 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Local breeds represent an underestimated resource in terms not only of their important cultural and economical role in marginal areas, but also because they often own a potential genetic pool well adapted to extreme conditions. This fact is of increasing interest, especially when considering climate global challenges where peculiar and uncommon traits could be advantageous. In this study, we genotyped 24 individuals belonging to the small residual Pagliarola sheep population using the OvineSNP50K array, in order to compare its genomic architecture with other 21 Italian local breeds. Moreover, we performed the fixation index (FST) outlier analysis to identify genes most differentiated between Pagliarola and Merino-derived Italian breeds. All population genetic analyses highlighted that Pagliarola breed represents a distinct genetic unit showing a genetic relationship with Merino-derived breeds underlying the close cultural connection between these breeds related to transhumance. However, Pagliarola breed resulted to be divided into two different sub-populations, named Pagliarola 1 (PAG-1) and Pagliarola 2 (PAG-2). Genetic diversity indices and inbreeding estimated from runs of homozygosity (FROH) indicated that while PAG-1 showed among the highest values, PAG-2 turned out strongly inbred, probably as a consequence of a founder effect. The FST outlier analysis identified the 5 most differentiated single nucleotide polymorphisms, two of which mapping within known genes (MACF1 and GLIS1) reported to be linked to feed efficiency and local adaption. All these information strongly suggest that proper conservation measures should be implemented in order to recover the Pagliarola population and its fundamental local products.
All population genetic analyses highlighted that Pagliarola breed represent a distinct genetic unit, and clusters with the Merino-derived Italian breeds.
The breed resulted to be divided into two different sub-populations.
For the two Pagliarola sub-populations, the genetic diversity indices suggest a very different genetic makeup.
Results from the Bayesian approach identified two know genes (MACF1and GLIS1) related to feed efficiency in livestock species and local adaptation.
The information generated in this study strongly suggests that proper conservation measures should be implemented in order to recover the Pagliarola population.
HIGHLIGHTS
Introduction
During the last century, erosion of livestock genetic resources was observed as the result of a massive replacement of low-productivity local breeds with highly productive ones. These local breeds are often reduced to populations having a small effective size, implying difficulties to manage inbreeding and within-breed genetic diversity. In recent years, an increasing interest in recovering and preserving local breeds has taken place (e.g. Ciani et al. Citation2014; Mastrangelo et al. Citation2018). Local breeds are the result of a combination of natural and artificial selection carried on over centuries which shaped animals to adapt and produce in a variety of different environments (Ajmone-Marsan Citation2010; Ajmone-Marsan et al. Citation2014). In fact, local populations often possess valuable traits such as disease resistance, high fertility, good maternal qualities, longevity and adaptation to harsh conditions and poor-quality feed, all desirable qualities for low-input, sustainable agriculture. In addition, local breeds are a reservoir of diversity that may turn out useful to face the upcoming climate change and disease outbreaks and contribute to conserve local culture and traditional products (Ciani et al. Citation2013).
With a total of 263, Italy is one of the European countries with the highest number of local livestock breeds. Among these 84 are sheep breeds, 17 of which are already extinct (Bittante Citation2011). Italy has a long history of sheep breeding and, despite a dramatic contraction in numbers, still raise several local sheep breeds that may represent a unique source of genetic diversity (Ciani et al. Citation2013, Citation2014; Mastrangelo et al. Citation2017).
The Pagliarola is an ancient sheep population farmed in central Italy, mainly in the Abruzzo region (Marchi and Mascheroni Citation1925; Fabrizio Citation1928; Bonadonna Citation1976), together with other small local populations such as Bariscianese, Pomarancina, Perugina, Pecora dell’Amiata, Todina and Vissana. This sheep finds its origins in the Apennines mountain range, and is identified as the ancestral Appenninica sheep population. Even if these small local populations were never selected to improve productive traits such as meat (Marchi and Mascheroni Citation1925; Bonadonna Citation1976; Sarti et al. Citation2002), they were raised for their adaptation to the Apennines environment and for their low nutritional requirements. In particular, Pagliarola was resident in the mountain villages all year round (Renieri et al. Citation1984). During the severe winter season, the breed was usually supplemented with straw (‘‘paglia’’) and this derived their actual name. Pagliarola is a dual-purpose breed; it is reared both for its milk and the resultant local dairy products (‘Pecorino di Farindola’, ‘Pecorino del Matese’, and ‘Formaggio di Pietracatella’) and for its meat (‘Agnello del Centro Italia IGP’, made from the meat of the lambs of the local sheep breeds of Apennines, including Pagliarola). Ceccobelli et al. (Citation2016) reported that only a few small Pagliarola flocks were still surviving in very hard environmental conditions, thus, an appropriate breeding program in order to recover Pagliarola population would be desirable. More recently, this population has been counted for a total of 25 individuals (21 ewes and 4 rams) in the territory of Barisciano (AQ) and samples of semen from two Pagliarola rams were collected and preserved in order to establish a genetic bank of the population (Anzalone et al. Citation2018). This is the last estimation of the population consistency.
