Abstract
Climate change has the potential to adversely affect the health of livestock, with consequences to animal welfare, greenhouse gas emissions, productivity, human health and livelihoods. Phenotypic plasticity is the ability of a genotype to produce different phenotypes, depending on environmental, biotic or abiotic conditions; it is a factor influencing and modifying the genes of animal and plant organisms, to adaptation to climate change. Among the various climate variables, heat stress has been reported to be the most detrimental factor to the economy of the livestock industry. There are a number of candidate genes that are associated with adaptation of ruminants, monogastric and poultry to heat stress. For instance, the genes encoding leptin, thyroid hormone receptor, insulin growth factor-1, growth hormone receptor, are associated with the impacts of heat stress on the physiological pathways of domestic animals such as dairy cows, beef cattle, buffaloes, poultry, pigs and horses. This review aims to highlight genes and traits that are involved with thermo-tolerance of domestic animals to sustain production and to cope with climate change. Selection and experimental evolution approaches have shown that plasticity is a trait that can evolve when under direct selection and has a correlated response to some specific traits. Therefore, new breeding goals should be defined for the potential of livestock species to acquire plasticity for adaptation to the current climate changing conditions.
Heat stress compromises feed intake, growth, milk and meat quality and quantity, resulting in a significant financial burden to global livestock.
Genetic selection and nutritional intervention are key strategies to consider in Animal Genetic Resources in hot environments.
Information from gene expression or genome-wide association studies can be used to further improve the accuracy of selection.
Highlights
The climate change
Climate change is defined as spatial (regional, national, continental, global) and temporal (yearly, quinquennial, decennial, millennial) variations of environmental climatic parameters on Earth. These climatic parameters include temperature, humidity, solar radiation, precipitation, cloudiness and temperature of water with melting glaciers (Hoffmann Citation2010).
Climate may change due to natural causes (Merilä et al. Citation2014) together with human activities (Alexander et al. Citation2016).
Many climatologists maintain that global warming is the ‘climate change’ which has developed throughout the 20th century and still ongoing (Houghton et al. Citation2016). It occurs in presence of excessive concentration of carbon dioxide (CO2) (63%) together with other gases such as methane (CH4), nitrous oxide (N2O), nitric oxide (NO), nitrogen dioxide (NO2) (37%) due to human activities (Gerber et al. Citation2013; Herrero et al. Citation2013).
Carbon dioxide, one of the most common environmental pollutant, has a negative effect on the climate and causes harmful changes for many life forms living on the planet (Solomon et al. Citation2009; Opio et al. Citation2013).
Global warming produces measurable effects by physical indicators such as the variation of the seasonal trend, less frequent but more intense precipitations and melting glaciers resulting rising sea level and increasing water temperature (Larsen and Per Citation2008).
Since 1880, year 2018 has been the hottest year in Europe with a rise in average temperature by more than 1.5° C (Rust Citation2019). This temperature increasing is above the limit chosen not to be exceeded as written in ‘COP21 of Paris’, an agreement signed by the European Commission on Climate Change. The earth’s climatic data are expected to worsen significantly in the future (Hume et al. Citation2011).
Climate change has also adverse effects on agricultural sector and may damage the other ones (commercial, economic and energy sectors). Indeed, it is estimated that from 1880 to 2015, loss of 433 billion euro in primary, secondary and quaternary sectors was caused by crop damages in agriculture (Rust Citation2019).
Adverse effects of harsh environmental conditions such as those presented by climate change can lead to adopting mechanisms of resilience (Canale and Henry Citation2010) such as phenotypic plasticity, or rather, the ability of a genotype to produce different phenotypes according to environmental pressure.
Livestock systems and environmental pollution
Worldwide, a large amount of greenhouse gases (GHG) (17.2%) is produced by primary sector in which intensive ruminant-based production system, causing methane and nitrous oxide emissions, is preferred (Collier et al. Citation2019).
However, the application of good management practices on agricultural land, avoiding degraded land, favouring the use of pastures and meadows, can transform the production system into carbon accumulators sequestering carbon from the atmosphere (Cassandro et al. Citation2013b; Cassandro Citation2020).
Methane emission represents 62% of GHG produced by agriculture-sector (Hume et al. Citation2011; Grossi et al. Citation2019). In terms of pollution, methane is the second responsible gas for the greenhouse effect, after carbon dioxide (Alexander et al. Citation2016). Moreover, methane affects degradation of the ozone layer. Atmospheric methane concentration is lower than the carbon dioxide, but its potential effects on global warming are much higher (Swain et al. Citation2008).
The greatest part (75%) of methane emitting from the agricultural sector is produced by enteric fermentation and 16% by manure management (Grossi et al. Citation2019). In 2015, the enteric fermentation was responsible for 32% of the total methane emissions in the world, 57% and 43% of which were attributed to dairy and beef cattle, respectively (Collier et al. Citation2019).
In 2015, the largest part of N20 emission (61%) was attributed to agricultural sector (Collier et al. Citation2019) due to use of fertilisers and livestock management practices (Herd et al. Citation2014).
Nitrous oxide is a greenhouse effect that affects depletion of the ozone layer. It can be found in small amounts in the atmosphere and it is the third most abundant GHG after carbon dioxide and methane (Muchenje et al. Citation2018).
Ammonia (NH3) originating from animals and chemical fertilisers affects the phenomena ‘acid rain’ or ‘acid depositions’ (Bernabucci Citation2019; Pasquì and Di Giuseppe Citation2019).
Increasing in temperatures and thermo-tolerance
Variations in climatic factors, including rising temperature, could impact negatively growth, reproduction and production in livestock species (Osei-Amponsah et al. Citation2019; Figure ). Almost in all regions of the world, climate change leads to increased temperatures, altered photoperiod and decreased precipitation which causes reduced feed quality and quantity, less water availability and, high disease susceptibility (Angel et al. Citation2018). In livestock, body temperature is controlled by a balance between metabolic heat production and heat loss from the body (Berman Citation2011). Rising temperature leads to heat stress (HS) taking place when an animal is unable to adequately dissipate the excess of endogenous heat to maintain homeothermy (Bernabucci et al. Citation2014; Lacetera Citation2019). In order to adapt to new environmental conditions, animals can change their physiology and behaviour (Marai et al. Citation2007). For example, shadow seeking behaviour has been reported in animals raised in hot geographic areas during summer (Osei-Amponsah et al. Citation2019).
Figure 1. Impacts of climate change on livestock health and pathogens (Özkan et al. Citation2016).
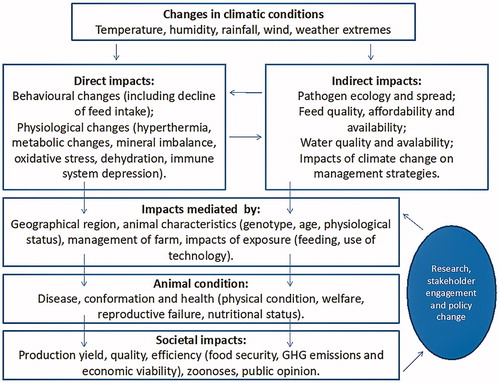
A thermo-tolerant animal can maintain thermal balance under conditions of heat load (Carabaño et al. Citation2019). HS compromises feed intake, growth, milk yield and quality, and meat quality, resulting in a significant financial burden to global animal agriculture (Dunshea et al. Citation2013).
The causes of the temperature increase and global livestock distribution
The most recent climate change has been analysed in detail the last 50 years during which the observation of the upper troposphere has become possible, and human activities have grown exponentially (Van Vliet et al. Citation2013).
All the factors connected with the temperature rise are linked to human activities including agriculture and animal husbandry.
These factors are:
increasing concentration of the GHG in the atmosphere;
changes on the Earth’s surface (i.e. deforestation);
multiplying aerosol;
farm methane emissions.
A report of the ‘Intergovernmental Panel on Climate Change’ (Pachauri et al. Citation2014) concludes that most of the temperature increase observed since the mid-twentieth century is likely to be due to the increase in human-made GHG; while it is very unlikely (it is estimated below 5%) that the climatic change can be explained by resorting only to natural causes (Stocker et al. Citation2013). Many negative effects of climate change arise from increased severity and frequency of drought, rainfall, floods and high temperatures with huge consequences for the sustainability of global agriculture, producer incomes, producer livelihood and food security (Lipper et al. Citation2014).
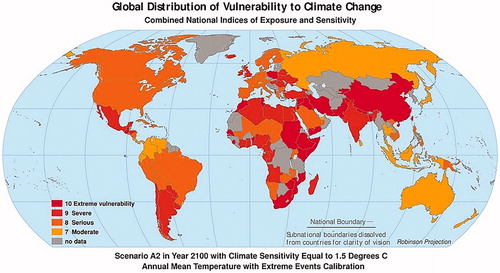
According to climatic data, global warming is estimated to increase at unusual rates (5.5 °C) by 2050 (Figure ) and average surface temperature is predicted to increase by 1.5 °C by 2100 (Figure ). There is also an exponential increase in the number of hot days per year from 90 to 132 in ‘extreme vulnerability’ areas, from 90 to 117 in ‘severe’ areas and from 90 to 109 in ‘moderate’ areas (Figures and ; Hollings et al. Citation2018).
Figure 2. Scenario in year 2050 with climate sensitivity equal to 5.5 °C annual mean temperature with extreme events calibration (Source: figure taken from https://sedac.ciesin.columbia.edu/mva/ccv/… and adapted for illustrative purpose only).
