caledonia) (Randall & Kulbicki Citation2006), and Vanuatu (present study; new record), at 36–175 m depth; U. seychellensis is endemic to the Seychelles and occurs there at 60 m depth (Uiblein & Heemstra Citation2011b); and U. vanuatu is known only from the Vanuatu archipelago at 191–321 m depth (Uiblein & Causse Citation2013).
When compared intraspecifically, no population differences in morphology and colour patterns between adult Upeneus taeniopterus from the Pacific and the Indian Ocean were found (, and 2). Subadults from the Pacific showed shallower bodies and slightly larger eyes than similar-sized conspecifics from the Indian Ocean (, ), but do not differ in any other characteristics. Subadult U. taeniopterus differ from adults in having slightly less developed gill rakers on the lower limb, larger eyes, a longer caudal peduncle and fewer and narrower caudal-fin bars (, ). In addition, Pacific subadults have a shallower body than co-occurring adults.
Remarks
Among the comparative material examined for this study is a paratype of Upeneus randalli (ZMUC 49161), which was erroneously referred to as coming from the Persian Gulf in the original description (Uiblein & Heemstra Citation2011a). During the course of re-examination for this study it was discovered to have been collected from the same locality as a specimen of U. suahelicus (reported in Uiblein & Gouws Citation2015) from off Chahabar, southern Iran, Gulf of Oman. This revised locality represents the first record of this species outside of the Persian Gulf.
The maximum size information for U. taeniopterus and U. vittatus in is derived from the literature (Fischer & Bianchi Citation1984; Randall Citation1996; later followed by e.g., Randall Citation2007 and Uiblein & Gouws Citation2015) and could not be verified based on specimens we have examined. For U. taeniopterus an even larger maximum size of 35 cm has been indicated in the FAO guide by Fischer & Whitehead (Citation1974), who state that the maximum size for U. vittatus is 27 cm. However, it remains unclear whether SL or total length was referred to.
Some of the examined photographs of fresh fish encountered in situ or shortly after capture indicate considerable variation in colour patterns that would deserve further investigations to fully understand the causes: the vertical bands on the specimen captured by flyfishing off Hawaii (F) may have emerged as a result of handling stress; the marbled colour pattern encountered on the upper caudal-fin lobe of the specimen from off Kapa Island, Vava’u, Tonga (H) may reflect irregular development of the bar pattern; and the rather dark, black mid-body stripe on the specimen photographed in situ off Vava’u is most certainly the consequence of insufficient illumination and/or the absence of a flash when the photograph was taken (G; Eric Clua, personal communication, 2014).
Genetic studies
The 25 Upeneus specimens included in the genetic analysis represented 13 species from all six species groups (see above). The aligned sequences, once trimmed to equal length, provided 598 characters for analysis. The model selected for the ML analysis included unequal base frequencies (A = 0.243, C = 0.306, G = 0.159 and T = 0.292), equal transition (R[A↔G] = R[C↔T] = 9.322) and two transversion rates (R[A↔C] = R[G↔T] = R[C↔G] = 1.000, and R[A↔T] = 1.616), a proportion of invariant sites (I = 0.648) and a gamma-distribution of among-site rate variation (α = 1.760). The ML phylogram (−lnL = 3023.803) obtained in this analysis is presented in A. The 182 variable characters included 168 parsimony informative characters and the parsimony analysis yielded a single most-parsimonious tree (B) of 487 steps (CI = 0554, RI = 0.796, Rescaled CI = 0.441). The two trees were largely congruent (an SH test revealed no significant difference in the likelihood of the two topologies: ΔlnL = 3.446, P = 0.183) and revealed identical relationships among all included species, with one notable exception (see below). Expectedly, multiple representatives of the same species grouped together across both analyses, although these relationships did not always receive strong bootstrap support (e.g., U. heemstra, U. tragula and U. suahelicus). In terms of the Upeneus species groups represented, only the suahelicus group (represented by Upeneus suahelicus and U. supravittatus) was retrieved as monophyletic, with strong bootstrap support (93% in ML and 94% in UP). The japonicus and stenopsis groups could not be evaluated as each was represented by single species only: U. guttatus and U. mascareinsis, respectively.
