 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Staphylococcus aureus is a gram-positive bacterium, commonly found in the nostrils, on the skin and on the hair of warm-blooded animals, including humans. It can produce a wide variety of virulence factors, including staphylococcal enterotoxins (SEs). In literature, 24 different SEs and many variants have been identified; among these, only five (the so-called classic enterotoxins) have been well-defined. Due to their emetic activity, SEs are frequently responsible for staphylococcal food poisoning, when consumers ingest contaminated food. SEs are proteins with a high tolerance of denaturing and can maintain their activity, even when the vegetative form of the bacteria is inactivated during food processing. The enterotoxin encoding genes are found in a variety of different genetic elements and, as a result, enterotoxin production varies widely between different populations of S. aureus. SEs production is modulated by multiple, and often overlapping, regulatory pathways, which are influenced by environmental factors. Furthermore, complex food matrices possess many characteristics (storage temperature, pH, sugar or salt concentration, presence of competitive microorganisms, etc.) that have a high impact on S. aureus behaviour. The multiple factors influencing S. aureus growth in food matrices and the production of SE complicates risk assessment procedures. In this review, we focus on enterotoxin production by S. aureus in food of animal origin, its regulation and detection and on the most recent developments in predictive microbiology and risk assessment models.
Staphylococcus aureus produces several virulence factors that contribute to the pathogenesis of several serious human diseases: among these staphylococcal enterotoxins (SEs) have emetic activity, and are responsible for staphylococcal food poisoning (SFP).
SEs are proteins that maintain their activity even though the vegetative form of the bacteria is inactivated during food processing.
Enterotoxin encoding genes are found in a variety of genetic elements and enterotoxin production varies and is regulated by multiple regulatory pathways, which are influenced by environmental factors.
Highlights
Introduction
Bacterial toxins are considered the third most significant cause of food-borne outbreaks worldwide. In 2015, over half of all the food-borne outbreaks (n = 434) associated with bacterial toxins were caused by staphylococcal enterotoxins (SEs; EFSA Citation2016). In the annual summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks, the European Food Safety Authority (EFSA) reported that in 2017, approximately 1800 samples (cheeses and dairy products) were tested within the context of national monitoring and surveillance and/or surveys by Cyprus, the Czech Republic, Italy, Romania and Spain. In total, 23 samples tested positive (1.2%), mainly from Italy (cheese and pasteurised milk) and Spain (milk). Data on SEs in other food was submitted by five member states. Out of the 645 samples tested, 40 were positive. These included two samples from Bulgaria (potato-based dishes and ready-to-eat (RTE) pig meat), seven from Italy (meat preparation, other processed food), 12 from Slovakia (from sandwiches, RTE food, confectionery products and frozen desserts) and 19 from Spain (in bakery products, sauces and dressings, meat-based dishes, vegetable-based dishes, RTE salads, other prepared dishes). Suspect samples were collected by Ireland (ice-cream, pre-cut vegetables, prepared dishes) and Hungary (noodles and fermented sausages; EFSA Citation2018). The Centres for Disease Control in the United States estimated that 240,000 cases of staphylococcal food poisoning (SFP) occur each year, leading to 1000 hospitalisations and six fatalities (Scallan et al. Citation2011). Food contamination often occurs either as a direct result of the presence of the bacteria in food producing animals, or is due to poor hygiene during the production, storage or sale of food products (Aydin et al. Citation2011; Kotzekidou Citation2013). Humans and livestock are considered major reservoirs for the transmission of enterotoxin-producing S. aureus (Fisher et al. Citation2018). The severity of the illness depends on the amount of toxin ingested and the general health of the consumer. Patients report symptoms, e.g. nausea and vomiting, shortly after the oral intake of the toxins, often accompanied by watery diarrhoea, prostration and fever. Symptoms usually subside within 24 hours, but in higher risk categories, e.g. children or the elderly, a fatality rate of 4.4% was reported (Doyle and Beuchat Citation2007). The amount of enterotoxin required to cause the illness can be as little as 20–100 ng (Asao et al. Citation2003). Of 24 SEs identified and reported in literature, only five (SEA, SEB, SEC, SED and SEE, the so-called classic enterotoxins) have been well-defined, and are detectable using commercially available assays or in-house developed methods (Nia et al. Citation2016). SEs are highly stable proteins, resistant to the environmental conditions that can easily inactivate the S. aureus vegetative form. SEs are stable at high temperatures: e.g. they are not completely destroyed during pasteurisation (15 s at 72 °C) and crude enterotoxin A remains active at 100 °C for 2 h in broth and at 121 °C for 28 min in mushrooms (Hennekinne et al. Citation2012), and high or low pH. They are also resistant to proteolytic enzymes; hence they retain their activity in the digestive tract after ingestion (Argudín et al. Citation2010). As a result of these characteristics, SEs are considered a serious threat to public health and the notification of food poisoning outbreaks caused by them has been mandatory since 2005 [Commission Regulation (EC) No. 1441/2007 of 5 December 2007].
The prevention of the growth of S. aureus and consequent enterotoxin production is of major importance to minimise the risk of food-poisoning. However, studies have demonstrated that assessing the SFP risk by in vitro models (liquid cultures with bacteria in a planktonic state) or based solely on the count of colony-forming units is unreliable (Zeaki et al. Citation2014). In fact, food-related factors (temperature, pH, sugar or salt concentration, presence of competitive microorganisms, etc.) have a high impact on the growth rate, lag phase duration and virulence of the gene expression of S. aureus (Schelin et al. Citation2011). Understanding the behaviour of enterotoxin producing S. aureus in authentic, complex food matrices and the effect of stressor factors on enterotoxin production is crucial for improving risk assessment models, consumer safety and food product quality. In this review, we aim to present and discuss the current knowledge along with novel findings regarding the growth and production of enterotoxins by S. aureus in foods and the advances made in risk assessment.
The organism and enterotoxins
S. aureus is a Gram-positive coccus, occurring in single, paired or grape-like clusters (staphylé means grape in Greek). Due to the production of a carotenoid pigment, colonies of these bacteria appear gold in colour, giving rise to the species name aureus (meaning golden). They are facultative, anaerobic, non-motile, non-sporing, catalase and coagulase positive bacteria (Willey and Prescott Citation2008). S. aureus can be found in the nostrils, on the skin and on the hair of warm-blooded animals, including humans (Le Loir et al. Citation2003). It is able to grow in a wide range of temperatures (7–48.5 °C with optimum 30–37 °C), pH (from 4.2 to 9.3 with an optimum of 7 to 7.5) and sodium chloride concentrations (NaCl concentrations up to 15%; Schmitt et al. Citation1990). These characteristics, combined with its particular ecological niche, can easily explain the relevance of these bacteria for the safety of foods, and, more specifically, those that require manipulation during processing. S. aureus can produce many virulence factors, e.g. toxins, invasiveness and antibiotic resistance, and can produce a wide variety of symptoms ranging from furuncles to toxic shock syndrome, based on different combinations of those virulence factors. On the other hand, SFP relies on one single type of virulence factor, the SEs.
In literature, 24 different SEs and many variants have been identified, based on their antigenicity, and named from SEA to SElY in chronological order of discovery (Ono et al. Citation2015; Denayer et al. Citation2017; Table ). This superfamily of proteins shares many common characteristics: they are non-glycosylated, single-chain proteins with a homologous, globular structure and a low molecular weight (19–29 kDa; Thomas et al. Citation2006). They can be divided into two groups: the true SEs, which comprise the toxins with demonstrated emetic potential in monkeys, and the enterotoxin-like toxins (SEl-), which comprise those lacking emetic activity or which have not been tested in non-human primates (Fisher et al. Citation2018). Based on their nucleotide and amino acid sequences, SEs and SEls can be divided into several groups: the SEA group (SEA, SED, SEE, SElJ, SEH, SEN, SEO, SEP, SES), the SEB group (SEB, SECs, SEG, SER, SElU, SElW, previously known as SElU2), the SEI group (SEI, SEK, SEL, SEQ, SEM, SElV) and the SElX group (TSST-1, SET, SElX, SElY and members of another group of staphylococcal exotoxins, called superantigen-like (SSL) toxins; Ono et al. Citation2015).
Table 1. Characteristics of staphylococcal enterotoxins.