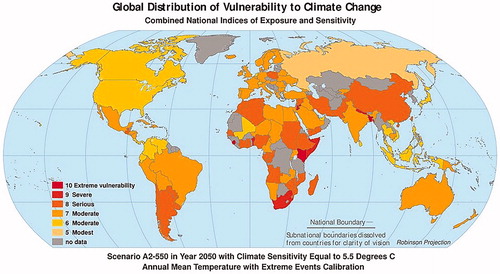
Figure 3. Scenario in year 2100 with climate sensitivity equal to 1.5 °C annual mean temperature with extreme events calibration (Source: figure taken from https://sedac.ciesin.columbia.edu/mva/ccv/… and adapted for illustrative purpose only).
Global data sets on the geographic distribution of livestock are essential for diverse applications in agricultural socio–economics, environmental impact assessment, food security and climate change (Gilbert et al. Citation2018). Environmental variables, such as temperature, are important determinants of the distributions of many species, such as dairy and beef cattle (Figure ), goats (Figure ), sheep (Figure ), chickens, pigs, ducks, horses and buffaloes (Scharlemann et al. Citation2008).
Figure 4. (a) Overview of the livestock of the world: data set for dairy and beef cattle (Source: figure taken from http://www.fao.org/livestock-systems/global-distributions/cattle/en/ and adapted for illustrative purpose only). (b) Overview of the livestock of the world: data set for goat (Source: figure taken from http://www.fao.org/livestock-systems/global-distributions/goats/en/ and adapted for illustrative purpose only). (c) Overview of the livestock of the world: data set for sheep (Source: figure taken from http://www.fao.org/livestock-systems/global-distributions/sheep/en/ and adapted for illustrative purpose only).
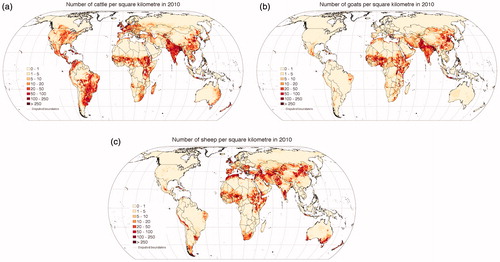
The following maps (Figure ) provide geographical information and resources to global livestock systems.
The world temperature scenarios (Figures and ) and livestock distribution (Figure and ) show that livestock will be affected by higher temperature all over the world in the next century (Gilbert et al. Citation2018). Therefore, priority must be given to the selection of adapted animals, depending on the level of vulnerability of the geographical area. In fact, priority should be given to animals rearing in the ‘extreme vulnerability’ areas (China, Ethiopia, India, and South Africa), subsequently to the ‘severe’ areas (Europe and Central America) and finally to the ‘moderate’ areas (Russia and Oceania) (Hollings et al. Citation2018).
The relationship between temperature increase and phenotypic plasticity
The phenotypic plasticity is the ability of a genotype to produce different phenotypes, depending on environmental, biotic or abiotic conditions (Alford et al. Citation2006). The phenotypic plasticity is a factor influencing and modifying animal and plant organisms by increasing their adaptation to climate change (Lacetera et al. Citation2009). Polyphenism is defined as discrete phenotype induced by differences in environment (Price et al. Citation2003; Kelly et al. Citation2012).
Fundamental to the way in which organisms cope with environmental variation, phenotypic plasticity encompasses all types of environmentally induced changes (e.g. morphological, physiological, behavioural, phenological) that may or may not be permanent throughout an individual’s lifespan. The term was originally used to describe developmental effects on morphological traits; today, however, it is more broadly used to describe all phenotypic responses to environmental change such as acclimation or acclimatisation as well as learning (Kelly et al. Citation2012).
The temperature increase is one of the factors altering health state and it is expected to exert an overwhelming negative effect on animal health (Chevin and Hoffmann Citation2017). It has also been demonstrated that the temperature increases significantly heightens mortality and/or worsens animal health and welfare in geographical areas characterised by temperate climate and standard cold during the year (Collier et al. Citation2008; Stocker et al. Citation2013; Bernabucci Citation2019).
In dairy cattle, the summer season affects health status of animals together with decreasing production performances in both quantity and quality in hot climates due to HS (Windig et al. Citation2005). It is known that body temperature is maintained between 38.6 °C and 39.3 °C in dairy cows by thermoregulation mechanism allowing continuously balance between the amount of endogenous heat produced and the amount of heat dispersed towards the external environment (Sartori et al. Citation2002).
The upper limit of ambient temperatures at which Holstein Friesian cattle may maintain a stable body temperature is from 25 to 26 °C and relative humidity between 50 and 80% (Berman et al. Citation1985; West Citation2003). At a temperature of 29 °C with 40% relative humidity the milk production of Holstein Friesian, Jersey and Brown Swiss cows was 97%, 93%, and 98% of normal, but when relative humidity was increased to 90%, yield was reduced to 69, 75, and 83% respectively (West Citation2003).
Moreover, Mateescu et al. (Citation2020) estimates the plasticity in body temperature during HS in six crossbred groups ranging from 100% Angus to 100% Brahman. They concluded that effective strategies will be required for the identification of the genes conferring the superior thermo-tolerance in Brahman cattle.
It is known that each livestock species could face temperature changes by reacting differently at different times. The different abilities vary according to several factors (species, breed, sex, age, etc.), depending on morphological traits (coat colour, presence or absence of hair, skin texture, colour of the limbs distal parts, mucous and genitals), rearing system (intensive, semi-extensive, extensive) and geographical area (altitude, latitude, longitude) (Schefers and Weigel Citation2012).
The climate change is expected to have an increasing impact on animal production systems in the world. In some regions, farmers need to adapt their practices in order to fight temperatures increase, against the onset of new animal diseases, for instance, and the negative repercussions on grazing lands (Collier et al. Citation2008; Nardone et al. Citation2010).
Livestock biodiversity
Worldwide, the rural development policies described in the ‘Kyoto protocol’ were initially formulated based on different aims compared to the climate change mitigation. The sub-division of the globe into macro-areas identifies the competitiveness, the environmental protection and the development of rural areas as priority intervention goals. This implied that some of the measures and actions planned under the Rural Development Programmes (RDPs) are characterised by aims referring to climate change mitigation adaptation of agriculture ecosystems, forest and livestock (Drucker et al. Citation2007).
Climate change which negatively affects almost all farms are mainly highlighted through three phenomena, including increase in temperatures and precipitation intensity and decrease in amount of precipitation (Liu et al. Citation2000).
In the last decade, in Europe and in America, breeding management of dairy and beef cattle has been influenced above all by the sudden and exceptional increase in temperatures, the drought and the consequent low availability of water for irrigation (Hollings et al. Citation2018). The greater disease incidence and parasitic attacks found both in crops and animals have been also quite significant. Erosion and the deterioration of soil quality are the less impactful phenomena (Hill et al. Citation2008).
Adverse effects of climate change have a non-negligible impact on production, because the farmers must face an unexpected economic cost. In a study conducted by the University of North Carolina, the increase in production costs of U.S. dairy products, is a circumstance mentioned by more than three quarters of respondents because of the adverse climatic events occurrence (Steinfeld et al. Citation2006).
In the last five years, challenges were also encountered in the performance of daily farming practices and the qualitative and quantitative reduction of fodder production which had negative consequences on the livestock feed availability (Knapp et al. Citation2014).
Measures adopted by breeders to face climate change
The fact of negative effects of climate change has forced the breeders to take mitigation and adaptation measures. One of the main ‘mitigation measures’ (O’Brien et al. Citation2020) well-adopted by breeders (Table ) is structural investments to improve the effluent management. Additionally, using effluent distribution techniques could reduce ammonia emissions (Thornton et al. Citation2007).
Table 1. Mitigation measures adopted by breeders to reduce ammonia emission (O’Brien et al. Citation2020).
Indeed, it is reported that globally, more than the 70% of the breeders have already implemented these interventions or would do so in future (Rojas-Downing et al. Citation2017). These measures of mitigation, adaptation and animal breeding will allow breeders to make medium-long term investments by decreasing their economic losses (Sejian et al. Citation2019).
The ‘adaptation measures’ (Table ) in the vast majority of the ‘next generation’ farms have already been implemented or must be carried out to improve the microclimate of livestock (for example the insulation of stables roofs or the cooling systems) (Pirlo and Caré Citation2013; O’Brien et al. Citation2020). In many farms, new insurance coverage about capital, machinery and installations have already been activated to face the losses due to extreme weather events; however, the tendency to ensure even the production in case of extreme weather events was lower (Thornton et al. Citation2007).
Table 2. Adaptation measures adopted by breeders to improve the microclimate of livestock (Pirlo and Caré Citation2013; O’Brien et al. Citation2020).
In addition to the physiological effects of high temperatures on animals, the consequences of climate change are likely to affect negatively rare livestock breeds reared in limited regions (Hoffman Citation2010).
Indirect effects of climate change may alter the distribution of animal diseases or affect the supply of feed. Breeding aims may be selected by considering higher temperature, lower quality food resources and more disease problems in the future (Oluwatayo and Oluwatayo Citation2018). Species and breeds well-adapted may be preferred by breeders to face these problems. Today, however, increased demand for food, forces breeders to raise high producing livestock breeds which convert animal feed into meat, milk and eggs (Pirlo and Caré Citation2013). Furthermore, these high producing breeds are very often held responsible of GHG emissions, even if several studies demonstrated the possibility for using GHG traits as large-scale indicator traits for genetically improving the accuracy of feed efficiency such as in dairy cows (Difford et al. Citation2020), in beef cattle (Barwick et al. Citation2019), in pigs (Alfonso Citation2019), and in poultry (Willems et al. Citation2013). This may lead to the negligence of local breeds adapted in developing countries (Mathias and Mundy Citation2005).