Figure 4. (A) Maximum-likelihood phylogram (–lnL = 3023.803) and (B) the single most parsimonious tree (487 steps: CI = 0554, RI = 0.796, Rescaled CI = 0.441) of relationships among the 25 included Upeneus individuals, representing 13 species, derived from the analyses of a 598 bp fragment of mitochondrial COI sequence data. Mulloidichthys vanicolensis and Parupeneus barberinus were used as outgroups. Nodal support from 1000 bootstrap replicates are indicated on the branches. Only bootstraps > 75% are shown. The terminal names include the GenBank accession numbers or BOLD Process IDs, and sampling regions. Terminal names are shaded and colour-coded, as per the key, to indicate species group membership (see also ). The japonicus (U. guttatus) and stenopsis (U. mascareinsis) groups, each represented by a single species in the analyses, are not indicated.
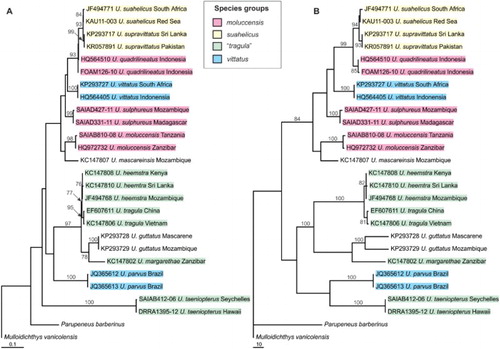
Across both analyses, the moluccensis, tragula and vittatus groups were retrieved as poly- and/or paraphyletic. Upeneus taeniopterus, the one species whose placement was not consistent across the analyses, was not placed within or closely associated with the remainder of the tragula group, which was paraphyletic with respect to U. guttatus (japonicus group). In the ML analysis, U. taeniopterus was retrieved basally and was the sister taxon to all included Upeneus species. In the parsimony analysis, U. taeniopterus was the sister-taxon to U. parvus, with this clade then sister to the remaining tragula group species (U. heemstra, U. margarethae and U. tragula) and U. guttatus. None of these relationships or alternative arrangements of the deeper nodes were supported. A topology constrained to reflect a monophyletic tragula group, including U. taeniopterus, and with U. guttatus sister to this (−lnL = 3035.977) was significantly less likely (ΔlnL = 12.174, P < 0.05) than the likely topology obtained above, rejecting the notion of a monophyletic tragula group, as currently defined. Interestingly, including U. guttatus as nested within the tragula group (as above), with U. taeniopterus as this group’s immediate sister taxon, was only marginally less likely (−lnL = 3025.628, ΔlnL = 1.825, P = 0.207) than the obtained topology. A much wider tragula group, retaining U. taeniopterus and including U. guttatus, cannot be rejected with the present data.
K2P sequence divergences () indicated the substantial divergence of U. taeniopterus from all Upeneus species included. Mean sequence divergences obtained in comparisons between U. taeniopterus and the other species ranged from 19.9 to 21.4%, with individual specimens being 19.8–21.7% divergent. This divergence is greater than observed among other species pairs, where the next greatest divergence was 18.6% (U. mascareinsis vs. U. margarethae), and with the majority (50 of 66, 75.8%) of the remaining (mean) divergences being ≤16%. In terms of group membership, U. taeniopterus showed no greater genetic similarity to the remaining members of the tragula group (mean divergences of 20.6–20.9%) than to the other species groups, as presently constituted (vs. japonicus group: 21.4%; vs. moluccensis group: 20.3–20.7%; vs. mascareinsis group: 21.4%; vs suahelicus group: 20.0–20.3%; and vs. vittatus group: 19.9–20.8%). Despite the vast geographic separation over which the two U. taeniopterus specimens included were collected (Seychelles vs. Hawaii), the sequence divergence among them was only 0.2%. This is comparable to most intraspecific divergences observed in the present study, some of which represent comparisons over much smaller geographic scales.
Table IV. Kimura (Citation1980) 2-parameter (K2P) sequence divergences among representatives of the Upeneus species included in the present study.