Most of the enterotoxin genes are found, alone or in groups, on a wide variety of mobile segments of DNA (mobile genetic elements, MGEs), including prophages, plasmids, transposons, S. aureus pathogenicity islands (SaPIs) and enterotoxin gene clusters (egc; Malachowa and DeLeo Citation2010). For example, among the classic SEs, sea is carried by a family of temperate phages (Wallin-Carlquist et al. Citation2010), seb and sec are located on SaPIs (Novick et al. Citation2001; Novick Citation2003), sed is carried by a plasmid and see is carried by a defective phage (Le Loir et al. Citation2003). The egc contains an operon of genes encoding SEG, SEI, SEM, SEN, SEO and two pseudogenes, φent1 and φent2 (Jarraud et al. Citation2001). The deletion, duplication and recombination in this tract of genomes often lead to the generation of new types of enterotoxins and variants (Thomas et al. Citation2006). Two toxin genes, selx (Wilson et al. Citation2011) and sely (Ono et al. Citation2015) are found exclusively on the genome. Considering the wide spectrum of different genetic bases for the production of enterotoxins, it is easy to understand that S. aureus isolates vary considerably: 80% of isolates carry an average of 5–6 SE genes (Umeda et al. Citation2017).
The regulation of enterotoxin production
Current knowledge of the regulation of enterotoxin production is still incomplete: whereas it is known that S. aureus is able to respond to environmental changes, using a combination of quorum-sensing and at least another 16 two-component systems and many trans-acting regulatory proteins (Haag and Bagnoli Citation2016). All those systems allow the bacteria to quickly adapt to stress factors by regulating the expression of genes associated with important physiological features, including enterotoxin production. Each system can directly or indirectly control the transcription of specific sets of genes and a single gene can be influenced by multiple systems, leading to many layers of regulation (Fisher et al. Citation2018; Figure ).
Figure 1. Direct and potential regulatory pathways of the regulation of SEs transcription. The individual regulators and pathways are described in the text. AIP, autoinducing peptide; agr, accessory gene regulator; SaPI, S. aureus pathogenicity island.
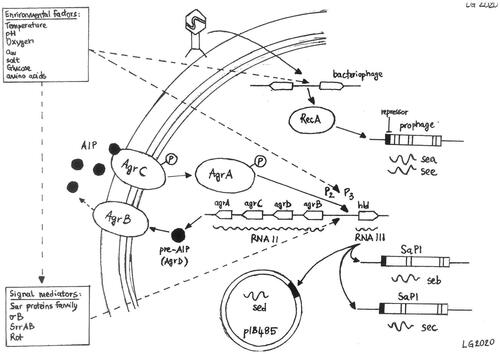
Regulation of classic enterotoxins (SEA-SEE)
The production of bacteriophage-associated enterotoxins (SEA and SEE) is generally constitutive, even though strains with distinct low and high levels of SEA production have been described in literature (Thomas et al. Citation2007; Wallin-Carlquist et al. Citation2010). The sea gene is carried by a polymorphic family of temperate bacteriophages inserted in the bacterial genome, which usually behave as prophages (Betley and Mekalanos Citation1985). It has been demonstrated that the transcription of sea is tied to the life-cycle of the encoding prophage (Cao et al. Citation2012) and is inducible by bacterial stress (Zeaki et al. Citation2015b). Six different sea-carrying prophages (φ252B, φMu3, φMu50A, φNM3, φSa3ms and φSa3mw) have been identified and they all often possess the genes for enterotoxin A, staphylokinase and the complement inhibitor (Goerke et al. Citation2009). The different prophages lead to different levels of SEA production (Borst and Betley Citation1994). The analysis of the nucleotide sequence and of the neighbouring regions has indicated that SEA-producing strains can be divided into two major groups, SEA1 and SEA2 (Wallin-Carlquist et al. Citation2010) and that there are two promoter regions (P1 and P2). P1 is found immediately upstream of sea in both groups (Borst and Betley Citation1994), whereas the phage-related, latent promoter P2 appears to be linked to a stress-induced boost in SEA production (Sumby and Waldor Citation2003). The see gene is, on the contrary, situated on a defective prophage and its expression does not appear to be linked to bacterial growth (Derzelle et al. Citation2009).
Despite the different genetic backbones, the production of the three non-phage-associated, classic enterotoxins (SEB, SEC and SED) usually occurs between the exponential and the stationary phase of bacterial growth (Zhang and Stewart Citation2000) and is regulated by the accessory gene regulator (Agr) system (Betley and Mekalanos Citation1985; Regassa et al. Citation1991; Derzelle et al. Citation2009). The seb gene is found on the SaPI3 pathogenicity island of S. aureus (Novick et al. Citation2001), whereas sec exists in multiple variants (C1, C3, Cbov), found on different pathogenicity islands (SaPI4, SaPIn1/m1 and SaPIbov, respectively; Novick Citation2003). The sed gene, on the other hand, is situated on a 27.6 kb penicillinase plasmid (pIB485; Bayles and Iandolo Citation1989). The Agr system is a quorum sensing system, activated at high cell densities, and consists of two different transcripts: RNAII, which encodes for the structural genes agrA, agrB, agrC and agrD, and RNAIII, a regulatory RNA. These two transcriptional units are driven by the promoters P2 and P3, respectively, and are transcribed in opposite directions (Novick et al. Citation1995). AgrD contains the sequence of the autoinducing peptide (AIP), which is transported out of the cell by AgrB, a membrane-associated export protein (Kavanaugh et al. Citation2007). AIP is the ligand of AgrC, a membrane-bound, histidine kinase, which leads to the phosphorylation of AgrA. The activated form of AgrA up-regulates the promoters P2 and P3, leading to the generation of more RNAII and RNAIII, which is the intracellular effector of gene regulation in S. aureus, resulting in a positive feedback loop (Koenig et al. Citation2004). The expression of agr is modulated by many trans-activating modulators, e.g. the Sar family of proteins, σB, SrrAB (Staphylococcal respiratory response AB) and Rot (repressor of toxins, a member of the Sar family; Pragman et al. Citation2004). When the Agr system is induced, RNAIII mediates the translational repression of rot (Boisset et al. Citation2007). Importantly, all these regulatory elements respond to various environmental stresses and stimuli, e.g. high temperature, alkaline pH, high salinity, catabolites, anaerobic and hypoxic conditions (Mashruwala and Boyd Citation2017). All these mechanisms lead to an increased transcription of secreted virulence factors, such as enterotoxins, and to a reduced transcription of a set of genes encoding for cell wall proteins (Dunman et al. Citation2001). It must be noted that SEB, SEC and SED are only partially regulated by the Agr system and can also be produced independently (Yarwood and Schlievert Citation2003).
Regulation of non-classic enterotoxins
Our knowledge of the regulation of non-classic enterotoxins is only beginning to emerge. It appears that, unlike the majority of classic enterotoxins, regulation of the newer enterotoxins is often Agr-independent, with the exception of SEH and SElJ (Derzelle et al. Citation2009). The production of the enterotoxins encoded in the ecg operon (SEG, SEI, SEM, SEN, SEO and SElU) is highest in the initial stages of the exponential growth phase and is dependent on σB (Kusch et al. Citation2011). On the other hand, the production of SEH is predominant in the late exponential phase of bacterial growth and appears to be positively regulated by Rot, via direct binding to the seh promoter, σB, several Sar homologs and SaeR (Sato'o et al. Citation2015). SaeRS also appears to have a positive impact on the production of SElX (Langley et al. Citation2017).
Impact of food-related factors on enterotoxin production
SFP is usually associated with the consumption of protein-rich, manipulated foods, e.g. meat preparations, creams and dairy products (De Buyser et al. Citation2001) that are vehicles of amino acids and low-molecular-weight peptides that support the survival and growth of S. aureus (Peles et al. Citation2007). All these products possess highly complex matrices in terms of microbial content, storage temperature, pH, salt, oxygen and nutrient availability compared with broth (Valero et al. Citation2009). S. aureus survivability and growth in every environment are linked to the presence of several nutrients (e.g. branched-chain amino acids; Grispoldi et al. Citation2019a). In foods, S. aureus often forms biofilm, where molecular signalling and communication between cells are highly relevant compared to the planktonic state found in laboratory culture broths. All these parameters can influence enterotoxin production by S. aureus in foods.