Local livestock breeds contributing to world animal genetic resources have unique genetic structure and genetic diversity of local breeds must be conserved in order to face climatic changes in the future (Bett et al. Citation2017). Indeed, Cassandro (Citation2013a) showed that a reduction of 10% of daily methane emissions per kg of metabolic body weight is expected for local breeds compared with cosmopolitan ones; they concluded that animal genetic resources needs to be evaluated not only per unit of output, but for other direct and indirect units of output related to social and human returns. This requires effective in situ and ex situ conservation programmes after characterisation of local breeds. Hence, improved mechanism to monitor and respond to threats to genetic diversity as well as genetic improvement programmes aimed for adaptation of high‐output local breeds is needed. Moreover, increased support for developing countries for management of animal genetic resources and wider access to genetic resources and associated knowledge are needed (Thornton et al. Citation2007).
Plasticity and genetics
There are several models used to explain the genetic basis of plasticity responses. The main and not mutually exclusive ones are:
over-dominance: the plasticity is an inverse function of the number of heterozygous loci (Sato and Stryker Citation2008);
pleiotropy: the plasticity is a function of the differential expression of a gene in different environments due to some pleiotropic effect: influence of a gene on multiple and partially unrelated traits (Des Marais and Juenger Citation2010);
epistasis: the plasticity may be due to the epistatic interaction between genes determining the degree of response to environmental influences and others causing the average expression of a trait (Remold and Lenski Citation2004).
Several studies on plasticity responses have demonstrated that the heterozygosity (over-dominance model) has lower effects on plasticity, while the pleiotropic and epistatic phenomenon have greater impacts (Scheiner Citation1993; Pigliucci Citation2005).
Molecular technologies on large-scale gene expression, such as the heterologous DNA hybridisation (micro-arrays), the next-generation sequencing technologies applied to the transcriptome (RNAseq) and the techniques to study non-coding small RNAs functions and proteomic tools may help to clarify plasticity at molecular level (You et al. Citation2015).
The pleiotropy effect has been investigated in chicken by Ng et al. (Citation2012). The authors highlighted the chicken frizzle feather is due to an KRT75 (α-Keratin) gene mutation that causes a defective rachis.
Moreover, Lafuente and Beldade (Citation2019) showed two emblematic examples of developmental plasticity in body size and pigmentation both in Drosophila melanogaster flies and Bicyclus anynana butterflies.
Epistatic gene interaction has been investigated in livestock. Knaust et al. (Citation2016) reported the identification of three-locus interaction that underlies ‘rat-tail’ syndrome in cattle, furnishing the first example of epistatic interaction between several independent loci that is required for the expression of a distinct phenotype. In pigs, Noguera et al. (Citation2009) described the existence of multiple epistatic quantitative traits loci (QTL) affecting phenotypic variance of swine prolificacy traits.
Genetic evolution of plasticity
The phenotypic plasticity is of importance because it expands the existing geocentric evolutionary theory, indicating the change within a population of inheritable characteristics with the generation succession (Wess Citation2003).
The phenotypic plasticity complements the role of mutations in evolution. Natural selection chooses not between genotypes, but between phenotypes. For this reason, phenotypes and the variation among them play the main role in evolution. Furthermore, since the environment in which an individual grows determines the phenotypes, environment plays an important role in phenotypic variation. This is because mutations are not only rare, but they are also usually deleterious (Krašovec et al. Citation2014).
In contrast, the environmental conditions change constantly and act at the same time on all the individuals of the population. Moreover, the mutations generally appear randomly without a real correlation with a specific environment. Instead, the plasticity induction of a phenotype by the environments correlate with the specific conditions that determine it, allowing a positive selection on the aforementioned phenotype (De Jong and Bijma Citation2002).
According to traditional theory of evolution by natural selection, the environment acts after occurring of phenotypic variation. Thus, the role of phenotype is simply to express the genetic variation for selection. Instead, thanks to the phenotypic plasticity, the environment seems to play a double role including creating phenotypic variation and selecting between the different variants (Mohn and Dirk Citation2009).
Plasticity indicators (GxE interaction, adaptability, acclimation of plasticity)
In the second half of the last century, the investigation of the genetic basis and the transmission of the phenotype have dominated the biological studies. In last twenty years, the role of plastic responses in both adaptation and evolutionary species history has been recognised (Pigliucci et al. Citation2006).
Any type of organism’s trait (chemical or biochemical, physiological, morphological and behavioural) may show plasticity (Merilä et al. Citation2014). Therefore, the processes regulating the expression of plasticity responses are important for the knowledge of physiological, morphological and behavioural characteristics of the species, as well as the evolutionary dynamics and the influence of the global climate change on the organisms (Hendry and Taylor Citation2004). In population genetic studies, phenotypic variance can be used as a trait for phenotypic plasticity (VandeHaar et al. Citation2016). Several statistical models for phenotypic plasticity have been considered in animal breeding: 1) the reaction norms or random regression model; 2) the character state or multi-trait model; 3) the infinite-dimensional or covariance-function model. These models differ in the way phenotypic plasticity is dissected into quantitative traits, but all of them are based on the general expressions for the change in mean values of a number of quantitative traits that undergo simultaneous selection. A model of phenotypic plasticity may be used as a tool to select animals for robustness, or can be used in breeding programmes that produce genetic material for a multiple of production environments (De Jong and Bijma Citation2002).
Reaction norms model
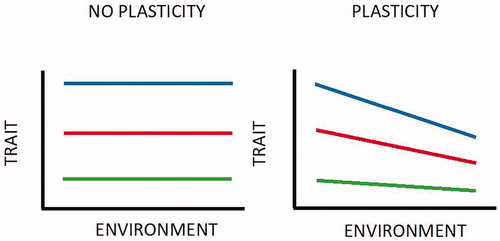
Several genotypes exhibit different reaction norms that differ in the expressed phenotype and in the degree of plasticity (Lu et al. Citation2013). The reaction norms are often represented in graphs, not necessarily by straight lines, in which the environmental parameters (biotic and abiotic) are reported on the abscissa axis and the phenotypic ones (morphological, behavioural or other) on the ordinate axis (Gautier and Naves Citation2011). The elevated plasticity in a trait results in a reaction norm with a high slope, which describes a considerable effect by environment on phenotype (Figure ). On the contrary, the non-plastic traits will give a substantially flat reaction norm (Dikmen et al. Citation2012).
Figure 5. The ability of one genotype to produce more than one phenotype when exposed to different environments (each of the coloured lines is a ‘reaction norm’) (Kelly et al. Citation2012).
Character state model
‘Character state’ is defined as the expression of a trait in different environments (Falconer Citation1952). The character state model, for instance, is an analogy of a multi-trait model of weight at two different ages. Phenotypic plasticity is equivalent to a difference in phenotypic level between the character states. The total additive genetic variance of the plasticity trait over both or all environments can be split into the genetic variance of the trait states within each environment and the genetic covariance between environments. The genetic covariance between environments is associated to the genotype by environment interaction variance (De Jong and Bijma Citation2002).
Infinite-dimensional model
In the infinite-dimensional models, a mean function gives the average value of the trait over environments, and an additive covariance function is specified. These models, also called regression models, have been extensively applied to analyse ontogeny traits (e.g. growth curves – Kirkpatrick and Heckman Citation1989; Wilson et al. Citation2005 – and milk yield in dairy cattle – Jamrozik and Schaeffer Citation1997). The infinite-dimensional approach is a very powerful tool in the quantitative genetics of continuous traits (De Jong and Bijma Citation2002).
Identified genes associated with climatic variables
Climatic variables include many factors; in Flori et al. (Citation2019) six factors were considered including annual average temperature (Bio 1), temperature range (Bio 7), quantity and intensity of rain (Bio 12), irradiation (Bio 20), humidity (Bio 28) and relation index between humidity and temperature (THI) of a specific geographical area. On a sample of 21 Mediterranean cattle breeds a genome – wide association analyses with covariables discriminating the different Mediterranean climate subtypes was carried out. The authors reported 55 genes related to one or more climatic variables. Biological function of the candidate genes associated with the climatic variables were subdivided in three categories (Table ). The same authors also defined the genes with four climatic covariates (using the most important principle components named PC1, PC2, PC3, PC4) which summarise the 35 climatic parameters expressed by the ‘CliMond database’ (Flori et al. Citation2019).
Table 3. Functions of candidate genes associated with at least one climatic covariable (Flori et al. Citation2019).