Discussion
Both the alpha-taxonomic and the barcoding analyses (albeit based on a limited sample size, including only two Upeneus taeniopterus individuals) revealed a high distinctiveness of U. taeniopterus that does not allow the authors to place it in any taxonomic group of Upeneus. Phenotypically, this species reaches together with U. vittatus the largest size known for this genus, differs clearly in the combination of meristic and colour characters from all congeners, and shows a species-specific distribution and habitat selection pattern. While showing the typical generic characters for Upeneus species, like villiform jaw teeth, teeth present on vomer and palatines, small scales on basal portion of second dorsal fin and anal fin, and snout length shorter than or subequal to postorbital length, its distribution pattern most closely resembles that of Mulloidichthys pfluegeri (Steindachner, 1900) among the Mullidae. The latter species occurs also almost exclusively around oceanic islands (Hawaiian Archipelago, Marquesas Islands, Society Islands, Tonga, Marshall Islands, Mariana Islands, Ogasawara Islands, Ryukyu Islands, Réunion and E Indonesia (Randall Citation2007; Uiblein Citation2011)); it differs, however, from U. taeniopterus – apart from the generic characters – in a larger maximum size (at least 40 cm SL) and by occurring mostly at greater depths of 20 m to at least 110 m (with a single record at 7 m; Randall Citation2007).
The genetic analyses, although based on limited species representation, small sample sizes for each species and including data from only one mitochondrial gene region, provide evidence for the genetic distinction of U. taeniopterus and its exclusion from the tragula species group. Topologically, a strictly monophyletic tragula group was rejected. The inclusion of U. taeniopterus within the tragula group was statistically likely only under a much less restrictive arrangement of the group in which U. guttatus (japonicus group) was subsumed and U. taeniopterus was included as a basally derived and, thus, genetically distant member. Upeneus taeniopterus was not more clearly aligned, in terms of genetic similarity, to the tragula group than to any of the other species groups (as currently defined and as represented in the present study). Although limited by sample sizes, species representation and the use of a single mitochondrial marker, there is no compelling genetic evidence for its membership in any of the six groups represented.
Genetic distances revealed U. taeniopterus to be the most divergent of the species examined, with higher divergence values obtained in species comparisons involving U. taeniopterus than in comparisons among other species. In fact, these values exceed the typical divergences among congeneric species recorded for the Mullidae (Lakra et al. Citation2011) and lie within the range documented for intergeneric comparisons (Zhang Citation2011). Collectively, this genetic evidence suggests a basal position for the highly divergent U. taeniopterus among the remaining Upeneus species groups. The species’ phenotypic distinctiveness (now supported by genetic data), with size, colour and varying combinations of meristic characters (see above) differentiating the species from congeners, including members of the tragula group to which it was previously assigned (Uiblein & Heemstra Citation2010), precludes it from clear membership of the existing species groups; this reflects the situation around U. filifer (Uiblein & Heemstra Citation2011a, Citation2011b; Uiblein & McGrouther Citation2012). By adopting this comprehensive alpha-taxonomic and genetic approach, it should be possible to further disentangle the phylogenetic relationships, when more genetic tissue samples of yet to be barcoded Upeneus species become available.
Many goatfishes, including those of the genus Upeneus, are valuable food fishes in tropical or subtropical countries and island areas of the Indo-Pacific (Uiblein Citation2007). This applies in particular to U. vittatus (Rawlinson et al. Citation1995) which can reach large sizes like U. taeniopterus. These two species, as well as goatfishes of the genera Mulloidichtys and Parupeneus, are frequently found in fish markets as important components of oceanic islands fisheries (e.g., NMFS Citation2011). However, in some areas like the Hawaiian Archipelago, U. taeniopterus is less appreciated as a food fish, as it has been found to create hallucinations after consumption (‘nightmare weke’) due to so-called ‘ciguatera’ poisoning (Halstead et al. Citation1990; Kaneko et al. Citation2005). While these effects have not been reported for any other Upeneus species, they may apply to some Mulloidichthys species and recommendations have been made to not eat these fishes (Kaneko et al. Citation2005). However, to our knowledge, the supposed biotoxin on which these recommendations are based has not yet been identified and the intoxication effects have not been well studied. Also, no comparative biochemical studies among goatfishes have been carried out to search for inter- or intraspecific variation in biotoxin content.