Temperature appears to affect both prophage-related and non-prophage-related enterotoxin synthesis more than bacterial growth in milk, ham and egg products (Schelin et al. Citation2011). It has been demonstrated that the effect of temperature on SEA production can vary, depending on the prophage variant present. In general, SEA production in foods was detected between 10 °C and 45 °C, with an increase of toxin production linked to an increase in temperature (Tsutsuura et al. Citation2013).
S. aureus appears to have a higher tolerance to pH variation under aerobic compared to anaerobic growth conditions. The pH range for SEs production in foods is 5–9.6, with an optimum between 7 and 8. Lactic acid appears to inhibit the formation of Agr-dependant toxins (SEB, SEC and SED) in particular (Schelin et al. Citation2011). It has been demonstrated that treating pork sausage with lactic acid solution (1–2%) can reduce SEA formation (Zeaki et al. Citation2014), whereas the use of non-dissociated lactic acid was reported to increase SEA production in Brain Heart Infusion broth (Rosengren et al. Citation2013). One study has demonstrated that mild, acetic acid stress (pH 5.5–7) can lead to prophage induction and a consequent increase of sea expression in ham products, whereas no or very low SEA was detected at lower pH levels (4.5–5; Wallin-Carlquist et al. Citation2010). Another study reported little or no effect of sorbic acid (0.15%, pH5) on phage induction (Zeaki et al. Citation2015a). Sihto et al. (Citation2016) reported that lactic acid stress had no significant effect on sed expression.
The lower limit of water activity for the production of enterotoxins in food by S. aureus reported in literature is 0.86. SEB and SEC appear to be the more sensitive to this parameter. On the other hand, the upper limit of NaCl concentration is 12% (Schelin et al. Citation2011). Mild NaCl stress (2%) was able to lead to phage induction and a consequent increase in SEA production in food. Sihto et al. (Citation2017) showed that mild stress conditions, similar to those encountered by S. aureus in many foods (NaCl 4.5%, nitrite 150 mg/L, glucose 30%, pH 6), can strongly influence seb expression, with glucose and NaCl reducing seb promoter activity. The expression of sec was also reported to be influenced by glucose and NaCl stress: a 1.2 M concentration of NaCl led to a 16-fold reduction of both sec mRNA and SEC protein (Regassa and Betley Citation1993). Another study demonstrated that NaCl (4.5%) and glucose stress can decrease sed expression (Sihto et al. Citation2015). It has been reported that nitrites (food additives often used as a preservative and colour fixative in cured meat) have little to no inhibitory effect on the growth and enterotoxin production by S. aureus at the concentration permitted by food regulations (Grispoldi et al. Citation2019c).
The production of SEs is optimal under aerobic conditions. However, it has been demonstrated that 10% dissolved oxygen in vacuum-packed ham and sausages can lead to a 10-fold increase in SEB production (Schelin et al. Citation2011).
The effect of co-culturing of Lactococcus lactis and S. aureus has been investigated in different environments. A mixed culture with L. lactis at a constant pH of 6.6 showed no effect on S. aureus growth, but strongly reduced the expression of sec. The same study reported a slight increase in sea expression during the stationary growth phase. On the other hand, expression of other SEs was impaired (sel) or unaffected (sek, seg, seh; Even et al. Citation2009). Studies have demonstrated the inhibition potential against S. aureus growth and SEs production by selected starter cultures (Lactococcus spp., Lactobacillus spp. and Enterococcus faecalis; De-Xian et al. Citation2020) in Italian dry sausage (Cenci-Goga et al. Citation2012; Cenci-Goga et al. Citation2015; Cenci-Goga et al. Citation2016; Cenci-Goga et al. Citation2018) and fresh cheeses (Nogueira Viçosa et al. Citation2019).
Enterotoxigenic Staphylococcus aureus in farm animals
Most of the data available in literature on the prevalence and characterisation of enterotoxins’ producing S. aureus in farm animals are focussed on milk producing animals and are often associated with the diagnosis of mastitis. S. aureus is a common causative agent of inflammation of the mammary gland, leading to risk of culling or death of the affected animals, increased antimicrobial treatments and severe economic losses for the dairy industries due to decreased milk production and additional costs for medical treatments (Francoz et al. Citation2017). There are many studies that analyse the presence of SEs’ producing S. aureus in milk samples from mastitic cows, ewes and goats around the world. Among the most recent ones, De-Xian et al. (Citation2020) reported the presence of S. aureus in 12% of the milk samples (56/462) obtained from clinical and subclinical mastitic cows in the Lianoning province of China. Twenty-three out of 56 isolates (41%) were found to have one or more genes that encode for enterotoxins. Isolates with seg (27%) were the most prevalent followed by those with sea (16%), seb (7%) and sec (7%). Algammal et al. (Citation2020) reported a prevalence in Ismailia province (Egypt) of S. aureus in milk samples from individual quarters of 36% (53/146) in cattle and 35% (31/88) in buffaloes. Thirty-six % of the isolates were enterotoxigenic, with 27% of them positive for sea gene, 7% positive for sec gene and none positive for seb and sed genes. Fursova et al. (Citation2020) investigated 21 S. aureus isolates from bovine mastitis in seven regions of Western Russia using whole-genome sequence analysis. None of the investigated strains contained genes encoding the classic enterotoxins from sea to see. However, 38% of the isolates were classified as toxinogenic by enterotoxins seg and sei. Genes encoding the SE-like proteins (sel), namely, selm, seln, selo and selv, were also identified in this group. Grispoldi et al. (Citation2019b) analysed S. aureus isolates from 12 cattle dairy farms in central Italy. The isolates presented a prevalence ranging from 47% for see to 6% for seb and sec. Khemiri et al. (Citation2019) reported the presence of S. aureus in 47 out of 150 bovine milk samples and in 9 out of 100 ovine milk samples from nine small family farms distributed in five regions in Tunisia. Sixteen enterotoxin-encoding genes were detected, with a prevalence ranging from 47% for sed to 2% for ser and selp.
Less data is available about enterotoxins producing S. aureus isolated from non-milk samples. Zhou et al. (Citation2017) reported that of 74 nasal swabs of goats collected from 10 herds in the Chongqing municipality (China), 43% were positive for S. aureus. Enterotoxin genes were detected in the majority (53%) of the strains, and sej gene was found in 22% of them, followed by seb, sec and see in 19% isolates each, sei (9%) and seg (3%). In a study published by (Zhang et al. Citation2018), 130 S. aureus isolates from samples associated with pork production were tested for the presence of 18 SEs genes. Ninety-five percent of isolates from different stages of pork meat production harboured one or more SEs genes forming 37 different enterotoxin gene profiles. Seb was present in 60% of the isolates, the highest among the genes tested. The genes sed, sej, seo, sep, ser and seu were not found. The five classical SEs genes had lower prevalence than the egc gene cluster. Velasco et al. (Citation2018) analysed S. aureus isolated from the pork chain supply in Chile. Samples from living animals were taken from the nostrils and the skin. The overall prevalence of S. aureus was 34%. A higher prevalence was detected on carcases (57%), in pigs sampled at farms (41%) than in pigs sampled at slaughterhouses (23%) and in non-packaged retail meat (43%), than packaged retail meat (5%). The presence of SEA, SEB, SEC and SED was determined by agglutination test. Only one strain isolated from a meat sample was positive for SEB.
The wide variety of data that can be observed in different studies reflects the differences in the S. aureus populations around the world. These differences must be taken into account during the development of SFP risk assessment models because the production of SEs can vary significantly.
Prevalence and detection of SEs in food matrices
The most commonly reported food categories involved in SFP outbreaks in the EU are mixed food (29.7%), meat and meat products (20.8%), cheese and dairy products (14.4%), bakery products (8.4%) and fish and fish products (6.5%; Nia et al. Citation2016). Even though EC Regulation 2073/2005 defines a criterion for SEs only for cheese and dairy products, this data highlights the involvement of other food categories, especially mixed food and meat products. The official method in the EU to detect SE types SEA to SEE in all categories of food is the European Screening Method (ESM) developed by the European Union Reference Laboratory for Coagulase Positive Staphylococci (EURL CPS; Ostyn et al. Citation2020). ESM is based on an extraction step with dialysis concentration, followed by qualitative detection using either of two validated commercial assays: immunoenzymatic Vidas® SET2 (bioMérieux, Marcy l’étoile, France) and/or RIDASCREEN® SET Total (R biopharma, Darmstadt, Germany; Hennekinne et al. Citation2007; Ostyn et al. Citation2011). The EURL CPS annually organises Inter-Laboratory Proficiency Testing Trials (ILPT) to evaluate the competence of the European Countries’ National Reference Laboratories to analyse SE content in food matrices. Eight food matrices were used for ILPT over the period 2013–2015, including cheese, freeze-dried cheese, tuna, mackerel, roasted chicken, ready-to-eat food, milk and pastry. Food samples were spiked with four SE types (SEA, SEC, SED and SEE) at various concentrations. The analysis of results obtained by participants for a total of 155 blank and 620 contaminated samples indicated sensitivity (>98%) and specificity (100%) of ESM (Nia et al. Citation2016). These immunological detection methods of the five classic SEs have demonstrated that SEA is the major contributor (almost 80%) to SFP outbreaks, followed by SED, SEB, SEC and SEE (Hu and Nakane Citation2014; Denayer et al. Citation2017; Grispoldi et al. Citation2019b).