The genes mentioned in Table , express their phenotypes in different ways by adapting to the involved variable (Bohmanova et al. Citation2007). Some authors reported that the TCF7 gene (Transcription Factor 7 – T Cell Specific) is associated, in cattle, not only with the annual average temperature of a specific geographical area, but also with rainfall, radiation, humidity and the relation index between temperature and humidity (THI) (Green Citation2009; Liu et al. Citation2009). In a study on two independent populations from different cattle breeds, BFGL-NGS-139, BFGL-NGS-89500, BFGL-BAC-38208, BFGL-NGS-30169 markers linked to sensitivity of milk production to THI have been identified on BTA29 (Hayes et al. Citation2009). These markers could help in genetic selection for high milk production under HS conditions (Hayes et al. Citation2009). Also, ANTXR2 (Anthrax toxin receptor 2) and GLDC (Glycine decarboxylase) located in different chromosomes have been reported to be associated with Bio1 and THI (Gautier and Naves Citation2011). A limited number of genes have been reported to significantly determine the phenotypic plasticity in animals (Harris et al. Citation2013). Three of the genes, VDAC1 (Voltage dependent anion channel 1), TCF7 and SKP1 (S-phase kinase associated protein 1) are located in the same chromosome (BTA7), and are associated with at least three climatic variables. The VDAC1 gene is associated with the variables PC1, Bio12 and Bio20, the TCF7 and SKP1 genes are associated with the variables PC1, Bio1, Bio12, Bio20, Bio28 and THI (Flori et al. Citation2019). Another gene located in BTA16, the KCNH1 (Potassium voltage-gated channel subfamily H member 1), compared to the previous ones, is associated with more than two variables (PC2, Bio7, Bio12 and Bio28). The HSPB3 (Heat shock protein family B (small) member 3) gene located in chromosome BTA20, is associated with the variables Bio12 and Bio28; the TRAM2 (Translocation associated membrane protein 2) in chromosome BTA23, is associated with the variables PC2, Bio20 and Bio28 (Joost et al. Citation2007; Sigdel et al. Citation2020).
Other genes, ANTXR2 and GLDC located in BTA6 and BTA8 respectively, are associated with the same climatic variables Bio1 and THI (Ho et al. Citation2008; Flori et al. Citation2019).
The RETSAT (Retinol saturase) and the ELMOD3 (ELMO domain containing 3) genes located in the BTA11 as well as the LAMC1 gene located in the BTA16 are equally associated with two climatic variables (PC2 and Bio7; Flori et al. Citation2019; ).
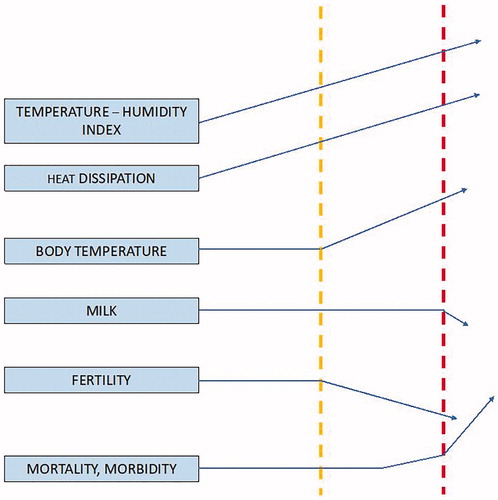
Among the climatic parameters, temperature increase has dramatic negative effects such as HS in livestock species. HS is associated with several mechanisms including different genes. It is reported that during the summer fertility and milk yield decrease in cows reared in Thailand due to HS (Boonkum et al. Citation2011; Figure ).
Figure 6. Hypothetical response of a dairy cow to increased heat stress (Source: Misztal Citation2017). In yellow and red two different temperature levels (in red the hottest temperature).
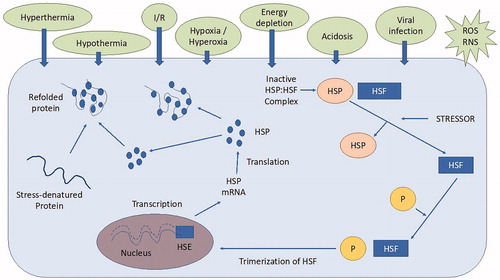
Under HS condition, anti-stress mechanisms, including heat shock proteins (HSPs; Figure ) and other thermo-tolerant genes, are activated at the cellular level in livestock species (Aleena et al. Citation2016).
Figure 7. Molecular mechanism of heat shock protein (HSP) synthesis: the heat shock factors (HSF) present in the cytosol are bound with HSP to maintain an inactive state. The wide variety of heat stressors (HS) can activate the HSF causing them to get detached from HSP. Then, HSF are phosphorylated (P) by protein kinases to form trimers in the cytosol. This HSF trimer complex enters the nucleus and bind with heat shock elements (HSE) in the promoter region of the HSP gene. HSP70 mRNA is then transcribed and leaves the nucleus into the cytosol where new HSP70 is synthesised. The newly synthesised HSP protects the cells by acting as a molecular chaperone facilitating the assembly and translocation of newly synthesised proteins within the cell and repairing and refolding damaged proteins in heat stressed subjects (Source: Sejian et al. Citation2019). I/R: ischemia/reperfusion; ROS: reactive oxygen species; RNS: reactive nitrogen species.
Today, thanks to developing techniques, such as genome-wide SNP scans, several candidate genes related to HS have been reported in livestock (Archana et al. Citation2017). The importance of indigenous livestock as model organisms for investigating selection sweeps and genome-wide association mapping was highlighted also by Kim et al. (Citation2016) both in goats and sheep.
THR is considered as an important gene affecting HS tolerance in livestock, because the animals reduce the thyroid activity in order to limit the metabolic heat production during HS as an important adaptation strategy (Collier et al. Citation2019). The LEP (Leptin) controls the energy metabolism by activating the hypothalamic-pituitary-adrenal axis and subsequently affects the growth in animals during HS (Tian et al. Citation2013). Lower expression of LEP was reported during HS in Osmanabadi goats (Bagath et al. Citation2016). In contrast, a higher expression of LEP in vitro (in 3T3-L1 rat cells) was reported during HS of dairy cows in transition period (Bernabucci et al. Citation2007).
Under HS conditions, heat shock factor 1 (HSF1), heat shock protein gene 60 (HSP60), Heat Shock Protein gene 70 (HSP70) and Heat Shock Protein gene 90 (HSP90) were associated with the resilient capacity of small ruminants. Among these genes, HSP70 is the well-known genetic marker for thermo-tolerance in small ruminants (McManus et al. Citation2011); also, HSP70.1 gene is reported as biomarker in Holstein lactating cows population (Basiricò et al. Citation2011). HSP70 family and other HSP genes are in fact biomarker used to select heat and cold tolerant genotypes in the context of climate change. Higher HSP70 gene expression was recorded in cold-adapted goats during summer and heat-adapted goats during winter (Das et al. Citation2015; Kumar et al. Citation2015).
Relevant variability in the expression of the HSP70 and other HSP genes family in different seasons has potential strategies for better adaptability in cattle (Kumar et al. Citation2015). Maibam et al. (Citation2017) reported higher HSP70 mRNA level in heat stress tolerant (HST) dairy cows than in heat tolerance susceptible (HSS) individuals in Tharparkar Zebù and Karan Fries breeds.
A major gene (SLICK) was associated with heat tolerance (HT) with a dominant inheritance determining ‘slick hair’ type in dairy cattle (Olson et al. Citation2003). This gene was introduced into Holstein Friesian cattle in order to obtain thermo-tolerant cows (Olson et al. Citation2006; Berman Citation2011; Dikmen et al. Citation2014).
KPNA4 (Karyopherin subunit alpha 4), MTOR (Mechanistic target of rapamycin kinase), SH2B1 (SH2B adaptor protein 1) and MAPK3 (Mitogen-activated protein kinase 3) genes were involved in the HSPs’ regulation of stress in PBMC (Peripheral blood mononuclear cell) purified from Karamojong goats’ blood, after exposure to 40 °C (Onzima et al. Citation2018; Li et al. Citation2020).
Lower growth hormone (GH) mRNA expression was reported in the hepatic tissue of heat stressed Malabari goats (Angel et al. Citation2018).
Genetic association study revealed possibility of ATP1A1 candidate genes for selection of crossbred Jersey cows for better HT (Das et al. Citation2015). One genotype at locus C1787061T (Intron 10) was reported to improve HT in Sahiwal cows indicating association with the HSP90ab1 gene (Sailo et al. Citation2015).
Finally, several studies have identified target gene expression responses associated, in dairy cows, with HS in mammary immune cells and liver (Tao et al. Citation2013). The hepatic transcriptome of transition dairy cows is strongly affected by season of calving: in particular, HS not only alters energy metabolism in liver, but also induces an inflammatory and intracellular stress response (Shahzad et al. Citation2015).
Relationship between phenotypic plasticity and selection signatures in livestock
Many genes and/or genomic regions related to ecological traits (e.g. hot-arid environment), may be identified via selection signatures (Gouveia et al. Citation2014). This approach can be used in QTL mapping experiments for meat, milk and eggs production (Nielsen Citation2001; Schlötterer Citation2003; Hayes et al. Citation2008).
Domestication greatly changed the morphological and behavioural features of modern domestic animals and, along with breed formation and selection schemes, allowed the development of different modern breeds (Gouveia et al. Citation2014). These features, along with extensive knowledge about genes and/or genomic regions, provide an opportunity for identifying loci to undergo selection and for the development of new methods to detect selection signatures (Flori et al. Citation2009).
In beef cattle, for example, using the FST approach, selection signatures were found in the regions of BTA2 and BTA7 chromosomes (The Bovine HapMap Consortium Citation2009; Gouveia et al. Citation2014). These regions are associated with feed efficiency and THI index (Barendse et al. Citation2009; Flori et al. Citation2009) and contains the R3HDM1 (R3H Domain Containing 1), ZRANB3 (Zinc Finger, RAN Binding Domain Containing 3) and TCF7 genes, which have been suggested to be involved in animal’s growth processes and in response to HS (The Bovine Genome Sequencing and Analysis Consortium Citation2009; Gouveia et al. Citation2014).