Although data from only two specimens of U. taeniopterus were available for the present genetic analysis, there was no intraspecies divergence among them, despite the vast distance separating the two sampling localities, the Seychelles and Hawaii. The geographic comparisons involving subadults also need to be regarded with some caution, as they are based on rather low sample sizes and are not representative for all recorded areas. The results of these latter comparisons suggest population differences in allometric growth and developmental trajectories. Certainly, more detailed investigations involving more samples and a fine-scaled phylogeographic approach would be required to investigate these questions.
The disruption of the currently known distribution of U. taeniopterus in the central part of its range may be due to data deficiency, but may to some extent also be related to the large shelf regions extending from the Malacca Strait to the western South China Sea and southwards to Bali as well as around Australia. While being mostly found off oceanic islands and atolls, this species has been also encountered off continental or large island shelves, especially if those areas are in close vicinity to deep oceanic waters, such as Pinda Bank off northern Mozambique, Trincomalee off northeastern Sri Lanka (type locality) and Biak Island, Papua, easternmost Indonesia. The latter location is in close proximity to deep oceanic waters, which also applies to the adjacent coast of eastern Papua New Guinea. For the latter, however, U. taeniopterus has not yet been reported (e.g., Fricke et al. Citation2014).
The lack of a biodiversity research focus on shallow sandy habitats of oceanic islands and atolls may have contributed to U. taeniopterus not being previously recorded or at least not verified for rather large oceanic island areas such as Fiji, Palau, Samoa, Tonga and Vanuatu. In a comprehensive annotated checklist of the fishes of Tonga (Randall et al. Citation2003), as well as in the more recently published biodiversity assessment for the Vava’u Archipelago, Tonga (Atherton et al. Citation2015), this species is not listed. The latter study lists 10 goatfish species (Mulloidichthys flavolineatus (Lacepède, 1801), M. vanicolensis, Parupeneus barberinoides (Bleeker, 1852), P. barberinus, P. ciliatus (Lacepède, 1802), P. crassilabris (Valenciennes, 1831), P. cyclostomus (Lacepède, 1801), P. indicus (Shaw, 1803), P. multifasciatus (Quoy & Gaimard, 1824) and P. pleurostigma (Bennett, 1831)), which are usually common on or close to coral reefs. In recent years, biodiversity research, ecological monitoring and conservation efforts in tropical coastal areas have increasingly targeted coral reefs (e.g., Williams et al. Citation2011; Mora Citation2015) and fish taxa closely linked to corals such as butterfly fishes (Chaetodontidae) (e.g., Palaki et al. Citation2005; Pratchet et al. Citation2013). Subtidal sandy beaches and lagoons of oceanic island areas have not yet received similar attention in biodiversity studies (e.g., Barboza & Defeo Citation2015).
A good example of the lack of focused biodiversity research and management of commercially important nearshore species such as U. taeniopterus comes from Tonga. Within the Vava’u Archipelago, as in other areas of this country, Mullidae are a targeted family for domestic fisheries and subsistence utilization. Common fishing methods are spearfishing during both day and night, fish traps and gill nets which are also common in adjacent oceanic island areas (Kronen Citation2004; Raubani Citation2006). The economic value of coastal fishes such as goatfishes to community fishermen is high and an emphasis is needed on improved management, including ecological monitoring of coastal habitats and species assemblages. The Special Managed Areas (SMAs) are a community-managed reef programme under the Ministry of Fisheries of Tonga that has the potential to further management practices for shoreline habitats (Gillett Citation2009). Within the SMAs, small no-take areas are allocated by the community and often include sandy habitats. At present there are, however, no protected areas or larger-scale managed areas off Vava’u and sandy habitats are still undervalued and overlooked as important ecological areas.
When it comes to the potential role of goatfishes as key and indicator species (Uiblein Citation2007), the bandtail goatfish U. taeniopterus may play a distinct ecological role in shallow sandy areas of oceanic islands and atolls. To fully understand this role, the autecology of this species would need to be studied in detail. In parallel, the overall biodiversity of its habitat should be investigated considering ecological vulnerability in response to a number of factors such as, for instance, climate induced sea-level rise and water temperature increase, fisheries (e.g., lining, beach seining, gill-netting, spearing, trapping; see also Dalzell et al. Citation1996), tourism, habitat modification, eutrophication and pollution.