There are no immunological detection kits available in commerce for newer SEs, even though a wide variety of diagnostic methods have been developed in order to understand their potential role in SFP. To date, molecular analysis to detect SE encoding genes remains the most common method used: in the last two decades, many studies have reported the presence of non-classic, enterotoxin encoding genes (in particular egc-encoded genes) in SFP outbreaks around the world (Blaiotta et al. Citation2004; Grumann et al. Citation2008; Yan et al. Citation2012; Viçosa et al. Citation2013; Chao et al. Citation2015; Johler et al. Citation2015; Cheng et al. Citation2016; Song et al. Citation2016; Shen et al. Citation2017; Umeda et al. Citation2017). In addition, an increasing number of immunological-assays has been presented in literature for SEG (Nagaraj et al. Citation2016), SEH (Su and Wong Citation1996), SEI (Zhao et al. Citation2016), SEK (Aguilar et al. Citation2014), SEM (Zhao et al. Citation2017) and SEQ (Hu et al. Citation2017). All this data indicates that multiple SEs are most likely present and can contribute to each SFP outbreak. Some authors have suggested that the expansion of the existing multiple assays would be the most efficient strategy to detect all SEs. However, some concerns regarding the high risk of cross-reaction have been raised and vigorous testing is required to develop this idea (Liang et al. Citation2015; Adhikari et al. Citation2016; Nia et al. Citation2016).
Predictive microbiology and risk assessment models
Given the wide variety of SEs and food-related factors that can influence SE production, it is difficult to estimate human exposure to the hazard and evaluate the safety of a food production process. Predictive, microbiological models, if validated in the food under examination, are precious tools to estimate the growth and production of enterotoxins by S. aureus (Castillejo-Rodriguez et al. Citation2002; Ross and McMeekin Citation2003). In literature, kinetic models of S. aureus response to food-related factors can be found. These models predict growth and lag time on the basis of various input values, e.g. temperature, aerobic/anaerobic conditions, water activity, pH and nitrite concentration. They can be built by using free software, including the growth model in the ComBase modelling toolbox and the growth and survival models in the US FDA pathogen modelling programme (PMP; McMeekin et al. Citation2006). In addition, many growth and inactivation curve studies are available online. This data can be useful to design food processes to prevent the growth and production of SEs (Obeso et al. Citation2010; Grispoldi et al. Citation2019c). The main challenges in the development of predictive models are that food contamination is often incidental with a difference of just a few cells, together with a highly variable strain level. Some studies have pointed out highly variable behaviour of S. aureus in foods at low levels of contamination and at single cell level. These differences were masked when studies were performed at population level (Vora et al. Citation2003; Lindqvist Citation2006). This emphasises the usefulness of probabilistic approaches to predictive modelling, using distributions of parameters instead of single, fixed values. On the other hand, there is a lack of predictive models of the kinetics of different SE production and expression of enterotoxin genes in foods. Most of the models that can be found in literature are fitted to the formula by Baranyi and Roberts (Citation1994), which developed a dynamic approach for the prediction of bacterial growth in food, and to the modified equation of Gompertz (Gibson et al. Citation1987). A study by Fujikawa and Morozumi (Citation2006) described an extended logistic model shown by the following equation:
where N is the bacterial population (CFU/g) at time t (h), r is the rate costant of growth (1/h), Nmax is the maximum population (CFU/g) and Nmin the initial populations (CFU/g). Parameters m and n (>0) are related to the curvatures of the deceleration and lag phase of growth, respectively. This model was shown to describe microbial growth with performance very similar to the Baranyi model. The model validation was performed with root mean square error (RMSE). They described a linear increase of SEA production in sterile milk between 15 °C and 32 °C with the following equation:
where p is the amount of SEA (ng/ml h) and t is the temperature (°C). Higher levels of SEA were produced above 32 °C, although correlation was no longer present. The same author (Fujikawa Citation2021) applied the previously described equation to predict the detection time of SE A formed in hydrated batter mix. The results of this study suggested that the kinetic model could well predict the SEA production from the temperature history.
A study by (Soejima et al. Citation2007) developed the following equation to describe the correlation between the toxin production (SEA) and the cell count in milk:
where Tox is the toxin production (log ng/ml) and C is the number of cells (log cfu/ml).
Castillejo-Rodriguez et al. (Citation2002) analysed the growth of S. aureus in commercially available vacuum-packaged cooked ham, turkey breast meat and chicken breast meat at different temperatures (2.3 to 17.7 °C). Growth rates observed were compared with those predicted on the basis of various growth models found in literature by using the mean square error (MSE), the bias factor (Bf) and the accuracy factor (Af). Their results indicate that only two models (Dengremont and Membrè model) accurately reflected S. aureus behaviour while the others overestimated the growth.
Mansur et al. (Citation2016) developed a predictive model for S. aureus growth in raw pork, ham and sausage at isothermal conditions of different temperatures (10–40 °C). Data obtained were fitted to the Baranyi and Roberts model (Baranyi and Roberts Citation1994) and the Ratkowsky’s square root model (Ratkowsky et al. Citation1983) was then used to describe the S. aureus maximum growth rate (µmax, log CFU/h) as a function of temperature with the following equation:
where c is the regression coefficient, T is the temperature (°C) and T0 the theoretical minimum temperature for bacterial growth. The model was then compared with the ComBase predictor. Results showed that this model seems suitable for the prediction.
Gunvig et al. (Citation2018) developed a dynamic mathematical model to predict formation of SE during heating and fermentation of meat products. A total of 78 experiments were carried out in a meat model system, covering a range of different temperatures (10–40 °C), pH (4.6–6.0), water phase salt (WPS; 2.2–5.6%) and sodium nitrate concentrations (0–150 ppm). The system was inoculated with a cocktail consistent of three different SEs producing strains at a concentration of approximately 103 CFU/g. The SEs production and growth models were validated on separate data sets including both static and dynamic conditions. According to authors, the model is dependable and the prediction errors are comparable to laboratory reproducibility. The models are available at www.dmripredict.dk.
Hazard identification in risk assessments varies from S. aureus in general to enterotoxigenic strains and/or enterotoxin. Poor personal hygiene, incorrect food handling practices and inadequate refrigeration of foods are identified as the main factors contributing to SFP. Although it is well known that temperatures below 7 °C and 10 °C can reduce the risk of S. aureus growth and production of enterotoxin, respectively, many foods are stored at higher temperatures for sensory reasons, and many processes include steps under growth permissive conditions (Schelin et al. Citation2011).
The hazard characterisation for a toxigenic microorganism should focus on dose-response relations and threshold models (Buchanan et al. Citation2000; Schelin et al. Citation2011). Due to lack of knowledge, this step for S. aureus is often based on food poisoning symptoms and cell counts in foods used as a proxy for potentially hazardous doses of enterotoxins. For example, (Lindqvist et al. Citation2002) expressed the threshold level as the number of S. aureus cells between 5 and 8 log CFU per g. On the basis of outbreak data, threshold levels of 20 ng (Kim et al. Citation2009), 94 ng (Heidinger et al. Citation2009) and 100 ng (Soejima et al. Citation2007) per serving were used.
Exposure assessment based on measurements of SEs in food has been reported. However, it relies basically on data of S. aureus prevalence, initial level of contamination, growth and inactivation due to cooking or processing the food (Schelin et al. Citation2011). On the other hand, consumer handling and consumption or the exposure of susceptible groups of population have rarely been described.