Plasticity and epigenetics in livestock
Epigenetics is a branch of science dealing with changes influencing the phenotype without altering the genotype (Murren et al. Citation2015). It consists of a series of molecular pathways through which the DNA transcription is altered without modification of the underlying DNA sequence (Bonamour et al. Citation2019). Genes are influenced by various environment patterns including the type and level of nutrients, the toxins and stress level during the animal’s lifespan. Moreover, several studies showed the effects of social experience in epigenetic pathways influencing the stress response and the reproductive behaviour (Scheiner et al. Citation2017). Epigenetic modifications may lead to up or down regulation of the DNA transcription through a variety of pathways including DNA methylation, histone modifications and microRNAs – miRNAs (Snell-Rood Citation2013).
The environmental impact
Although epigenetic mechanisms may cause a high degree of plasticity in gene expression, their flexibility in response to environmental signals is limited at the first stages of embryo development (Carvalho et al. Citation2019).
It is reported that the first external environmental effects experienced during childhood in sheep, can change gene expressions at individual level (Yurchenko et al. Citation2019).
A study on twins in Holstein cattle, born in a ‘Frisia’ farm (Southern region of the Netherlands) and rearing in different environments (both geographically and structurally), showed potential of epigenetic plasticity in phenotype (Barwick et al. Citation2019). In fact, Barwick et al. (Citation2019) showed that the DNA methylation and acetylation patterns were highly concordant between twins under seven years old, but they diverged significantly between ‘adult’ twins, leading to speculate that the strength of limbs and body size, emerged in response to environmental exposure unique for each animal (Carvalho et al. Citation2019).
On the contrary, similar phenotype was reported in ‘Bruna Alpina’ calves born in completely different environments (Spain, Switzerland and Greece) with different breeding systems (one extensive and two intensive). At the age of three months, in fact, calves transferred with their respective mothers to an extensive Normandy herd (Northern France), were very similar in terms of hardiness (size and robustness of the limbs), resistance to climate change (temperatures and humidity), temperament between other calves and with farmers (Kahiluoto et al. Citation2019).
The epigenetic changes do not take place only in the early stages of development, but also in the stage of adult life, as a response to adaptation to the external environment (Barwick et al. Citation2019).
Even during the development, the influences of the other animals can contribute to construct specific epigenetic patterns (Kahiluoto et al. Citation2019). Mehla et al. (Citation2014) showed the association between the quality of the social environment and the neurobiological and behavioural consequences on animals. This study indicated that epigenetic effects are primarily induced by the mother-child interaction and the managing systems (intensive/extensive, free housing or fixed post, cage or ground).
The epigenetic effects have also the potential to be used in animal breeding due to providing information related to the inheritance of traits and complex diseases (Triantaphyllopoulos et al. Citation2016).
Limitation to the application of phenotypic plasticity in animal breeding
There is a wide range of factors leading to constraints in the evolution of plasticity (Cipollini Citation2004). Evolution of plasticity may be limited by the lack of genetic variability as well as allometric relationship among the traits in which a certain plasticity could limit another plastic trait, environmental covariance expressing certain types of plasticity. Additionally, constraints deriving from evolutionary history of species could limit the expression of some traits (DeWitt et al. Citation1998; Auld et al. Citation2010). The plasticity responses are limited due to the expression of a sub-optimal phenotype by a plasticity individual under certain environmental conditions. Environmental conditions are considered as evident when an additional development cannot produce a target trait while fixed development can. Therefore, a plasticity limitation is detected when a plastic response is observed which cannot express the same phenotype as a non-plastic genotype (Cipollini Citation2004). The plasticity limits (DeWitt et al. Citation1998) can be divided into: 1) the reliability of information: reduced reliability of the signals used by individuals to evaluate environmental conditions can lead to phenotype-environment mismatch; 2) the time: due to the time lapse between the perception of the signal and the production of response, during which the environmental conditions can change; 3) the range of development: usually individuals with fixed phenotype are more efficient in producing extreme phenotypes than plastic individuals; 4) epi-phenotype problem: a phenotype produced through a mechanism of progressive addition of elements, in response to environmental signals, may be less efficient than one produced entirely during development.
Future goals
Depending on the results of the ‘COP 21’ in Paris 2015, the European Union’s commitment to fighting climate change goes on with increasingly ambitious emission reduction targets. As regards the agricultural sector, the overall GHG reduction target by 2030 is 30% (Hume et al. Citation2011).
Moreover, even if agriculture is a sector in which ‘decarbonisation’ in the long term is not conceivable (due to the intrinsic nature of GHG emissions in the production processes considered), there are margins for improving the performance of the sector in terms of decreasing GHG which could still be exploited (Opio et al. Citation2013).
In the future, livestock production will probably be increasingly characterised by differences between developed and developing countries and between high-intensity production and small-scale or agro-pastoral systems. However, the way in which the various driving forces will take place in different regions of the world in the future, is highly unclear (Hume et al. Citation2011). First of all, the future demand for animal products will be met through sustainable intensification in a low-carbon economy. The growing demand for livestock products will generate considerable land competition for food and feed production. The increase in the industrialisation of livestock production may lead to problems of air and water pollution. The greatest impacts of climate change will be seen in cattle and mixed systems in developing countries (Nardone et al. Citation2010). The need to adapt to climate change and to mitigate GHG emissions will undoubtedly increase production costs that vary from place to another. Furthermore, the expected biofuel growth demand could have further significant impacts on competition for land and food security (Green Citation2009). Thanks to selection based on developing molecular techniques, animals that are of lower environmental impact could be identified in terms of feed intake and reduction of methane emissions into the atmosphere (De Haas et al. Citation2011).
Conclusions
Plasticity is usually thought to be an evolutionary adaptation to environmental variation. As plasticity allows individuals to ‘fit’ their phenotype to different environments, it may occur in different times within the lifespan of an organism. Plasticity is a key mechanism with which organisms can cope with a changing climate, as it allows individuals to respond to change within their lifetime. Therefore, plasticity is of importance for livestock species with long generation interval, as evolutionary responses via natural selection may not produce change fast enough to mitigate the effects of a warmer climate.
The benefits of plasticity can be limited by the energetic costs of plasticity responses as the synthesising new proteins and the maintaining sensory systems to detect changes.
Given the ecological importance of temperature and its predictable variability over large spatial and temporal scales, adaptation to thermal variation has been hypothesised to be the key mechanism for the new livestock species. Hence, the selection of animals adapted to thermal variations is needed.
Species evolving in the warm and constant climate of the tropics have a lower capacity for plasticity compared to those living in variable temperate habitats, as the magnitude of thermal variation is thought to be directly proportional to plasticity capacity.
This is an excellent opportunity for animal breeding of livestock populations reared in European areas which have diverse climate conditions with variable temperatures. Selection and experimental evolution approaches have shown that plasticity is a trait that can evolve when under direct selection and as a correlated response to select specific traits. Therefore, new breeding goals should be defined in order to use the potential plasticity accumulated in livestock species, especially for species as ruminants with a long generation interval. Finally, it should be underscored that the largest increase in the demand for livestock products is coming from the developing countries. As livestock contributes to increase GHG, more emphasis should be directed to these emergent areas in using aspects of phenotypic and genetic plasticity to promote sustainable mitigation strategies.
Acknowledgements
The work was undertaken as part of the ASPA study commission: Adaptability of Livestock Production Systems to Climate Change. The authors thank the two anonymous referees for their valuable comments on the manuscript and their constructive suggestions.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Aleena J, Pragna P, Archana PR, Sejian V, Bagath M, Krishnan G, Manimaran A, Beena V, Kurien EK, Varma G, et al. 2016. Significance of metabolic response in livestock for adapting to heat stress challenges. Asian J Anim Sci. 10:224–234.
- Alexander LV, Zhang X, Peterson TC, Caesar J, Gleason B, Klein Tank AMG, Haylock M, Collins D, Trewin B, Rahimzadeh F, et al. 2016. Global observed changes in daily climate extremes of temperature and precipitation. J Geophys Res Atmos. 111:D05109.
- Alfonso L. 2019. Impact of incorporating greenhouse gas emission intensities in selection indexes for sow productivity traits. Livest Sci. 219:57–61.
- Alford AR, Hegarty RS, Parnell PF, Cacho OJ, Herd RM, Griffith GR. 2006. The impact of breeding to reduce residual feed intake on enteric methane emissions from the Australian beef industry. Aust J Exp Agric. 46:813–820.
- Angel SP, Amitha JP, Rashamol VP, Vandana GD. 2018. Climate change and cattle production – impact and adaptation. J Med Res. 5:1134.
- Archana PR, Aleena J, Pragna P, Vidya MK, Niyas APA, Bagath M, Krishnan G, Manimaran A, Beena V, Kurien EK, et al. 2017. Role of heat shock proteins in livestock adaptation to heat stress. International J Dairy Sci. 12:1–11.
- Auld JR, Agrawal AA, Relyea RA. 2010. Re-evaluating the costs and limits of adaptive phenotypic plasticity. Proc Biol Sci. 277:503–511.
- Bagath M, Sejian V, Archana SS, Manjunathareddy BG, Parthipan S, Selvaraju S, Mech A, David ICG, Ravindra JP, Bhatta R. 2016. Effect of dietary intake on somatotrophic axis-related gene expression and endocrine profile in Osmanabadi goats. J Vet Behav. 13:72–79.
- Barendse W, Harrison BE, Bunch RJ, Thomas MB, Turner LB. 2009. Genome wide signatures of positive selection: the comparison of independent samples and the identification of regions associated to traits. BMC Genomics. 10:178.
- Barwick SA, Henzell AL, Herd RM, Walmsley BJ, Arthur PF. 2019. Methods and consequences of including reduction in greenhouse gas emission in beef cattle multiple-trait selection. Genet Sel Evol. 51:18.
