Abstract
The short-haired Segugio Italiano and rough-haired Segugio Italiano are two breeds of Italian scenthound. The two breeds were subjected to divergent selection based on the type (long versus short) and pigmentation (fawn versus black & tan) of the hair, leading to the development of the two above-mentioned phenotypes. Microsatellites markers from 120 unrelated dogs were used to estimate the genetic variability within the two Segugio Italiano breeds and to assess if divergent selection for the two phenotypes has possibly led to the development of subpopulations distinguished by coat type and pigmentation. Finally, we also estimated the genetic distance between the two Segugio Italiano breeds and two other related breeds, Segugio Maremmano and Segugio dell'Appennino. The mean Fis, Fit and Fst values indicated a moderate level of inbreeding. In both breeds the mean observed heterozygosity was lower than the expected heterozygosity. The AMOVA, PCoA and STRUCTURE analyses showed lack of differentiation between the two breeds and the absence of substructuring in the population. Cavalli-Sforza chord distance, Nei's genetic distance and Reynolds–Weir Cockerham’s genetic distance between the two Segugio Italiano breeds were low. Low genetic distances were also found between the Segugio Italiano and the others two related breeds indicating that there is no difference in terms of genetic distance among the three breeds despite the important phenotypic differences among them. This paper provides a basis to change the breeding guidelines of the four breeds in the light of these informations.
Introduction
The short-haired (SH) Segugio Italiano and rough-haired (RH) Segugio Italiano are two breeds of Italian scenthound classified by the Ente Nazionale Cinofilo Italiano (ENCI, Italian Kennel Club) in the group 6; these dogs are widespread throughout Italy and mainly used for hunting hares and wild boars. As of 2015, 4753 dogs (1106 RH and 3647 SH) were registered in the ENCI stud book.
Concerning general appearance, the Segugio Italiano is a dog of medium proportion and robust construction that is well adapted to the most varied terrains and able to work alone or in a pack. In the past, the two breeds were subjected to strong divergent selection based on the type (long versus short) and pigmentation (fawn versus black & tan) of the hair, leading to the development of the two above-mentioned phenotypes: the SH Segugio Italiano, which is characterised by a short-haired coat all over the body, and the RH Segugio Italiano, which is characterised by a hair length of no more than 5 cm. For both breeds, the Italian Kennel Club officially recognise two pigmentations: fawn and black & tan.
Although recently researches was performed studying the genetic state of popular Italian hunting dog breeds (Ciampolini et al. Citation2011; La Manna et al. Citation2015), no study has investigated the genetic structure of the Segugio Italiano population until now. Because the use of pedigree analysis for the evaluation of genetic parameters could lead to an incorrect estimation of the actual genetic variability present in dog populations (Leroy et al. Citation2009; Cecchi et al. Citation2013) microsatellite markers have been proven to be one of the most informative marker types for unveiling population genetic structures.
In this study, 21 microsatellite markers were used to estimate the genetic variability within the two Segugio Italiano breeds and to assess if divergent selection for the two phenotypes has possibly led to the development of subpopulations distinguished by coat type and pigmentation, the latter to the habit of the breeders to avoid the mating between dogs with different coat colour.
Finally, we also estimated the genetic distance between the two Segugio Italiano breeds and two other related breeds, Segugio Maremmano and Segugio dell'Appennino.
Materials and methods
Sampling design, blood collection and microsatellite analysis
A total of 120 unrelated dogs aged no older than 2 years were sampled at 11 meetings organised by the Societá Italiana Prosegugio (SIPS) over the course of the study (from March to November 2015), with the aim of collecting a representative sample of the Italian population. The sample was structured to include approximately equal numbers of both RH (51 dogs) and SH (69 dogs). The structure of the sample with the amount of animals for each category is given in Table . All the sampled animals were registered in the Italian Kennel Club stud book.
Table 1. Sample structure: numbers are referred to the amount of animals for each category.
Blood samples were taken from the radial vein of the foreleg, spotted on a Whatman FTA Nucleic Acid Collection card (Sigma-Aldrich, Milano, Italy) and stored at room temperature. Samples were shipped to the Genefast Srl Laboratory (Valsamoggia, BO, Italy) for the genotyping of a panel of 21 microsatellite markers proposed by the International Society of Animal Genetics (ISAG) for parentage analysis (http://www.isag.us/Docs/consignmentforms/2005ISAGPanelDOG.pdf).