The main limitation of the risk characterisation step is linked to the complexity of the dynamics that occur between bacterial growth, gene expression and enterotoxin production in foods. As also expressed in this manuscript, the potential uncoupling between cell count and the level of enterotoxin produced should be taken in account.
Conclusion
SFP is a major public health concern worldwide. Consumers’ increasing demand for ready-to-eat foods, together with the high stability and resistance of SEs to the majority of the conditions that inactivate the vegetative form of S. aureus and its wide distribution in animals and humans make SFP a serious threat. The traditional approach is to consider SE production correlated with bacterial growth (more cells equal more toxin), and the colony count is the parameter considered to determine whether or not a food product is safe for human consumption. However, explanations have been given of the way in which different parameters found in foods can influence S. aureus survivability, growth and enterotoxin production and the extent to which the behaviour of these bacteria in actual food environments can differ from what can be observed in liquid cultures. In the light of these discoveries, the importance of developing new risk assessment models for S. aureus based on real foods in order to improve consumer safety and food product quality is clear.
Compliance with ethics requirements
This article does not contain any studies with human or animal subjects.
Author contributions
LG: Writing – original draft; AA: Writing- Reviewing and Editing; BC-G: Conceptualisation, Writing- Reviewing and Editing, Supervision, Project administration, Funding acquisition.
Acknowledgements
The authors express sincere appreciation to members of Polyglot (Perugia, Italy) for a careful reading and comments on the article. The findings and conclusions in this paper are those of the authors and do not necessarily represent the views of the University of Perugia.
Disclosure statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Funding
References
- Adhikari RP, Haudenschild C, Sterba PM, Sahandi S, Enterlein S, Holtsberg FW, Aman MJ. 2016. Development of a novel multiplex electrochemiluminescent-based immunoassay for quantification of human serum IgG against 10 Staphylococcus aureus toxins. J Immunol Methods. 430:33–42.
- Aguilar JL, Varshney AK, Wang X, Stanford L, Scharff M, Fries BC. 2014. Detection and measurement of staphylococcal enterotoxin-like K (SEl-K) secretion by Staphylococcus aureus clinical isolates. J Clin Microbiol. 52(7):2536–2543.
- Algammal AM, Enany ME, El-Tarabili RM, Ghobashy MOI, Helmy YA. 2020. Prevalence, antimicrobial resistance profiles, virulence and enterotoxins-determinant genes of MRSA isolated from subclinical bovine mastitis in Egypt. Pathogens. 9(5):362.
- Argudín MA, Mendoza MC, Rodicio MR. 2010. Food poisoning and Staphylococcus aureus enterotoxins. Toxins (Basel). 2(7):1751–1773.
- Asao T, Kumeda Y, Kawai T, Shibata T, Oda H, Haruki K, Nakazawa H, Kozaki S. 2003. An extensive outbreak of staphylococcal food poisoning due to low-fat milk in Japan: estimation of enterotoxin A in the incriminated milk and powdered skim milk. Epidemiol Infect. 130(1):33–40.
- Aydin A, Muratoglu K, Sudagidan M, Bostan K, Okuklu B, Harsa S. 2011. Prevalence and antibiotic resistance of foodborne Staphylococcus aureus isolates in Turkey. Foodborne Pathog Dis. 8(1):63–69.
- Baranyi J, Roberts TA. 1994. A dynamic approach to predicting bacterial growth in food. Int J Food Microbiol. 23(3–4):277–294.
- Bayles KW, Iandolo JJ. 1989. Genetic and molecular analyses of the gene encoding staphylococcal enterotoxin D. J Bacteriol. 171(9):4799–4806.
- Bergdoll MS, Borja CR, Avena RM. 1965. Identification of a new enterotoxin as enterotoxin C. J Bacteriol. 90(5):1481–1485.
- Bergdoll MS, Borja CR, Robbins RN, Weiss KF. 1971. Identification of enterotoxin E. Infect Immun. 4(5):593–595.
- Betley MJ, Mekalanos JJ. 1985. Staphylococcal enterotoxin A is encoded by phage. Science. 229(4709):185–187.
- Blaiotta G, Ercolini D, Pennacchia C, Fusco V, Casaburi A, Pepe O, Villani F. 2004. PCR detection of staphylococcal enterotoxin genes in Staphylococcus spp. strains isolated from meat and dairy products. Evidence for new variants of seg and sei in S. aureus AB-8802. J Appl Microbiol. 97(4):719–730.
- Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, et al. 2007. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 21(11):1353–1366.
- Borst DW, Betley MJ. 1994. Phage-associated differences in staphylococcal enterotoxin A gene (sea) expression correlate with sea allele class. Infect Immun. 62(1):113–118.
- Buchanan RL, Smith JL, Long W. 2000. Microbial risk assessment: dose-response relations and risk characterization. Int J Food Microbiol. 58(3):159–172.
- Cao R, Zeaki N, Wallin-Carlquist N, Skandamis PN, Schelin J, Rådström P. 2012. Elevated enterotoxin A expression and formation in Staphylococcus aureus and its association with prophage induction. Appl Environ Microbiol. 78(14):4942–4948.
- Castillejo-Rodriguez AM, Gimeno RM, Cosano GZ, Alcala EB, Perez MR. 2002. Assessment of mathematical models for predicting Staphylococcus aureus growth in cooked meat products. J Food Prot. 65(4):659–665.
- Cenci-Goga BT, Karama M, Sechi P, Iulietto MF, Grispoldi L, Selvaggini R, Ceccarelli M, Barbera S. 2018. Fate of selected pathogens in spiked «SALAME NOSTRANO» produced without added nitrates following the application of NONIT™ technology. Meat Sci. 139:247–254.
- Cenci-Goga BT, Karama M, Sechi P, Iulietto MF, Novelli S, Selvaggini R, Barbera S. 2016. Effect of a novel starter culture and specific ripening conditions on microbiological characteristics of nitrate-free dry-cured pork sausages [Article]. Ital J Anim Sci. 15(3):358–374.
- Cenci-Goga BT, Karama M, Sechi P, Iulietto MF, Novelli S, Selvaggini R, Mattei S. 2015. Growth inhibition of selected microorganisms by an association of dairy starter cultures and probiotics [Article]. Ital J Anim Sci. 14(2):3250–3745.
- Cenci-Goga BT, Rossitto PV, Sechi P, Parmegiani S, Cambiotti V, Cullor JS. 2012. Effect of selected dairy starter cultures on microbiological, chemical and sensory characteristics of swine and venison (Dama dama) nitrite-free dry-cured sausages [Article]. Meat Sci. 90(3):599–606.
- Chao G, Bao G, Cao Y, Yan W, Wang Y, Zhang X, Zhou L, Wu Y. 2015. Prevalence and diversity of enterotoxin genes with genetic background of Staphylococcus aureus isolates from different origins in China. Int J Food Microbiol. 211:142–147.
- Cheng J, Wang Y, Cao Y, Yan W, Niu X, Zhou L, Chen J, Sun Y, Li C, Zhang X, et al. 2016. The distribution of 18 enterotoxin and enterotoxin-like genes in Staphylococcus aureus strains from different sources in East China. Foodborne Pathog Dis. 13(4):171–176.
- Couch JL, Soltis MT, Betley MJ. 1988. Cloning and nucleotide sequence of the type E staphylococcal enterotoxin gene. J Bacteriol. 170(7):2954–2960.
- De Buyser ML, Dufour B, Maire M, Lafarge V. 2001. Implication of milk and milk products in food-borne diseases in France and in different industrialised countries. Int J Food Microbiol. 67(1–2):1–17.
- Denayer S, Delbrassinne L, Nia Y, Botteldoorn N. 2017. Food-borne outbreak investigation and molecular typing: high diversity of Staphylococcus aureus strains and importance of toxin detection. Toxins. 9(12):407.
- Derzelle S, Dilasser F, Duquenne M, Deperrois V. 2009. Differential temporal expression of the staphylococcal enterotoxins genes during cells growth. Food Microbiol. 26(8):896–904.
- De-Xian Z, Yao L, Xiao-Qing Y, Hong-Yu S, Wang Q, Ze-Hui Z, Yao-Chuan L, Chun-Lian T, Can-Can C, Ming-Chun L. 2020. In vitro antibiotic susceptibility, virulence genes distribution and biofilm production of Staphylococcus aureus isolates from bovine mastitis in the Liaoning Province of China. Infect Drug Resist. 13:1365–1375.