Acknowledgements
We thank the following colleagues for hospitality and assistance during visits to collections or for providing other collection-related favours: Mark Sabaj Perez (ANSP); Mark McGrouther, Amanda Hay and Sally Reader (AMS); James Maclaine (BMNH); John E. Randall, Arnold Suzumoto and Loreen O’Hara (BPBM); Dave Catania, Jon Fong and Mysi Hoang (CAS); Alastair Graham, William T. White, Carlie Devine, Peter Last, John Pogonoski and Robert Ward (CSIRO); Caleb McMahan and Marc Westneat (FMNH); Rupert Wienerroither (HIFIRE); Hisashi Imamura and Kazuhiro Nakaya (HUMZ); Rick Feeney and Christine Thacker (LACM); Romain Causse, Patrice Pruvost and Gabsi Zora (MNHN); Dimitri A. Pavlov (Moscow State University and Russian-Vietnamese Tropical Center); Renny Hadiaty (MZB); Pak Fahmi, Inayat Al Hakim, Wanwan Kurniawan and Selvia Oktaviyani (NCIP); Dianne Bray and Martin Gomon (NMV); Sven Kullander, Bo Delling and Bodil Kajrup (NRM); Jeffrey T. Williams, David G. Smith and the NMNH (USNM) collection staff; Gavin Dally and Michael Hammer (NTM); Jeffrey W. Johnson (QM); Ronald de Ruiter (RMNH); Roger Bills, Bafo Konobe and the SAIAB National Fish Collection staff; Elaine and Phil C. Heemstra, Wouter Holleman and Alan Whitfield (SAIAB); Michael Bougaardt and Dylan Clarke (SAM); Ronald Fricke (SMNS); Eric Hilton and Sarah K. Huber (VIMS); Nguyen Tien Tao, Pham Hong Thai and Hoan Anh Tuan (VNMN); Sue Morrison (WAM); and Peter Rask Møller, Jørgen Nielsen, Marcus Krag and Tammes Menne (ZMUC). Furthermore, we thank Carole Baldwin (NMNH) and Ross Robertson (STRI) for assistance in acquiring barcoding data from Hawaii and Robert Myers (Wellington FL, USA) and Terry Donaldson (Guam) for communication regarding IDs of fish. We acknowledge Bob Ward (CSIRO) and Tilman Alpermann (SMF) for the barcoding data used in this study and those cited above. For photographs of fresh fish and/or assistance with compiling the colour plate we thank Clay Ching (Hallelujah Hou Fishing, Hawaii), Eric Clua (Papeete, French Polynesia), Carlie Devine, Phil C. Heemstra, Robert Myers, John E. Randall and Jeff Williams. For logistical support and invaluable assistance during research visits, the first author thanks Ludi Parwadani Aji, Technical Implementation Unit for Marine Life Conservation, Indonesian Institute of Sciences (LIPI), Bosnik Raya, Biak, Papua, Indonesia, and Ian and Vanessa Jones, Vava’u, Tonga. For comments on former versions of the manuscript we thank Michael Mincarone and two anonymous referees.
Disclosure statement
No potential conflict of interest was reported by the authors.
ORCiD
Franz Uiblein http://orcid.org/0000-0002-5642-0384
Additional information
Funding
References
- Allen G, Bailey S. 2011. Reef fishes of the Phoenix Islands, Central Pacific Ocean. Atoll Research Bulletin 589:83–118. doi:10.5479/si.00775630.589.83
- Atherton JN, McKenna SA, Wheatley A. 2015. Rapid Biodiversity Assessment of the Vava’u Archipelago, Kingdom of Tonga. Apia, Samoa: SPREB. 312 pages.
- Barboza FR, Defeo O. 2015. Global diversity patterns in sandy beach macrofauna: a biogeographic analysis. Scientific Reports 5:14515. 9 pages. doi:10.1038/srep14515
- Boldsystems. 2015. The Barcode of Life Systems. http://www.boldsystems.org (accessed 26 January 2016).