Software and statistical analysis
GenAlEx 6.5 software (Peakall & Smouse Citation2012) was used to estimate the following parameters: mean number of alleles per locus (Na), F-statistics (Fis, Fit and Fst), observed heterozygosity (Ho), expected heterozygosity (He), number of private alleles in the two breeds, deviation from H–W equilibrium, analysis of molecular variance (AMOVA) and principal coordinate analysis (PcoA). Genepop 4.2 software (Raymond & Rousset Citation1995) was used to calculate p values across all loci according to the Fisher's exact probability test applying Markov chain with default setting. The significances of the fixation indices were calculated with ARLEQUIN 3.5.2.2. software (Excoffier & Lischer Citation2010). GENEDIST, an application of PHYLIP software version 3.69 (Felsenstein Citation1993), was used for the computation of the Cavalli-Sforza chord distance, Nei's genetic distance and Reynolds–Weir–Cockerham genetic distance. Last, STRUCTURE software 2.3.4 (Pritchard et al. Citation2000) was used for the identification of genetically homogeneous groups of individuals. The most likely number of clusters (K) were identified according to Evanno method (Evanno et al. Citation2005) using the online software Structure Harvester (Earl Citation2012). The estimation of the genetic distances between Segugio Italiano and two other related breeds, Segugio Maremmano and Segugio dell' Appennino, was completed using the raw microsatellite data available from a previous study by La Manna et al. (Citation2015).
Results and discussion
All the analysed loci were polymorphic for both breeds, with a total of 157 and 165 alleles identified for RH and SH, respectively (Table ). The average number of alleles per locus was 7.76. For the RH, the AHTk211, AHTk253 and INU055 loci showed the lowest number of allelic variants (n = 5), while the highest number of allelic variants was found for the AHT121 (n = 11) and AHTh171 (n = 11) loci. The AHTk211 and AHTk253 loci showed the lowest number of allelic variants (n = 5) in the SH, whereas the AHTh171 locus showed the highest number of alleles (n = 12). The polymorphic information content (PIC) values ranged from 0.466 (REN247M23) to 0.843 (AHTh171), with a mean value of 0.69.
Table 2. Characterization of the 21 analysed microsatellite loci in the two dog breeds.
The mean Fis and Fit values (inbreeding coefficients) were 0.046 and 0.061, respectively, while the mean Fst value was 0.013. The Fis and Fit values were not equal to zero, indicating a lack of heterozygosity at both the individual level within the subpopulation and the total population level. The Fst values were low and not significant (Table ). Some factors such as the mating of close kin or the substructuring of the populations could lead to an overall heterozygosity deficiency; however, the positive value of the inbreeding coefficient suggests that inbreeding is the most likely cause. Moreover, the use of a small number of champion stud dogs is a breeding practice often used by breeders that inevitably led to a rise in homozygosity.
The observed and expected heterozygosities were similar in the two breeds, with mean Ho values of 0.680 and 0.689 for RH and SH, respectively (Table ). The He values were 0.722 for RH and 0.716 for SH. With regard to the results reported for other European dog breeds, our values are similar to those reported by Parra et al. (Citation2008) and Bigi et al. (Citation2015) but higher than those reported for other breeds raised in Italy (Ciampolini et al. Citation2011) and France (Leroy et al. Citation2009). However, the Ho and He values from our study are lower than those reported by La Manna et al. (Citation2015) for the related Italian breeds Segugio Maremmano and Segugio dell' Appennino, where the Ho value was 0.73 for both breeds and the He values were 0.78 and 0.77 for Segugio Maremmano and Segugio dell’ Appennino, respectively.
Table 3. Mean number of observed alleles, mean observed and expected heterozygosity, number of private alleles, markers deviated from the Hardy–Weinberg equilibrium per breed and inbreeding coefficient per breed.
The number of private alleles was different in the two breeds, with 12 and 20 private alleles appearing in RH and SH, respectively. Two loci (AHT121and REN247M23) were not in H–W equilibrium in RH, while three (AHTh260, INU055 and REN247M23) were not in H–W equilibrium in SH, indicating a deficiency in heterozygotes. The mean number of observed alleles per breed, mean observed and expected heterozygosity per breed, number of private alleles per breed, markers deviated from the Hardy–Weinberg equilibrium per breed and inbreeding coefficient per breed is given in Table .
The results of the analysis of molecular variance (AMOVA) are given in Table ; the larger portion of genetic variability was between individuals (93%) rather than between the two breeds (2%).
Table 4. Results from AMOVA for the SH and LH.
This fact is also supported by the low Fst value (0.018), which shows the absence of significant genetic differences in the RH and SH breeds. To better understand the distribution of the two breeds, we carried out principal coordinate analysis (PCoA) based on Nei's genetic distance. As shown in Figure , this analysis unveiled an almost complete overlap between the two populations and the first and the second axis explain 5.35% and 4.73% of the total variance.
Figure 1. Principal coordinate plots (PCoA) for the two Segugio Italiano breeds. Pop1: rough-haired Segugio Italiano; Pop2: short-haired Segugio Italiano.