- Doyle MP, Beuchat LR. 2007. Food microbiology: fundamentals and frontiers. 3rd ed. Washington (DC): ASM Press.
- Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg G, Wu S, Brown EL, Zagursky RJ, Shlaes D, Projan SJ. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol. 183(24):7341–7353.
- EFSA. 2016. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2015. EFSA J. 14(12):4634.
- EFSA. 2018. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2015. EFSA J. 16(12):5500.
- Even S, Charlier C, Nouaille S, Ben Zakour NL, Cretenet M, Cousin FJ, Gautier M, Cocaign-Bousquet M, Loubière P, Le Loir Y. 2009. Staphylococcus aureus virulence expression is impaired by Lactococcus lactis in mixed cultures. Appl Environ Microbiol. 75(13):4459–4472.
- Fisher EL, Otto M, Cheung GYC. 2018. Basis of virulence in enterotoxin-mediated staphylococcal food poisoning. Front Microbiol. 9:436.
- Fitzgerald JR, Monday SR, Foster TJ, Bohach GA, Hartigan PJ, Meaney WJ, Smyth CJ. 2001. Characterization of a putative pathogenicity island from bovine Staphylococcus aureus encoding multiple superantigens. J Bacteriol. 183(1):63–70.
- Francoz D, Wellemans V, Dupré JP, Roy JP, Labelle F, Lacasse P, Dufour S. 2017. Invited review: a systematic review and qualitative analysis of treatments other than conventional antimicrobials for clinical mastitis in dairy cows. J Dairy Sci. 100(10):7751–7770.
- Fujikawa H. 2021. Prediction of detection time of staphylococcal enterotoxin A formed in hydrated batter mix. Food Control. 121:107559.
- Fujikawa H, Morozumi S. 2006. Modeling Staphylococcus aureus growth and enterotoxin production in milk. Food Microbiol. 23(3):260–267.
- Fursova K, Sorokin A, Sokolov S, Dzhelyadin T, Shulcheva I, Shchannikova M, Nikanova D, Artem'eva O, Zinovieva N, Brovko F. 2020. Virulence factors and phylogeny of Staphylococcus aureus associated with bovine mastitis in russia based on genome sequences. Front Vet Sci. 7:135.
- Gibson AM, Bratchell N, Roberts TA. 1987. The effect of sodium chloride and temperature on the rate and extent of growth of Clostridium botulinum type A in pasteurized pork slurry. J Appl Bacteriol. 62(6):479–490.
- Goerke C, Pantucek R, Holtfreter S, Schulte B, Zink M, Grumann D, Bröker BM, Doskar J, Wolz C. 2009. Diversity of prophages in dominant Staphylococcus aureus clonal lineages. J Bacteriol. 191(11):3462–3468.
- Grispoldi L, Karama M, Ianni F, La Mantia A, Pucciarini L, Camaioni E, Sardella R, Sechi P, Natalini B, Cenci-Goga BT. 2019a. The relationship between S. aureus and branched-chain amino acids content in composite cow milk [Article]. Animals. 9(11):981.
- Grispoldi L, Massetti L, Sechi P, Iulietto MF, Ceccarelli M, Karama M, Popescu PA, Pandolfi F, Cenci-Goga BT. 2019b. Short communication: characterization of enterotoxin-producing Staphylococcus aureus isolated from mastitic cows. J Dairy Sci. 102(2):1059–1065.
- Grispoldi L, Popescu PA, Karama M, Gullo V, Poerio G, Borgogni E, Torlai P, Chianese G, Fermani AG, Sechi P, et al. 2019c. Study on the growth and enterotoxin production by Staphylococcus aureus in canned meat before retorting. Toxins. 11(5):291.
- Grumann D, Scharf SS, Holtfreter S, Kohler C, Steil L, Engelmann S, Hecker M, Völker U, Bröker BM. 2008. Immune cell activation by enterotoxin gene cluster (egc)-encoded and non-egc superantigens from Staphylococcus aureus. J Immunol. 181(7):5054–5061.
- Gunvig A, Andresen MS, Jacobsen T, Borggaard C. 2018. Staphtox predictor – a dynamic mathematical model to predict formation of Staphylococcus enterotoxin during heating and fermentation of meat products. Int J Food Microbiol. 285:81–91.
- Haag AF, Bagnoli F. 2016. The role of two-component signal transduction systems in Staphylococcus aureus virulence regulation. Curr Top Microbiol Immunol. 409:145–198.
- Heidinger JC, Winter CK, Cullor JS. 2009. Quantitative microbial risk assessment for Staphylococcus aureus and Staphylococcus enterotoxin A in raw milk. J Food Prot. 72(8):1641–1653.
- Hennekinne J-A, De Buyser M-L, Dragacci S. 2012. Staphylococcus aureus and its food poisoning toxins: characterization and outbreak investigation. FEMS Microbiol Rev. 36(4):815–836.
- Hennekinne JA, Guillier F, Perelle P, de Buyser ML, Dragacci S, Krys S, Lombard B. 2007. Intralaboratory validation according to the EN ISO 16 140 Standard of the Vidas SET2 detection kit for use in official controls of staphylococcal enterotoxins in milk products. J Appl Microbiol. 102(5):1261–1272.
- Hu DL, Nakane A. 2014. Mechanisms of staphylococcal enterotoxin-induced emesis. Eur J Pharmacol. 722:95–107.
- Hu D-L, Omoe K, Shimoda Y, Nakane A, Shinagawa K. 2003. Induction of emetic response to staphylococcal enterotoxins in the house musk shrew (Suncus murinus). Infect Immun. 71(1):567–570.
- Hu D-L, Ono HK, Isayama S, Okada R, Okamura M, Lei LC, Liu ZS, Zhang X-C, Liu MY, Cui JC, et al. 2017. Biological characteristics of staphylococcal enterotoxin Q and its potential risk for food poisoning. J Appl Microbiol. 122(6):1672–1679.
- Igarashi H. 1972. Staphylococcal enterotoxin D. Immunological identification with purified toxin. Jpn J Microbiol. 16(6):483–491.
- Jarraud S, Peyrat MA, Lim A, Tristan A, Bes M, Mougel C, Etienne J, Vandenesch F, Bonneville M, Lina G. 2001. egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J Immunol. 166(1):669–677.
- Johler S, Giannini P, Jermini M, Hummerjohann J, Baumgartner A, Stephan R. 2015. Further evidence for staphylococcal food poisoning outbreaks caused by egc-encoded enterotoxins. Toxins (Basel). 7(3):997–1004.
- Kavanaugh JS, Thoendel M, Horswill AR. 2007. A role for type I signal peptidase in Staphylococcus aureus quorum sensing. Mol Microbiol. 65(3):780–798.
- Khemiri M, Abbassi MS, Elghaieb H, Zouari M, Dhahri R, Pomba C, Hammami S. 2019. High occurrence of enterotoxigenic isolates and low antibiotic resistance rates of Staphylococcus aureus isolated from raw milk from cows and ewes. Lett Appl Microbiol. 68(6):573–579.
- Kim HJ, Griffiths MW, Fazil AM, Lammerding AM. 2009. Probabilistic risk model for staphylococcal intoxication from pork-based food dishes prepared in food service establishments in Korea. J Food Prot. 72(9):1897–1908.
- Koenig RL, Ray JL, Maleki SJ, Smeltzer MS, Hurlburt BK. 2004. Staphylococcus aureus AgrA binding to the RNAIII-agr regulatory region. J Bacteriol. 186(22):7549–7555.
- Kotzekidou P. 2013. Microbiological examination of ready-to-eat foods and ready-to-bake frozen pastries from university canteens. Food Microbiol. 34(2):337–343.
- Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, et al. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet. 357(9264):1225–1240.
- Kusch K, Hanke K, Holtfreter S, Schmudde M, Kohler C, Erck C, Wehland J, Hecker M, Ohlsen K, Bröker B, et al. 2011. The influence of SaeRS and σ(B) on the expression of superantigens in different Staphylococcus aureus isolates. Int J Med Microbiol. 301(6):488–499.
- Langley RJ, Ting YT, Clow F, Young PG, Radcliff FJ, Choi JM, Sequeira RP, Holtfreter S, Baker H, Fraser JD. 2017. Staphylococcal enterotoxin-like X (SElX) is a unique superantigen with functional features of two major families of staphylococcal virulence factors. PLoS Pathog. 13(9):e1006549.