The availability of sheep genome-wide SNP panels allows providing background information concerning genome structure in local and cosmopolite breeds (e.g. Kijas et al. Citation2012; Ciani et al. Citation2014). Genetic diversity is a key measure for the monitoring of genetic parameters that are important for the prevention of genetic erosion, inbreeding and other deleterious processes that may lead to population extinction. In addition, an investigation of genomic variation is an important prerequisite to maintain its integrity and to ensure appropriate conservation strategies in the presence of genetic structure that is not easily identifiable with morphological data. Moreover, considering the challenging of the climate changes, the identification of genetic stock which might harbour potential genes well adapted to extreme environment, should be a topic of increasing importance (Hoffmann Citation2010; Biscarini et al. Citation2015).
The aim of this work was to study the genetic diversity and population structure of Pagliarola sheep in the Italian context using data generated from Illumina OvineSNP50K array. Moreover, a genome-scan analytical approach was performed to identify genomic regions harbouring genes conceivably associated to the breed aptitude. This paper highlights how genomic survey is a valuable tool in order to advise possible reintroduction and conservation plans. Indeed, contrariwise to other local sheep for which conservation programs, supported by several authorities (PSRN, ASSONAPA), have a longstanding strategy to manage zootechnical biodiversity (genomic characterisation, semen collection and reconstitution), fewer efforts have been made on small residual local breeds that instead represent an important reservoir for genes adapted to specific environmental conditions.
Materials and methods
DNA sampling, genotyping and quality control
Blood samples were collected from 24 individuals of Pagliarola sheep. Genomic DNA was extracted using the Genelute Mammalian Genomic DNA Kit (Sigma). The concentration of extracted DNA was assessed with NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). All animals were genotyped for 54,241 single nucleotide polymorphisms (SNPs) using the Illumina OvineSNP50K array. The raw data have been merged using the software PLINK (Purcell et al. Citation2007) with the genotypic data of 21 Italian sheep breeds assembled by the Italian Sheep Consortium (BIOVITA) (Ciani et al. Citation2014). The genotypes of Merinizzata Italiana breed have been retrieved from the dataset used in Ciani et al. (Citation2015). In addition to these Italian breeds, the Ile de France sheep (IDF) was considered due to its likely cross-breeding with the Pagliarola breed. Genotypic data of IDF breed were available from Rochus et al. (Citation2018). After filtering for minor allele frequency (–maf 0.05), missing genotype call rate (–geno 0.01), linkage disequilibrium (r2 0.02) and missngness per individual (–mind 0.1), a final dataset consisting of 29,970 SNPs was obtained and one Pagliarola individual has been removed.
Analysis of genetic relationships and population structure among breeds
Genetic structure of breeds has been inspected by using the maximum likelihood clustering approach as implemented in the software ADMIXTURE (Alexander and Lange Citation2011) with K values ranging from 2 to 24. The results have been plotted using the membercoef.circos function in the R package BITE (Milanesi et al. Citation2017).
To explore the genetic differentiation among breeds, we performed a multidimensional scaling plot (MDS) based on pairwise identical-by-state (IBS) distances using the software PLINK (–cluster, mds-plot 3) and visualised using the R package ggplot2 (Wickham Citation2011).
In order to reconstruct genomic relationships among all populations and to investigate the presence of patterns of population splitting and migration events, a maximum likelihood tree has been generated using the software TREEMIX (Pickrell and Pritchard Citation2012). For this analysis we also used the genotypic data of Soay sheep (Kijas et al. Citation2012) as an outgroup, and allowing migration edges from 1 to 10. The best predictor for the number of migration edges was selected using the optM function in the R package OptM (Fitak Citation2018).
Genetic diversity indices
Genetic diversity indices such as observed (Ho) and expected (He) heterozygosity and minor allele frequency (MAF) have been estimated using PLINK, while trends in historical effective population size (Ne) for the 13th generation (presumed to be contemporary Ne), based on linkage disequilibrium (LD), have been estimated using the software SNeP v1.1. (Barbato et al. Citation2015).
In addition, to have an estimate of the inbreeding within and among breeds, we performed runs of homozygosity (ROH) using PLINK and the following parameters: a sliding window of 50 SNPs across the genome; the minimum number of consecutive SNPs included in an ROH was 50; the minimum length of an ROH was set to 4000 kb; the maximum gap between consecutive homozygous SNPs was 1000 kb; a density of one SNP per 100 kb; and a maximum of one SNP with missing genotype and up to one heterozygous genotype were allowed in an ROH. The inbreeding coefficient (F) on the basis of ROH (FROH) for each animal was calculated as follows:
where LROH is the total length of all ROH in the genome of an individual and Laut is the specified length of the autosomal genome covered by SNPs on the chip (2452.06 Mb).