- Dalzell P, Adams TJH, Polunin NVC. 1996. Coastal fisheries in the Pacific Islands. Oceanography and Marine Biology: An Annual Review 34:395–531.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9:772. doi:10.1038/nmeth.2109
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–91. doi:10.2307/2408678
- Fischer W, Whitehead PJW, editors. 1974. FAO Species Identification Sheets for Fishery Purposes. Eastern Indian Ocean (Fishing Area 57) and Western Central Pacific (Fishing Area 71). Vols. 1–4. Rome: FAO. Variable pagination.
- Fischer W, Bianchi G, editors. 1984. FAO Species Identification Sheets for Fishery Purposes. Western Indian Ocean (Fishing Area 51). Vols. 1–6. Rome: FAO. Variable pagination.
- Fisheries Division. 1983. The Fishery Resources of Rotuma. Suva, Fiji: Ministry of Agriculture and Fisheries. 31 pages.
- Fowler HW. 1928. The fishes of Oceania. Memoirs of the Bernice Pauahi Bishop Museum 10:1–540.
- Fricke R, Eschmeyer WN. 2016. A Guide to Fish Collections in the Catalog of Fishes. http://researcharchive.calacademy.org/research/ichthyology/catalog/collections.asp (accessed January 2016).
- Fricke R, Allen GR, Andréfouet S, Chen W-J, Hamel MA, Laboute P, et al. 2014. Checklist of the marine and estuarine fishes of Madang District, Papua New Guinea, western Pacific Ocean, with 820 new records. Zootaxa 3832:1–247. doi:10.11646/zootaxa.3832.1.1
- Froese R, Pauly D, editors. 2016. FishBase. World Wide Web Electronic Publication. www.fishbase.org (accessed 19 January 2016).
- GenBank. 2015. National Center for Biotechnology Information. Bethesda, MD: US National Library of Medicine. http://www.ncbi.nlm.nih.gov/genbank/ (accessed 5 August 2015).
- Gillett MN. 2009. Success of special management areas in Tonga. SPC Fisheries Newsletter 130:27–30.
- Halstead BW, Auerbach PS, Campbell DR. 1990. A Colour Atlas of Dangerous Marine Animals. Ipswich, UK: Wolfe Medical Publications, W.S. Cowell. 192 pages.
- Hebert PDN, Cywinska A, Ball SL, DeWaard JR. 2003. Biological identifications through DNA barcodes. Proceedings of the Royal Society of London B 270:313–21. doi:10.1098/rspb.2002.2218
- Jones S, Kumaran M. 1980. Fishes of the Laccadive Archipelago. Cochin, India: Mathrubhumi Press. 760 pages.
- Kaneko JJ, Takenaka B, Bartram P. 2005. Keeping Hawaii Seafood Safe to Eat. Project Report. Hawaii Seafood Safety Project. Honolulu, HI: PacMar. 24 pages.
- Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16:111–20. doi:10.1007/BF01731581
- Kronen M. 2004. Fishing for fortunes? A socio-economic assessment of Tonga’s artisanal fishing. Fisheries Research 70:121–34.
- Lachner EA. 1954. A revision of the goatfish genus Upeneus with descriptions of two new species. Proceedings of the United States National Museum 103:497–532. doi:10.5479/si.00963801.103-3330.497
- Lakra WS, Verma MS, Goswami M, Lal KK, Mohindra V, Punia P, et al. 2011. DNA barcoding Indian marine fishes. Molecular Ecology Resources 11:60–71. doi:10.1111/j.1755-0998.2010.02894.x
- Lobel PS, Lobel LK. 2004. Annotated checklist of the fishes of Wake Atoll. Pacific Science 58:65–90. doi:10.1353/psc.2004.0007
- Masuda H, Amaoka K, Araga C, Uyeno T, Yoshino T. 1984. The Fishes of the Japanese Archipelago. Vol. 1. Tokyo: Tokai University Press. 437 pages.
- McCormack G. 2007. Cook Islands Biodiversity Database. Version 2007.2. Rarotonga, Cook Islands: Cook Islands Natural Heritage Trust, http://cookislands.bishopmuseum.org (accessed 26 January 2016).