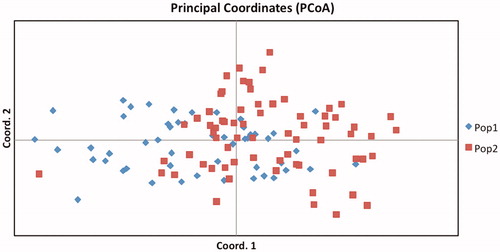
This result is in agreement with the high inbreeding coefficient. The Cavalli-Sforza chord distance, Nei's genetic distance and Reynolds–Weir–Cockerham genetic distance between the two Segugio Italiano breeds were 0.022, 0.072 and 0.026, respectively. All the three values were low, which further confirmed the absence of genetic differentiation between the two breeds previously identified by the AMOVA and the PCoA.
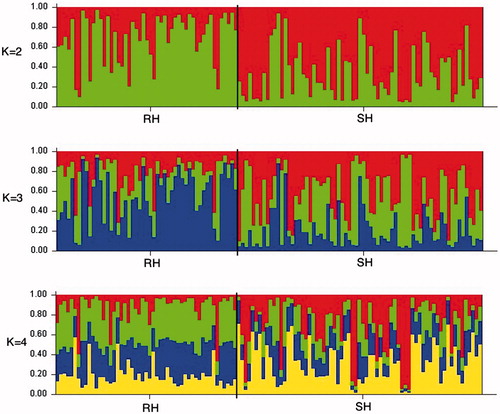
The presence of genetically homogeneous groups of individuals within the Segugio Italiano population was evaluated using STRUCTURE software. The burning period was set to 300,000; the repetition of the MCMC chain, to 600,000; and the K value, from K2 to K30. In the bar plots (Figure ), the two breeds did not tend to form clusters and it was not possible to assign a correct K value with the classic method by Pritchard et al. (Citation2000). Therefore, we used Structure Harvester to assign the right K value according to the Evanno method (2005). Even in this case, the software was unable to assign the right K values. The two results suggested the absence of substructuring in the Segugio Italiano population and were in agreement with a study carried out by Cadieu et al. (Citation2009) which showed that in the domestic dog only variants in three genes account for most coat variations, therefore, the genetic differences underlying the RH and the SH phenotypes is minimal.
Figure 2. Proportion of membership for rough-haired Segugio Italiano and short-haired Segugio Italiano. RH: rough-haired Segugio Italiano; SH: short-haired Segugio Italiano.
To evaluate the presence of subpopulations derived from the divergent selection based on coat colour, we analysed the dataset considering two distinct populations without regard for breed: one population was composed of 46 dogs with fawn coats (F) and the other was composed of 74 dogs with black & tan coats (Bt).
The observed heterozygosity was 0.672 for the Bt dogs and 0.707 for the F dogs, while the expected heterozygosity was 0.726 and 0.718 for Bt dogs and F dogs, respectively.
The AMOVA showed that 94% of the molecular variance was intra individual and 6% was inter-individual. No molecular variance was found between the two putative subpopulations. Moreover, we found low values for genetic distance, with results of 0.017, 0.037 and 0.014 for the Cavalli-Sforza chord distance, Nei's genetic distance and Reynolds–Weir–Cockerham genetic distance, respectively.
These results showed that the divergent selection according to coat colour did not led to any subpopulation in the two Segugio Italiano breeds.
In order to evaluate the genetic variance among RH Segugio Italiano, SH Segugio Italiano and the other related breeds Segugio Maremmano and Segugio dell'Appennino, we carried out the AMOVA. The analysis showed the absence of molecular variance between the four breeds. The proportion of variation among populations, among individuals and within individuals was 2%, 6% and 92%, respectively, indicating a lack of genetic differentiation among the four Segugio breeds. Next, we performed the principal coordinate analysis, running the programme with a dataset that considered the four populations. As shown in Figure , no clear cluster was found in this scenario; the individuals belonging to the four breeds occupied the same position in the diagram, with very few individuals showing an unexpected position with respect to the rest of the population. The percentage of variation explained by the first two axes was 4.13% and 3.60% for the first and the second, respectively.
Figure 3. Principal coordinate plots (PCoA) for the RH Segugio Italiano, SH Segugio Italiano, Segugio Maremmano and Segugio dell' Appennino. Pop1: RH Segugio Italiano; Pop2: SH Segugio Italiano; Pop3: Segugio Maremmano; Pop4: Segugio dell' Appennino.
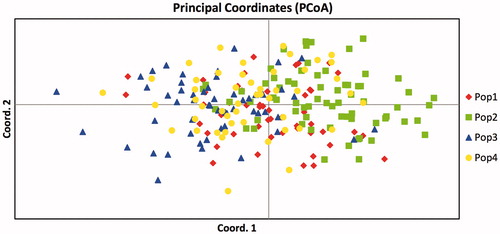
The PCoA result confirmed the absence of molecular variance as determined by AMOVA between the four breeds.