- Le Loir Y, Baron F, Gautier M. 2003. Staphylococcus aureus and food poisoning. Genet Mol Res. 2(1):63–76.
- Letertre C, Perelle S, Dilasser F, Fach P. 2003. Identification of a new putative enterotoxin SEU encoded by the egc cluster of Staphylococcus aureus. J Appl Microbiol. 95(1):38–43.
- Liang M, Zhang T, Liu X, Fan Y, Xia S, Xiang Y, Liu Z, Jinnian L. 2015. Development of an indirect competitive enzyme-linked immunosorbent assay based on the multiepitope peptide for the synchronous detection of staphylococcal enterotoxin A and G proteins in milk. J Food Prot. 78(2):362–369.
- Lindqvist R. 2006. Estimation of Staphylococcus aureus growth parameters from turbidity data: characterization of strain variation and comparison of methods. Appl Environ Microbiol. 72(7):4862–4870.
- Lindqvist R, Sylven S, Vagsholm I. 2002. Quantitative microbial risk assessment exemplified by Staphylococcus aureus in unripened cheese made from raw milk. Int J Food Microbiol. 78(1–2):155–170.
- Malachowa N, DeLeo FR. 2010. Mobile genetic elements of Staphylococcus aureus. Cell Mol Life Sci. 67(18):3057–3071.
- Mansur AR, Park JH, Oh DH. 2016. Predictive model for growth of Staphylococcus aureus on raw pork, ham, and sausage. J Food Prot. 79(1):132–137.
- Mashruwala AA, Boyd JM. 2017. The Staphylococcus aureus SrrAB regulatory system modulates hydrogen peroxide resistance factors, which imparts protection to aconitase during aerobic growth. PLoS One. 12(1):e0170283.
- McMeekin TA, Baranyi J, Bowman J, Dalgaard P, Kirk M, Ross T, Schmid S, Zwietering MH. 2006. Information systems in food safety management. Int J Food Microbiol. 112(3):181–194.
- Munson SH, Tremaine MT, Betley MJ, Welch RA. 1998. Identification and characterization of staphylococcal enterotoxin types G and I from Staphylococcus aureus. Infect Immun. 66(7):3337–3348.
- Nagaraj S, Ramlal S, Kingston J, Batra HV. 2016. Development of IgY based sandwich ELISA for the detection of staphylococcal enterotoxin G (SEG), an egc toxin. Int J Food Microbiol. 237:136–141.
- Nia Y, Mutel I, Assere A, Lombard B, Auvray F, Hennekinne J-A. 2016. Review over a 3-year period of European Union Proficiency Tests for detection of staphylococcal enterotoxins in food matrices. Toxins (Basel). 8(4):107.
- Nogueira Viçosa G, Vieira Botelho C, Botta C, Bertolino M, Fernandes de Carvalho A, Nero LA, Cocolin L. 2019. Impact of co-cultivation with Enterococcus faecalis over growth, enterotoxin production and gene expression of Staphylococcus aureus in broth and fresh cheeses. Int J Food Microbiol. 2:308.
- Novick RP. 2003. Mobile genetic elements and bacterial toxinoses: the superantigen-encoding pathogenicity islands of Staphylococcus aureus. Plasmid. 49(2):93–105.
- Novick RP, Projan SJ, Kornblum J, Ross HF, Ji G, Kreiswirth B, Vandenesch F, Moghazeh S. 1995. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol Gen Genet. 248(4):446–458.
- Novick RP, Schlievert P, Ruzin A. 2001. Pathogenicity and resistance islands of staphylococci. Microbes Infect. 3(7):585–594.
- Obeso JM, Garcia P, Martinez B, Arroyo-Lopez FN, Garrido-Fernandez A, Rodriguez A. 2010. Use of logistic regression for prediction of the fate of Staphylococcus aureus in pasteurized milk in the presence of two lytic phages. Appl Environ Microbiol. 76(18):6038–6046.
- Omoe K, Hu D-L, Ono HK, Shimizu S, Takahashi-Omoe H, Nakane A, Uchiyama T, Shinagawa K, Imanishi K. 2013. Emetic potentials of newly identified staphylococcal enterotoxin-like toxins. Infect Immun. 81(10):3627–3631.
- Omoe K, Hu DL, Takahashi-Omoe H, Nakane A, Shinagawa K. 2003. Identification and characterization of a new staphylococcal enterotoxin-related putative toxin encoded by two kinds of plasmids. Infect Immun. 71(10):6088–6094.
- Omoe K, Imanishi K, Hu D-L, Kato H, Fugane Y, Abe Y, Hamaoka S, Watanabe Y, Nakane A, Uchiyama T, et al. 2005. Characterization of novel staphylococcal enterotoxin-like toxin type P. Infect Immun. 73(9):5540–5546.
- Ono HK, Hirose S, Naito I, Sato'o Y, Asano K, Hu DL, Omoe K, Nakane A. 2017. The emetic activity of staphylococcal enterotoxins, SEK, SEL, SEM, SEN and SEO in a small emetic animal model, the house musk shrew. Microbiol Immunol. 61(1):12–16.
- Ono HK, Omoe K, Imanishi K, Iwakabe Y, Hu DL, Kato H, Saito N, Nakane A, Uchiyama T, Shinagawa K. 2008. Identification and characterization of two novel staphylococcal enterotoxins, types S and T. Infect Immun. 76(11):4999–5005.
- Ono HK, Sato'o Y, Narita K, Naito I, Hirose S, Hisatsune J, Asano K, Hu D-L, Omoe K, Sugai M, et al. 2015. Identification and characterization of a novel staphylococcal emetic toxin. Appl Environ Microbiol. 81(20):7034–7040.
- Ostyn A, Guillier F, Prufer AL, Papinaud I, Messio S, Krys S, Lombard B, Hennekinne JA. 2011. Intra-laboratory validation of the Ridascreen® SET Total kit for detecting staphylococcal enterotoxins SEA to SEE in cheese. Lett Appl Microbiol. 52(5):468–474.
- Ostyn A, Prufer A-L, Papinaud I, Hennekinne JA, Assere A, Lombard B. 2020. European Screening Method of the EURL for CPS including Staphylococcus aureus applicable to the detection of SEs in all types of food matrices including milk and milk products. http://eurl-staphylococci.anses.fr, accessed January 2021
- Peles F, Wagner M, Varga L, Hein I, Rieck P, Gutser K, Keresztúri P, Kardos G, Turcsányi I, Béri B, et al. 2007. Characterization of Staphylococcus aureus strains isolated from bovine milk in Hungary. Int J Food Microbiol. 118(2):186–193.
- Pragman AA, Yarwood JM, Tripp TJ, Schlievert PM. 2004. Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J Bacteriol. 186(8):2430–2438.
- Ratkowsky DA, Lowry RK, McMeekin TA, Stokes AN, Chandler RE. 1983. Model for bacterial culture growth rate throughout the entire biokinetic temperature range. J Bacteriol. 154(3):1222–1226.
- Regassa LB, Betley MJ. 1993. High sodium chloride concentrations inhibit staphylococcal enterotoxin C gene (sec) expression at the level of sec mRNA. Infect Immun. 61(4):1581–1585.
- Regassa LB, Couch JL, Betley MJ. 1991. Steady-state staphylococcal enterotoxin type C mRNA is affected by a product of the accessory gene regulator (agr) and by glucose. Infect Immun. 59(3):955–962.
- Reiser RF, Robbins RN, Noleto AL, Khoe GP, Bergdoll MS. 1984. Identification, purification, and some physicochemical properties of staphylococcal enterotoxin C3. Infect Immun. 45(3):625–630.
- Rosengren A, Lindblad M, Lindqvist R. 2013. The effect of undissociated lactic acid on Staphylococcus aureus growth and enterotoxin A production. Int J Food Microbiol. 162(2):159–166.
- Ross T, McMeekin TA. 2003. Modeling microbial growth within food safety risk assessments. Risk Anal. 23(1):179–197.
- Sato'o Y, Hisatsune J, Nagasako Y, Ono HK, Omoe K, Sugai M. 2015. Positive regulation of staphylococcal enterotoxin H by Rot (Repressor of Toxin) protein and its importance in clonal complex 81 subtype 1 lineage-related food poisoning. Appl Environ Microbiol. 81(22):7782–7790.
- Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States – Major pathogens. Emerg Infect Dis. 17(1):7–15.