- Basiricò L, Morera P, Primi V, Lacetera N, Nardone A, Bernabucci U. 2011. Cellular thermotolerance is associated with heat shock protein 70.1 genetic polymorphisms in Holstein lactating cows. Cell Stress Chaperon. 16:441–448.
- Berman A. 2011. Invited review: are adaptations present to support dairy cattle productivity in warm climates? J Dairy Sci. 94:2147–2158.
- Berman A, Folman Y, Kaim M, Mamen M, Herz Z, Wolfenson D, Arieli A, Graber Y. 1985. Upper critical temperatures and forced ventilation effects for high-yielding dairy cows in a subtropical climate. J Dairy Sci. 68:1488–1495.
- Bernabucci U. 2019. Climate change: impact on livestock and how can we adapt. Anim Front. 9:3–5.
- Bernabucci U, Biffani S, Buggiotti L, Vitali A, Lacetera N, Nardone A. 2014. The effects of heat stress in Italian Holstein dairy cattle. J Dairy Sci. 97:471–486.
- Bernabucci u, Basiricò L, Morera P, Lacetera M, Ronchi B, Nardone A. 2007. Heat shock induced changes of adipokines gene expression in 3T3-L1 adipocytes. Ital J Anim Sci. 6:254–256.
- Bett B, Kiunga P, Gachohi J, Sindato C, Mbotha D, Robinson T, Lindahl J, Grace D. 2017. Effects of climate change on the occurrence and distribution of livestock diseases. Prev Vet Med. 137:119–129.
- Bohmanova J, Misztal I, Cole JB. 2007. Temperature-humidity indices as indicators of milk production losses due to heat stress. J Dairy Sci. 90:1947–1956.
- Bonamour S, Chevin LM, Charmantier A, Teplitsky C. 2019. Phenotypic plasticity in response to climate change: the importance of cue variation. Philos Trans R Soc Lond B Biol Sci. 374:20180178.
- Boonkum W, Misztal I, Duangjinda M, Pattarajinda V, Tumwasorn S, Sanpote J. 2011. Genetic effects of heat stress on milk yield of Thai Holstein crossbreds. J Dairy Sci. 94:487–492.
- Canale CI, Henry PY. 2010. Adaptive phenotypic plasticity and resilience of vertebrates to increasing climatic unpredictability. Clim Res. 43:135–147.
- Carabaño MJ, Ramón M, Menéndez-Buxadera A, Molina A, Díaz C. 2019. Selecting for heat tolerance. Anim Front. 9:62–68.
- Carvalho EB, Gionbelli MP, Rodrigues RTS, Bonilha SFM, Newbold CJ, Guimarães SEF, Silva W, Verardo LL, Silva FF, Detmann E, et al. 2019. Differentially expressed mRNAs, proteins and miRNAs associated to energy metabolism in skeletal muscle of beef cattle identified for low and high residual feed intake. BMC Genomics. 20:501.
- Cassandro M. 2013a. Comparing local and commercial cattle breeds on added values for milk production and the predicted methane emissions. Anim Genet Resour. 53:129–134.
- Cassandro M. 2020. Animal breeding and climate change, mitigation and adaptation. J Anim Breed Genet. 137:121–122.
- Cassandro M, Mele M, Stefanon B. 2013b. Genetic aspects of enteric methane emission in livestock ruminants. Ital J Anim Sci. 12:450–458.
- Chevin LM, Hoffmann AA. 2017. Evolution of phenotypic plasticity in extreme environments. Phil Trans R Soc B. 372:20160138.
- Cipollini D. 2004. Stretching the limits of plasticity: can a plant defend against both competitors and herbivores? Ecology. 85:28–37.
- Collier RJ, Baumgard LH, Zimbelman RB, Xiao Y. 2019. Heat stress: physiology of acclimation and adaptation. Anim Front. 9:12–19.
- Collier RJ, Collier JL, Rhoads RP, Baumgard LH. 2008. Invited review: genes involved in the bovine heat stress response. J Dairy Sci. 91:445–454.
- Das R, Gupta ID, Verma A, Singh A, Chaudhari MV, Sailo L, Upadhyay RC, Goswami J. 2015. Genetic polymorphisms in ATP1A1 gene and their association with heat tolerance in Jersey crossbred cows. Indian J Dairy Sci. 68:50–54.
- De Haas Y, Windig JJ, Calus MPL, Dijkstra J, De Haan M, Bannink A, Veerkamp RF. 2011. Genetic parameters for predicted methane production and potential for reducing enteric emissions through genomic selection. J Dairy Sci. 94:6122–6134.
- De Jong G, Bijma P. 2002. Selection and phenotypic plasticity in evolutionary biology and animal breeding. Livest Prod Sci. 78:195–214.
- Des Marais DL, Juenger TE. 2010. Pleiotropy, plasticity, and the evolution of plant abiotic stress tolerance. Ann N Y Acad Sci. 1206:56–79.
- DeWitt TJ, Sih A, Wilson DS. 1998. Costs and limits of phenotypic plasticity. Trends Ecol Evol (Amst). 13:77–81.
- Difford GF, Løvendahl P, Veerkamp RF, Bovenhuis H, Visker MHPW, Lassen J, De Haas Y. 2020. Can greenhouse gases in breath be used to genetically improve feed efficiency of dairy cows? J Dairy Sci. 103:2442–2459.
- Dikmen S, Cole JB, Null DJ, Hansen PJ. 2012. Heritability of rectal temperature and genetic correlations with production and reproduction traits in dairy cattle. J Dairy Sci. 95:3401–3405.
- Dikmen S, Khan FA, Huson HJ, Sonstegard TS, Moss JI, Dahl GE, Hansen PJ. 2014. The SLICK hair locus derived from Senepol cattle confers thermotolerance to intensively managed lactating Holstein cows. J Dairy Sci. 97:5508–5520.
- Drucker AG, Hiemstra SJ, Louwaars N, Oldenbroek JK, Tvedt MW, Hoffmann I, Awgichew K, Abegaz Kehedé S, Bhat PN, da Silva Manante A. 2007. Back to the future. How scenarios of future globalisation, biotechnology, disease and climate change can inform present animal genetic resources policy development. Anim Genet Resour Inf. 41:75–89.
- Dunshea FR, Leury BJ, Fahri F, DiGiacomo K, Hung A, Chauhan S, Clarke IJ, Collier R, Little S, Baumgard L, et al. 2013. Amelioration of thermal stress impacts in dairy cows. Anim Prod Sci. 53:965–975.
- Falconer DS. 1952. The problem of environment and selection. Am Nat. 86:293–298.
- Flori L, Fritz S, Jaffrézic F, Boussaha M, Gut I, Heath S, Foulley JL, Gautier M. 2009. The genome response to artificial selection: a case study in dairy cattle. PLoS One. 4:e6595.
- Flori L, Moazami-Goudarzi K, Alary V, Araba A, Boujenane I, Boushaba N, Casabianca F, Casu S, Ciampolini R, Coeur D'Acier A, et al. 2019. A genomic map of climate adaptation in Mediterranean cattle breeds. Mol Ecol. 28:1009–1029.
- Gautier M, Naves M. 2011. Footprints of selection in the ancestral admixture of a New World Creole cattle breed. Mol Ecol. 20:3128–3143.
- Gerber PJ, Steinfeld H, Henderson B, Mottet A, Opio C, Dijkman J, Falcucci A, Tempio G. 2013. Tackling climate change through livestock – a global assessment of emissions and mitigation opportunities. Rome: Food and Agriculture Organization of the United Nations (FAO).
- Gilbert M, Nicolas G, Cinardi G, Van Boeckel TP, Vanwambeke SO, Wint GW, Robinson TP. 2018. Global distribution data for cattle, buffaloes, horses, sheep, goats, pigs, chickens and ducks in 2010. Sci Data. 5:180227.
- Gouveia JJDS, Silva M, Paiva SR, Oliveira SMPD. 2014. Identification of selection signatures in livestock species. Genet Mol Biol. 37:330–342.
- Green RD. 2009. ASAS Centennial Paper: future needs in animal breeding and genetics. J Anim Sci. 87:793–800.
- Grossi G, Goglio P, Vitali A, Williams AG. 2019. Livestock and climate change: impact of livestock on climate and mitigation strategies. Anim Front. 9:69–76.
- Harris GR, Sexton DM, Booth BB, Collins M, Murphy JM. 2013. Probabilistic projections of transient climate change. Clim Dyn. 40:2937–2972.
- Hayes BJ, Bowman PJ, Chamberlain AJ, Savin K, Van Tassell CP, Sonstegard TS, Goddard ME. 2009. A validated genome wide association study to breed cattle adapted to an environment altered by climate change. PLoS One. 4:e6676.
- Hayes BJ, Lien S, Nilsen H, Olsen HG, Berg P, Maceachern S, Potter S, Meuwissen THE. 2008. The origin of selection signatures on bovine chromosome 6. Anim Genet. 39:105–111.
- Hendry AP, Taylor EB. 2004. How much of the variation in adaptive divergence can be explained by gene flow? An evaluation using lake-stream stickleback pairs. Evolution. 58:2319–2331.
- Herd RM, Arthur PF, Donoghue KA, Bird SH, Bird-Gardiner T, Hegarty RS. 2014. Measures of methane production and their phenotypic relationships with dry matter intake, growth, and body composition traits in beef cattle. J Anim Sci. 92:5267–5274.