- Mora C, editor. 2015. Ecology of Fishes on Coral Reefs. Cambridge, UK: Cambridge University Press. 388 pages.
- Mundy BC, Wass R, Demartini E, Greene B, Zgliczynski B, Schroeder RE, Musberger C. 2010. Inshore fishes of Howland Island, Baker Island, Jarvis Island, Palmyra Atoll, and Kingman Reef. Atoll Research Bulletin 585:1–131. doi:10.5479/si.00775630.585
- Myers RF. 1999. Micronesian Reef Fishes. A Field Guide for Divers and Aquarists, 3rd edition. Guam: Coral Graphics. 216 pages.
- Myers RF, Donaldson TJ. 2003. The fishes of the Mariana Islands. Micronesica 35–36:594–648.
- NMFS. 2011. Environmental assessment for annual catch limit specifications and accountability measures for Pacific islands coral reef ecosystem fisheries in 2012 and 2013. December 13 2011. Honolulu, HI: National Marine Fisheries Service. 224 pages. http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.434.4480&rep=rep1&type=pdf (accessed 26 January 2016).
- Okamura O, Amaoka K. 1997. Sea Fishes of Japan. Tokyo: Yama-Kei. 784 pages.
- Palaki A, Samani T, Masi M. 2005. Dredging Operation on Toula Coastal Marine Resources. Nuku’alofa, Tonga: Technical and Sustainable Development Division, Department of Environment. 12 pages.
- Pratchet MS, Berumen ML, Kapoor BG. 2013. Biology of Butterflyfishes. Boca Raton, FL: CRC Press. 362 pages.
- Randall JE. 1996. Shore Fishes of Hawaii. Vida, OR: Natural World Press. 216 pages.
- Randall JE. 2007. Reef and Shore Fishes of the Hawaiian Islands. Honolulu, HI: Sea Grant College Program, University of Hawaii. 546 pages.
- Randall JE, Kulbicki M. 2006. A review of the goatfishes of the genus Upeneus (Perciformes: Mullidae) from New Caledonia and the Chesterfield Bank, with a new species, and four new records. Zoological Studies 45:298–307.
- Randall JE, Williams JT, Smith DG, Kulbicki M, Mou Tham G, Labross P, et al. 2003. Checklist of the shore and epipelagic fishes of Tonga. Atoll Research Bulletin 502:1–35.
- Raubani JJJ. 2006. Community Fisheries Management (CFM): Future Considerations for Vanuatu. Reykjavik, Iceland: UNU-Fisheries Training Programme. 47 pages.
- Rawlinson NJF, Milton DA, Blaber SJM, Sesewa A, Sharma SP. 1995. A survey of the subsistence and artisanal fisheries in rural areas of Viti Levu, Fiji. ACIAR Monograph 35:138.
- Ribeiro A, Caires R, Mariguela T, Pereira L, Hanner R, Oliveira C. 2012. DNA barcodes identify marine fishes of São Paulo State, Brazil. Molecular Ecology Resources 12:1012–20. doi:10.1111/1755-0998.12007
- Schultz LP. 1943. Fishes of the Phoenix and Samoan Islands. United States National Museum Bulletin 180:1–316.
- Seeto J, Baldwin WJ. 2010. A Checklist of the Fishes of Fiji and a Bibliography of Fijian Fishes. Division of Marine Studies Technical Report 1/2010. Suva, Fiji: The University of the South Pacific. 102 pages.
- Shimodaira H, Hasegawa M. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Molecular Biology and Evolution 16:1114–16. doi:10.1093/oxfordjournals.molbev.a026201
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–13. doi:10.1093/bioinformatics/btu033
- Swofford DL. 2002. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sunderland, MA: Sinauer Associates. Computer program.
- Uiblein F. 2007. Goatfishes (Mullidae) as indicators in tropical and temperate coastal habitat monitoring and management. Marine Biology Research 3:275–88. doi:10.1080/17451000701687129
- Uiblein F. 2011. Taxonomic review of Western Indian Ocean goatfishes of the genus Mulloidichthys (Family Mullidae), with description of a new species and remarks on colour and body form variation in Indo-West Pacific species. Smithiana Bulletin 13:51–73.