- Schelin J, Wallin-Carlquist N, Cohn MT, Lindqvist R, Barker GC, Rådström P. 2011. The formation of Staphylococcus aureus enterotoxin in food environments and advances in risk assessment. Virulence. 2(6):580–592.
- Schlievert PM, Jablonski LM, Roggiani M, Sadler I, Callantine S, Mitchell DT, Ohlendorf DH, Bohach GA. 2000. Pyrogenic toxin superantigen site specificity in toxic shock syndrome and food poisoning in animals. Infect Immun. 68(6):3630–3634.
- Schmitt M, Schuler-Schmid U, Schmidt-Lorenz W. 1990. Temperatures limit of growth, Tnase, and enterotoxin production of Staphylococcus aureus strains isolated from foods. Int J Food Microbiol. 11(1):1–19.
- Shen M, Li Y, Zhang L, Dai S, Wang J, Li Y, Zhang L, Huang J. 2017. Staphylococcus enterotoxin profile of China isolates and the superantigenicity of some novel enterotoxins. Arch Microbiol. 199(5):723–736.
- Sihto HM, Stephan R, Engl C, Chen J, Johler S. 2017. Effect of food-related stress conditions and loss of agr and sigB on seb promoter activity in S. aureus. Food Microbiol. 65:205–212.
- Sihto HM, Tasara T, Stephan R, Johler S. 2015. Temporal expression of the staphylococcal enterotoxin D gene under NaCl stress conditions. FEMS Microbiol Lett. 362:1–7.
- Sihto HM, Tasara T, Stephan R, Johler S. 2016. Growth behavior and temporal enterotoxin D expression of Staphylococcus aureus strains under glucose and lactic acid stress. Food Control. 62:69–73.
- Soejima T, Nagao E, Yano Y, Yamagata H, Kagi H, Shinagawa K. 2007. Risk evaluation for staphylococcal food poisoning in processed milk produced with skim milk powder. Int J Food Microbiol. 115(1):29–34.
- Song M, Shi C, Xu X, Shi X. 2016. Molecular typing and virulence gene profiles of enterotoxin gene cluster (egc)-positive Staphylococcus aureus isolates obtained from various food and clinical specimens. Foodborne Pathog Dis. 13(11):592–601.
- Su YC, Wong AC. 1995. Identification and purification of a new staphylococcal enterotoxin, H. Appl Environ Microbiol. 61(4):1438–1443.
- Su YC, Wong AC. 1996. Detection of staphylococcal enterotoxin H by an enzyme-linked immunosorbent assay. J Food Prot. 59(3):327–330.
- Sumby P, Waldor MK. 2003. Transcription of the toxin genes present within the staphylococcal phage phiSa3ms is intimately linked with the phage's life cycle. J Bacteriol. 185(23):6841–6851.
- Thomas D, Chou S, Dauwalder O, Lina G. 2007. Diversity in Staphylococcus aureus enterotoxins. Chem Immunol Allergy. 93:24–41.
- Thomas DY, Jarraud S, Lemercier B, Cozon G, Echasserieau K, Etienne J, Gougeon M-L, Lina G, Vandenesch F. 2006. Staphylococcal enterotoxin-like toxins U2 and V, two new staphylococcal superantigens arising from recombination within the enterotoxin gene cluster. Infect Immun. 74(8):4724–4734.
- Tsutsuura S, Shimamura Y, Murata M. 2013. Temperature dependence of the production of staphylococcal enterotoxin A by Staphylococcus aureus. Biosci Biotechnol Biochem. 77(1):30–37.
- Umeda K, Nakamura H, Yamamoto K, Nishina N, Yasufuku K, Hirai Y, Hirayama T, Goto K, Hase A, Ogasawara J. 2017. Molecular and epidemiological characterization of staphylococcal foodborne outbreak of Staphylococcus aureus harboring seg, sei, sem, sen, seo, and selu genes without production of classical enterotoxins. Int J Food Microbiol. 256:30–35.
- Valero A, Perez-Rodriguez F, Carrasco E, Fuentes-Alventosa JM, Garcia-Gimeno RM, Zurera G. 2009. Modelling the growth boundaries of Staphylococcus aureus: effect of temperature, pH and water activity. Int J Food Microbiol. 133(1–2):186–194.
- Velasco V, Vergara JL, Bonilla AM, Munoz J, Mallea A, Vallejos D, Quezada-Aguiluz M, Campos J, Rojas-Garcia P. 2018. Prevalence and characterization of Staphylococcus aureus strains in the pork chain supply in Chile. Foodborne Pathog Dis. 15(5):262–268.
- Viçosa GN, Le Loir A, Le Loir Y, De Carvalho AF, Nero LA. 2013. egc characterization of enterotoxigenic Staphylococcus aureus isolates obtained from raw milk and cheese. Int J Food Microbiol. 165(3):227–230.
- Vora P, Senecal A, Schaffner DW. 2003. Survival of Staphylococcus aureus ATCC 13565 in intermediate moisture foods is highly variable. Risk Anal. 23(1):229–236.
- Wallin-Carlquist N, Cao R, Marta D, da Silva AS, Schelin J, Radstrom P. 2010. Acetic acid increases the phage-encoded enterotoxin A expression in Staphylococcus aureus. BMC Microbiol. 10:147.
- Willey J, Prescott L. 2008. Prescott, Harley, and Klein's microbiology. New York, NY: McGraw-Hill Higher Education.
- Wilson GJ, Seo KS, Cartwright RA, Connelley T, Chuang-Smith ON, Merriman JA, Guinane CM, Park JY, Bohach GA, Schlievert PM, et al. 2011. A novel core genome-encoded superantigen contributes to lethality of community-associated MRSA necrotizing pneumonia. PLoS Pathog. 7(10):e100227.
- Yan X, Wang B, Tao X, Hu Q, Cui Z, Zhang J, Lin Y, You Y, Shi X, Grundmann H. 2012. Characterization of Staphylococcus aureus strains associated with food poisoning in Shenzhen, China. Appl Environ Microbiol. 78(18):6637–6642.
- Yarwood JM, McCormick JK, Paustian ML, Orwin PM, Kapur V, Schlievert PM. 2002. Characterization and expression analysis of Staphylococcus aureus pathogenicity island 3. Implications for the evolution of staphylococcal pathogenicity islands. J Biol Chem. 277(15):13138–13147.
- Yarwood JM, Schlievert PM. 2003. Quorum sensing in Staphylococcus infections. J Clin Invest. 112(11):1620–1625.
- Zeaki N, Cao R, Skandamis PN, Rådström P, Schelin J. 2014. Assessment of high and low enterotoxin A producing Staphylococcus aureus strains on pork sausage. Int J Food Microbiol. 182–183:44–50.
- Zeaki N, Radstrom P, Schelin J. 2015a. Evaluation of potential effects of NaCl and sorbic acid on staphylococcal enterotoxin A formation. Microorganisms. 3(3):551–566.
- Zeaki N, Susilo YB, Pregiel A, Rådström P, Schelin J. 2015b. Prophage-encoded staphylococcal enterotoxin A: regulation of production in Staphylococcus aureus strains representing different sea regions. Toxins (Basel). 7(12):5359–5376.
- Zhang S, Stewart GC. 2000. Characterization of the promoter elements for the staphylococcal enterotoxin D gene. J Bacteriol. 182(8):2321–2325.
- Zhang Y, Wang Y, Cai R, Shi L, Li C, Yan H. 2018. Prevalence of enterotoxin genes in Staphylococcus aureus isolates from pork production. Foodborne Pathog Dis. 15(7):437–443.
- Zhao Y, Zhu A, Tang J, Tang C, Chen J. 2017. Identification and measurement of staphylococcal enterotoxin M from Staphylococcus aureus isolate associated with staphylococcal food poisoning. Lett Appl Microbiol. 65(1):27–34.
- Zhao Y, Zhu A, Tang J, Tang C, Chen J, Liu J. 2016. Identification and measurement of staphylococcal enterotoxin-like protein I (SEll) secretion from Staphylococcus aureus clinical isolate. J Appl Microbiol. 121(2):539–546.
- Zhou Z, Zhang M, Li H, Yang H, Li X, Song X, Wang Z. 2017. Prevalence and molecular characterization of Staphylococcus aureus isolated from goats in Chongqing, China. BMC Vet Res. 13(1):352.