- Herrero M, Havlik P, Valin H, Notenbaert A, Rufino MC, Thornton PK, Blümmel M, Weiss F, Grace D, Oberstein M. 2013. Biomasse use, production, feed efficiencies, and greenhouse gas emissions from global livestock systems. Proc Natl Acad Sci USA. 110:20888–20893.
- Hill WG, William G, Michael E, Goddard ME, Peter MV. 2008. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 4:e1000008.
- Ho SY, Larson G, Edwards CJ, Heupink TH, Lakin KE, Holland PW, Shapiro B. 2008. Correlating Bayesian date estimates with climatic events and domestication using a bovine case study. Biol Lett. 4:370–374.
- Hoffmann I. 2010. Climate change and the characterization, breeding and conservation of animal genetic resources. Anim Genet. 41:32–46.
- Hollings T, Burgman M, Van Andel M, Gilbert M, Robinson T, Robinson A. 2018. How do you find the green sheep? A critical review of the use of remotely sensed imagery to detect and count animals. Methods Ecol Evol. 9:881–892.
- Houghton JT, Jenkins GJ, Ephraums JJ. 2016. Natural climate solutions. Earth Syst Sci Data. 82:605–649.
- Hume DA, Whitelaw CBA, Archibald AL. 2011. The future of animal production: improving productivity and sustainability. J Agric Sci. 149:9–16.
- Jamrozik J, Schaeffer L. 1997. Estimates of genetic parameters for a test day model with random regressions for yield traits of first lactation Holsteins. J Dairy Sci. 80:762–770.
- Joost S, Bonin A, Bruford MW, Després L, Conord C, Erhardt G, Taberlet P. 2007. A spatial analysis method (SAM) to detect candidate loci for selection: towards a landscape genomics approach to adaptation. Mol Ecol. 16:3955–3969.
- Kahiluoto H, Kaseva J, Balek J, Olesen JE, Ruiz-Ramos M, Gobin A, Kersebaum KC, Takáč J, Ruget F, Ferrise R, et al. 2019. Decline in climate resilience of European wheat. Proc Natl Acad Sci USA. 116:123–128.
- Kelly SA, Panhuis TM, Stoehr AM. 2012. Phenotypic plasticity: molecular mechanisms and adaptive significance. Compr Physiol. 2:1417–1439.
- Kim ES, Elbeltagy AR, Aboul-Naga AM, Rischkowsky B, Sayre B, Mwacharo JM, Rothschild MF. 2016. Multiple genomic signatures of selection in goats and sheep indigenous to a hot arid environment. Heredity (Edinb). 116:255–264.
- Kirkpatrick M, Heckman N. 1989. A quantitative genetic model for growth, shape, reaction norms, and other infinite-dimensional characters. J Math Biol. 27:429–450.
- Knapp JR, Laur GL, Vadas PA, Weiss WP, Tricarico JM. 2014. Invited review: enteric methane in dairy cattle production: quantifying the opportunities and impact of reducing emissions. J Dairy Sci. 97:3231–3261.
- Knaust J, Hadlich F, Weikard R, Kuehn C. 2016. Epistatic interactions between at least three loci determine the “rat-tail” phenotype in cattle. Genet Sel Evol. 48:26
- Krašovec R, Belavkin RV, Aston JAD, Channon A, Aston E, Rash BM, Kadirvel M, Forbes S, Knight CG. 2014. Mutation rate plasticity in rifampicin resistance depends on Escherichia coli cell-cell interactions. Nat Commun. 5:3742.
- Kumar A, Ashraf S, Goud TS, Grewal A, Singh SV, Yadav BR, Upadhyay RC. 2015. Expression profiling of major heat shock protein genes during different seasons in cattle (Bos indicus) and buffalo (Bubalus bubalis) under tropical climatic condition. J Therm Biol. 51:55–64.
- Lacetera N. 2019. Impact of climate change on animal health and welfare. Anim Front. 9:26–31.
- Lacetera N, Bernabucci U, Basiricò L, Morera P, Nardone A. 2009. Heat shock impairs DNA synthesis and down-regulates gene expression for leptin and Ob-Rb receptor in concanavalin A-stimulated bovine peripheral blood mononuclear cells. Vet Immunol Immunopathol. 127:190–194.
- Lafuente E, Beldade P. 2019. Genomics of developmental plasticity in animals. Front Genet. 10:720.
- Larsen TS, Per H. 2008. Single-sided natural ventilation driven by wind pressure and temperature difference. Energ Buildings. 40:1031–1040.
- Li R, Li C, Chen H, Li R, Chong Q, Xiao H, Chen S. 2020. Genome‐wide scan of selection signatures in Dehong humped cattle for heat tolerance and disease resistance. Anim Genet. 51:292–299.
- Lipper L, Thornton P, Campbell BM, Baedeker T, Braimoh A, Bwalya M, Caron P, Cattaneo A, Garrity D, Henry K, et al. 2014. Climate-smart agriculture for food security. Nature Clim Change. 4:1068–1072.
- Liu MF, Goonewardene LA, Bailey DRC, Basarab JA, Kemp RA, Arthur PF, Okine EK, Makarechian M. 2000. A study on the variation of feed efficiency in station tested beef bulls. Can J Anim Sci. 80:435–441.
- Liu Y, Qin X, Song X-ZH, Jiang H, Shen Y, Durbin KJ, Lien S, Kent MP, Sodeland M, Ren Y, et al. 2009. Bos taurus genome assembly. BMC Genomics. 10:180.
- Lu D, Miller S, Sargolzaei M, Kelly M, Vander Voort G, Caldwell T, Wang Z, Plastow G, Moore S. 2013. Genome-wide association analyses for growth and feed efficiency traits in beef cattle. J Anim Sci. 91:3612–3633.
- Maibam U, Hooda OK, Sharma PS, Mohanty AK, Singh SV, Upadhyay RC. 2017. Expression of HSP70 genes in skin of zebu (Tharparkar) and crossbred (Karan Fries) cattle during different seasons under tropical climatic conditions. J Therm Biol. 63:58–64.
- Marai IFM, El- Darawany AA, Fadiel A, Abdel-Hafez MAM. 2007. Physiological traits as affected by heat stress in sheep. Small Rumin Res. 71:1–12.
- Mateescu RG, Sarlo-Davila KM, Dikmen S, Rodriguez E, Oltenacu PA. 2020. The effect of Brahman genes on body temperature plasticity of heifers on pasture under heat stress. J Anim Sci. 98:126.
- Mathias E, Mundy P. 2005. Herd movements: the exchange of livestock breeds and genes between North and South. Ober-Ramstadt (Germany): League for Pastoral Peoples and Endogenous Livestock Development.
- McManus C, Louvandini H, Gugel R, Sasaki LCB, Bianchini E, Bernal FEM, Paiva SR, Paim TP. 2011. Skin and coat traits in sheep in Brazil and their relation with heat tolerance. Trop Anim Health Prod. 43:121–126.
- Mehla K, Magotra A, Choudhary J, Singh AK, Mohanty AK, Upadhyay RC, Srinivasan S, Gupta P, Choudhary N, Antony B, et al. 2014. Genome-wide analysis of the heat stress response in Zebu (Sahiwal) cattle. Gene. 533:500–507.
- Merilä J, Hendry F, Andrew P. 2014. Climate change, adaptation, and phenotypic plasticity: the problem and the evidence. Evol Appl. 7:1–14.
- Misztal I. 2017. Breeding and Genetics Symposium: resilience and lessons from studies in genetics of heat stress. J Anim Sci. 95:1780–1787.
- Mohn F, Dirk S. 2009. Genetics and epigenetics: stability and plasticity during cellular differentiation. Trends Genet. 25:129–136.
- Muchenje V, Mukumbo FE, Njisane YZ. 2018. Meat in a sustainable food system. SA J an Sci. 48:818–828.
- Murren CJ, Auld JR, Callahan H, Ghalambor CK, Handelsman CA, Heskel MA, Kingsolver JG, Maclean HJ, Masel J, Maughan H, et al. 2015. Constraints on the evolution of phenotypic plasticity: limits and costs of phenotype and plasticity. Heredity (Edinb). 115:293–301.
- Nardone A, Ronchi B, Lacetera N, Ranier MS, Bernabucci U. 2010. Effects of climate changes on animal production and sustainability of livestock systems. Livest Sci. 130:57–69.
- Ng CS, Wu P, Foley J, Foley A, McDonald ML, Juan WT, Huang CJ, Lai YT, Lo WS, Chen CF, et al. 2012. The chicken frizzle feather is due to an α-keratin (KRT75) mutation that causes a defective rachis. PLoS Genet. 8:e1002748.
- Nielsen R. 2001. Statistical tests of selective neutrality in the age of genomics. Heredity (Edinb). 86:641–647.
- Noguera JL, Rodríguez C, Varona L, Tomàs A, Muñoz G, Ramírez O, Barragàn C, Arqué M, Bidanel JP, Amills M, et al. 2009. A bi-dimensional genome scan for prolificacy traits in pigs shows the existence of multiple epistatic QTL. BMC Genomics. 10:636.
- O’Brien D, Herron J, Andurand J, Caré S, Martinez P, Migliorati L, Moro M, Pirlo G, Dollé JB. 2020. LIFE BEEF CARBON: a common framework for quantifying grass and corn-based beef farms’ carbon footprints. Animal. 14:834–845.