- Uiblein F, Causse R. 2013. A new deep-water goatfish of the genus Upeneus (Mullidae) from Vanuatu, South Pacific. Zootaxa 3666:337–44. doi:10.11646/zootaxa.3666.3.4
- Uiblein F, Gledhill D. 2015. A new goatfish of the genus Upeneus (Mullidae) from Australia and Vanuatu, with inter- and intraspecific comparisons. Marine Biology Research 11:475–91. doi:10.1080/17451000.2014.958088
- Uiblein F, Gouws G. 2014. A new goatfish species of the genus Upeneus (Mullidae) based on molecular and morphological screening and subsequent taxonomic analysis. Marine Biology Research 10:655–81. doi:10.1080/17451000.2013.850515
- Uiblein F, Gouws G. 2015. Distinction and relatedness – taxonomic and genetic studies reveal a new species group of goatfishes (Upeneus; Mullidae). Marine Biology Research 11:1021–42. doi:10.1080/17451000.2015.1064963
- Uiblein F, Heemstra PC. 2010. A taxonomic review of the Western Indian goatfishes of the genus Upeneus (Family Mullidae), with descriptions of four new species. Smithiana Bulletin 11:35–71.
- Uiblein F, Heemstra PC. 2011a. Description of a new goatfish species, Upeneus randalli n. sp. (Mullidae), from the Persian Gulf, with remarks and identification keys for the genus Upeneus. Scientia Marina 75:585–94. doi:10.3989/scimar.2011.75n3585
- Uiblein F, Heemstra PC. 2011b. A new goatfish, Upeneus seychellensis sp. nov. (Mullidae), from the Seychelles Bank, with remarks on Upeneus guttatus and a key to Western Indian Ocean Upeneus species. Marine Biology Research 7:637–50. doi:10.1080/17451000.2010.547202
- Uiblein F, Lisher M. 2013. A new goatfish of the genus Upeneus (Mullidae) from Angoche, northern Mozambique. Zootaxa 3717:85–95. doi:10.11646/zootaxa.3717.1.7
- Uiblein F, McGrouther M. 2012. A new deep-water goatfish of the genus Upeneus (Mullidae) from northern Australia and the Philippines, with a taxonomic account of U. subvittatus and remarks on U. mascareinsis. Zootaxa 3550:61–70.
- Uiblein F, White WT. 2015. A new goatfish of the genus Upeneus (Mullidae) from Lombok, Indonesia and first verified record of U. asymmetricus for the Indian Ocean. Zootaxa 3980:51–66. doi:10.11646/zootaxa.3980.1.3
- Uiblein F, Köhler C, Tian MC. 1998. Quantitative examination of morphological variability among goatfishes of the genus Upeneus from the Malayan Province (Pisces: Perciformes: Mullidae). Senckenbergiana Maritima 28:123–32. doi:10.1007/BF03043143
- Wass RC. 1984. An Annotated Checklist of the Fishes of Samoa. NOAA Technical Report SSRF-781. 43 pages.
- Williams ID, Richards BL, Sandin SA, Baum JK, Schroeder RE, Nadon MO, et al. 2011. Differences in reef fish assemblages between populated and remote reefs spanning multiple archipelagos across the Central and Western Pacific. Journal of Marine Biology 2011:826234. 14 pages. doi:10.1155/2011/826234
- Winterbottom R, Emery AR, Holm E. 1989. An annotated checklist of fishes of the Chagos Archipelago, Central Indian Ocean. Royal Ontario Museum Life Sciences Contributions 145:1–226.
- Yamashita Y, Golani D, Motomura H. 2011. A new species of Upeneus (Perciformes: Mullidae) from southern Japan. Zootaxa 3107:47–58.
- Zhang J. 2011. Species identification of marine fishes in China with DNA barcoding. Evidence-Based Complementary and Alternative Medicine 978253:1–10.
- Zug GR, Springer VG, Williams JT, Johnson GD. 1989. The vertebrates of Rotuma and surrounding waters. Atoll Research Bulletin 316:1–25. doi: 10.5479/si.00775630.316.1