The results of the evaluation of the genetic distances between RH and SH Segugio Italiano and the other two related breeds are shown in Table . Low values were found, with a slightly higher result for Nei's genetic distance between Segugio Maremmanno and the others two Segugio Italiano breeds. Additionally, in this case, the three values were low, indicating that there is no difference in terms of genetic distance among the four breeds despite the important phenotypic differences among them.
Table 5. Genetic distances between RH Segugio Italiano, SH Segugio Italiano, Segugio Maremmano and Segugio dell’ Appennino.
Conclusions
The genetic variability of the SH and RH was low. Moreover, the genetic differentiation between the two breeds was almost inexistent, suggesting that the variability between the RH and SH breed is in terms of the type of coat rather than their overall genetic pool as showed in the paper by Cadieu et al. (Citation2009). Indeed, the values generated from the computation of the three genetic distances were low, and it was not possible to clearly identify subgroups from the principal component analysis. The same situation was found when the two pigmentation types were considered putative populations; the divergent selection that occurred for generations did not lead to the development of a subpopulation that is genetically distinguishable from the entire Segugio Italiano population.
A lack of genetic differentiation was also found between the Segugio Italiano and the two related breeds Segugio Maremmano and Segugio dell’ Appennino, suggesting that a small number of common ancestors are shared among the three breeds.
The common breeding method of non-random mating (between close kin) and the use of a small number of sires for the selection of desirable phenotypic traits led the Segugio Italiano population to a moderate inbreeding value and overall heterozygote deficiency; this loss of genetic variability could reduce the possibility of implementing effective breeding programmes.
Our results open a debate about the concept of breed and variety in dog breeding. The information included in this study suggest that the four breeds belonged to the same genetic pool. In our case the “variety” category could explain better the relationship among the four breeds. The SIPS kennel club is revising the guidelines used for the definition of a new breed not only based on the morphological variation but also on the genetic structure of the population. Other breeds are interested in the same evaluation (Wijnrocx et al. Citation2014). An international review of the breed concept and the guideline for defining a new breed is needed.
Funding
The authors would like to thank the Societá Italiana Prosegugio [Italian Company Prosegugio] (SIPS) “Luigi Zacchetti” and the Italian Kennel Club (ENCI) for their financial support.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Bigi D, Marelli SP, Randi E, Polli M. 2015. Genetic characterization of four native Italian shepherd dog breeds and analysis of their relationship to cosmopolitan dog breeds using microsatellite markers. Animal. 9:1921–1928.
- Cadieu E, Neff MW, Quignon P, Walsh K, Chase K, Parker HG, Vonholdt BM, Rhue A, Boyko A, Byers A, et al. 2009. Coat variation in the domestic dog is governed by variants in three genes. Science. 326:150–153.
- Cecchi F, Paci G, Spaterna A, Ciampolini R. 2013. Genetic variability in Bracco Italiano dog breed assessed by pedigree data. Ital J Anim Sci. 12:e54.
- Ciampolini R, Cecchi F, Bramante A, Casetti F, Presciuttini S. 2011. Genetic variability of the Bracco Italiano dog breed based on microsatellite polimorphysm. Ital J Anim Sci. 10:267–270.
- Earl DA. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 4:359–361.
- Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 14:2611–2620.
- Excoffier L, Lischer HE. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 10:564–567.
- Felsenstein. 1993. PHYLIP: phylogenetic inference package, version 3.5 c.
- La Manna V, Lasagna E, Ceccobelli S, Di Lorenzo P, De Cosmo AM, Attard G, Sarti FM, Panella F, Renieri C. 2015. Genetic differentiation between Segugio dell’Appennino and Segugio Maremmano dog breeds assessed by microsatellite markers. Ital J Anim Sci. 14:3809.
- Leroy G, Verrier E, Meriaux JC, Rognon X. 2009. Genetic diversity of dog breeds: within breed diversity comparing genealogical and molecular data. Anim Genet. 40:323–332.
- Parra D, Méndez S, Canon J, Dunner S. 2008. Genetic differentiation in pointing dog breeds inferred from microsatellites and mitochondrial DNA sequence. Anim Genet. 39:1–7.
- Peakall R, Smouse PE. 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update. Bioinformatics. 28:2537–2539.
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics. 155:945–959.
- Raymond M, Rousset F. 1995. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered. 86:248–249.
- Wijnrocx K, Janssens S, Buys N. 2014. Effect of dog breed ‘varieties’ on population genetic structure. Commun Agric Appl Biol Sci. 9:206–211.