- Olson TA, Cc C, Jr Lucena C, Godoy E, Zuniga A, Collier RJ. 2006. Effect of hair characteristics on the adaptation of cattle to warm climates. Proceedings of the 8th World Congress on Genetics Applied to Livestock Production; 2006 Aug 13–18; Belo Horizonte, Minas Gerais, Brasil. Instituto Prociência; p. 16.07.
- Olson TA, Lucena C, Chase CC, Jr, Hammond AC. 2003. Evidence of a major gene influencing hair legth and heat tolerance in Bos taurus cattle. J Anim Sci. 81:80–90.
- Oluwatayo IB, Oluwatayo TB. 2018. Small ruminants as a source of financial security among women in rural southwest Nigeria. jard. 49:333–341.
- Onzima RB, Upadhyay MR, Doekes HP, Brito LF, Bosse M, Kanis E, Groenen MAM, Crooijmans RPMA. 2018. Genome-wide characterization of selection signatures and runs of homozygosity in Ugandan goat breeds. Front Genet. 9:318.
- Opio C, Gerber P, Mottet A, Falcucci A, Tempio G, MacLeod M, Vellinga T, Henderson B, Steinfeld H. 2013. Greenhouse gas emissions from ruminant supply chains–a global life cycle assessment. Rome (Italy): Food and Agriculture Organization (FAO).
- Osei-Amponsah R, Chauhan SS, Leury BJ, Cheng L, Cullen B, Clarke IJ, Dunshea FR. 2019. Genetic selection for thermotolerance in ruminants. Anim. 9:948.
- Özkan Ş, Vitali A, Lacetera N, Amon B, Bannink A, Bartley DJ, Blanco-Penedo I, De Haas Y, Dufrasne I, Elliott J, et al. 2016. Challenges and priorities for modelling livestock health and pathogens in the context of climate change. Environ Res. 151:130–144.
- Pachauri RK, Allen MR, Barros VR, Broome J, Cramer W, Christ R, Church JA, Clarke L, Dahe Q, Dasgupta P. 2014. Climate change 2014: synthesis report. In: Pachauri R, Meyer L, editors. Contribution of Working Groups I, II and III to the fifth assessment report of the Intergovernmental Panel on Climate Change. Geneva (Switzerland): IPCC; p. 151.
- Pasquì M, Di Giuseppe E. 2019. Climate change, future warming, and adaptation in Europe. Anim Front. 9:6–11.
- Pigliucci M. 2005. Evolution of phenotypic plasticity: where are we going now? Trends Ecol Evol (Amst). 20:481–486.
- Pigliucci M, Murren CJ, Schlichting CD. 2006. Phenotypic plasticity and evolution by genetic assimilation. J Exp Biol. 209:2362–2367.
- Pirlo G, Caré S. 2013. A simplified tool for estimating carbon footprint of dairy cattle milk. Ital J Anim Sci. 12:14–81.
- Price TD, Qvarnström A, Irwin DE. 2003. The role of phenotypic plasticity in driving genetic evolution. Proc R Soc Lond B. 270:1433–1440.
- Remold SK, Lenski RE. 2004. Pervasive joint influence of epistasis and plasticity on mutational effects in Escherichia coli. Nat Genet. 36:423–426.
- Rojas-Downing MM, Pouyan NA, Timothy H, Sean AW. 2017. Climate change and livestock: impacts, adaptation, and mitigation. Clim Risk Manag. 16:145–163.
- Rust JM. 2019. The impact of climate change on extensive and intensive livestock production systems. Anim Front. 9:20–25.
- Sailo L, Gupta ID, Verma A, Das R, Chaudhari MV. 2015. Association of single nucleotide polymorphism of Hsp90ab1 gene with thermotolerance and milk yield in Sahiwal cows. Afr J Biochem Res. 9:99–103.
- Sartori R, Sartor-Bergfelt R, Mertens SA, Guenther JN, Parrish JJ, Wiltbank MC. 2002. Fertilization and early embryonic development in heifers and lactating cows in summer and lactating and dry cows in winter. J Dairy Sci. 85:2803–2812.
- Sato M, Stryker MP. 2008. Distinctive features of adult ocular dominance plasticity. J Neurosci. 28:10278–10286.
- Scharlemann JP, Benz D, Hay SI, Purse BV, Tatem AJ, Wint GW, Rogers DJ. 2008. Global data for ecology and epidemiology: a novel algorithm for temporal Fourier processing MODIS data. PLoS One. 3:e1408.
- Schefers JM, Weigel KA. 2012. Genomic selection in dairy cattle: integration of DNA testing into breeding programs. Anim Front. 2:4–9.
- Scheiner SM. 1993. Genetics and evolution of phenotypic plasticity. Annu Rev Ecol Syst. 24:35–68.
- Scheiner SM, Barfield M, Holt RD. 2017. The genetics of phenotypic plasticity. XV. Genetic assimilation, the Baldwin effect, and evolutionary rescue. Ecol Evol. 7:8788–8803.
- Schlötterer C. 2003. Hitchhiking mapping–functional genomics from the population genetics perspective. Trends Genet. 19:32–38.
- Sejian V, Bagath M, Krishnan G, Rashamol VP, Pragna P, Devaraj C, Bhatta R. 2019. Genes for resilience to heat stress in small ruminants: a review. Small Rumin Res. 173:42–53.
- Shahzad K, Akbar H, Vailati-Riboni M, Basiricò L, Morera P, Rodriguez-Zas SL, Nardone A, Bernabucci U, Loor JJ. 2015. The effect of calving in the summer on the hepatic transcriptome of Holstein cows during the peripartal period. J Dairy Sci. 98:5401–5413.
- Sigdel A, Liu L, Abdollahi‐Arpanahi R, Aguilar I, Peñagaricano F. 2020. Genetic dissection of reproductive performance of dairy cows under heat stress. Anim Genet. 51:511–520.
- Snell-Rood EC. 2013. In overview of the evolutionary causes and consequences of behavioural plasticity. Anim Behav. 85:1004–1011.
- Solomon S, Plattner GK, Knutti R, Friedlingstein P. 2009. Irreversible climate change due to carbon dioxide emissions. Proc Natl Acad Sci USA. 106:1704–1709.
- Steinfeld H, Wassenaar T, Jutzi S. 2006. Livestock production systems in developing countries: status, drivers, trends. Rev Off Int Epizoot. 25:505–516.
- Stocker TF, Qin D, Plattner GK, Tignor M, Allen SK, Boschung J, Nauels A, Xia Y, Bex V, Midgley PM. 2013. Climate change 2013: the physical science basis. In: Stocker TF, Qin D, Plattner GK, Tignor M, Allen SK, Boschung J, Nauels A, Xia Y, Bex V, Midgley PM, editors. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge (UK): Cambridge University Press; p. 1535.
- Swain MR, Vasisht G, Tinetti G. 2008. The presence of methane in the atmosphere of an extrasolar planet. Nature. 452:329–331.
- Tao S, Connor EE, Bubolz JW, Thompson IM, Do Amaral BC, Hayen MJ, Dahl GE. 2013. Short communication: effect of heat stress during the dry period on gene expression in mammary tissue and peripheral blood mononuclear cells. J Dairy Sci. 96:378–383.
- The Bovine Genome Sequencing and Analysis Consortium. 2009. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Sci. 324:522–528.
- The Bovine HapMap Consortium. 2009. Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Sci. 324:528–532.
- Thornton PK, Herrero M, Freeman HA, Okeyo AM, Rege E, Jones PG, McDermott J. 2007. Vulnerability, climate change and livestock-opportunities and challenges for the poor. J SAT Agric Res. 4:1–23.
- Tian J, Zhao Z, Zhang L, Zhang Q, Yu Z, Li J, Yang R. 2013. Association of the leptin gene E2-169T>C and E3-299T>A mutations with carcass and meat quality traits of the Chinese Simmental-cross steers. Gene. 518:443–448.
- Triantaphyllopoulos KA, Ikonomopoulos I, Bannister AJ. 2016. Epigenetics and inheritance of phenotype variation in livestock. Epigenet Chromatin. 9:31.
- Van Vliet MT, Franssen WH, Yearsley JR, Ludwig F, Haddeland I, Lettenmaier DP, Kabat P. 2013. Global river discharge and water temperature under climate change. Global Environ Chang. 23:450–464.
- VandeHaar MJ, Armentano LE, Weigel K, Spurlock DM, Tempelman RJ, Veerkamp R. 2016. Harnessing the genetics of the modern dairy cow to continue improvements in feed efficiency. J Dairy Sci. 99:4941–4954.
- Wess R. 2003. Geocentric ecocriticism. Interdiscip Stud Lit Environ. 10:1–19.
- West JW. 2003. Effects of heat-stress on production in dairy cattle. J Dairy Sci. 86:2131–2144.
- Willems OW, Miller SP, Wood BJ. 2013. Aspects of selection for feed efficiency in meat producing poultry. Worlds Poult Sci J. 69:77–88.
- Wilson AJ, Kruuk LEB, Coltman DW. 2005. Ontogenetic patterns in heritable variation for body size: using random regression models in a wild ungulate population. Am Nat. 166:E177–E192.
- Windig JJ, Calus MPL, Veerkamp RF. 2005. Influence of herd environment on health and fertility and their relationship with milk production. J Dairy Sci. 88:335–347.
- You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al. 2015. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 18:603–610.
- Yurchenko AA, Deniskova TE, Yudin NS, Dotsev AV, Khamiruev TN, Selionova MI, Egorov SV, Reyer H, Wimmers K, Brem G, et al. 2019. High-density genotyping reveals signatures of selection related to acclimation and economically important traits in 15 local sheep breeds from Russia. BMC Genomics. 20:294.