FST-outlier analysis
Based on the genetic structure results, the FST-outlier approach as implemented in the BAYESCAN software (Foll and Gaggiotti Citation2008) was adopted to identify loci involved in the differentiation between the Pagliarola 1 (see Results and discussion section) and the other Merino-derived Italian breeds (GDP, SPV and MER), using the default parameters. To control the number of false positives, significant markers were defined by applying a q-value threshold of 0.05 and using the 0.9999 SNPs of FST percentile distribution (Moscarelli et al. Citation2021). Chromosomal coordinates for each SNP were obtained using the sheep reference genome Oar 3.1.
Results and discussion
High-throughput genotyping arrays have greatly facilitated the study of genetic structure in livestock species, but whereas much effort has been devoted to study population structure within dominant commercial breeds, local breeds, which are less widely used, are generally understudied (Beynon et al. Citation2015). This study provides the first overview of population structure estimates of the Pagliarola sheep from a genome-wide perspective.
Analysis of genetic relationships and population structure among breeds
The ADMIXTURE analysis, MDS and TREEMIX were used to visualise and explore the genetic relationships between Pagliarola and the other sheep breeds.
Population structure inferred using the software ADMIXTURE, with K ranging from 2 to 24, showed the lowest CV error at K = 18 (Supplementary Materials, Figure S1). At this K value, the ADMIXTURE components highlighted that all breeds have their own genetic identity, with the exception of Biellese and Bergamasca breeds which appear to be indistinguishable and Comisana and Pinzirita which share a large proportion of genetic components (Mastrangelo et al. Citation2014) (Figure ). At K = 2, Ile de France breed first differentiated while Merinizzata showed high level of admixture when compared with all the other Italian native breeds. At K = 3, Alpagota, Biellese and Bergamasca separated, while the Pagliarola breed resulted to be divided into two different sub-populations which are clearly distinguishable at K = 4, and hereinafter named Pagliarola 1 (PAG-1, n = 14) and Pagliarola 2 (PAG-2, n = 9). It should be noted that despite the number of individuals belonging to the two Pagliarola subgroups was quite small, it has been empirically demonstrated that for population structure analyses, the patterns observed using six randomly extracted animals per breed closely mirrors those inferred from 20 to 24 animals per breed (Gaouar et al. Citation2017).
Figure 1. ADMIXTURE analysis showing the genetic structure of the analysed breeds. For full definition of breeds see Table .
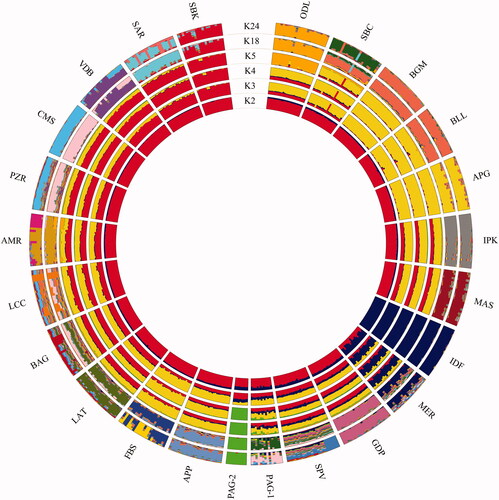
These two genetic clusters (PAG-1 and PAG-2) match with the two different farms where the samples were taken. Therefore, the genetic structure detected for Pagliarola sheep could be due to the introgression of genes from other breeds (PAG-1) and/or to geographical isolation for a long time with genetic drift (PAG-2). These results were also in agreement with a previous study conducted on the genetic structure and relationship using microsatellite markers (Ceccobelli et al. Citation2016).
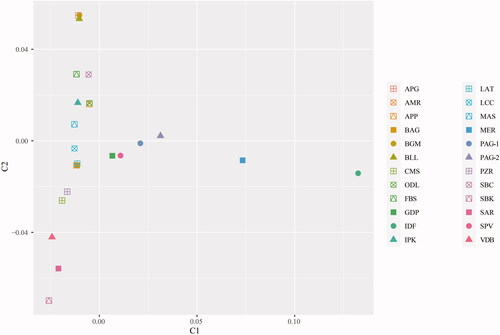
Component 1 of the MDS plot reveals a clear differentiation of breeds from central Italy (MER, PAG-1, PAG-2, SPV and GDP) including the Ile de France breed which as expected is shown as the most differentiated. On the other hand, component 2 reflects a latitudinal genetic cline with breeds from northern Italy located mostly in the upper portion of the plot, while breeds from southern Italy and Sardinia cluster in the lower quadrant of the plot (Figure ). This strong phylogeographic pattern of sheep breeds has been highlighted in previous studies at both worldwide (Kijas et al. Citation2009, Citation2012) and regional level (Ciani et al. Citation2014; Mastrangelo et al. Citation2017). Such a geographic structuring has been suggested to be the outcome of a combination of adaptive processes driven by natural selection and historical admixture prompted by human selection. Moreover, the observed phylogeographic pattern mirror the distribution of human genetic diversity (Brisighelli et al. Citation2012; Anagnostou et al. Citation2017) underling the close connection between livestock diversity and historical human migrations/barriers as highlighted in other livestock species (Senczuk et al. Citation2021). For the Pagliarola sheep, these results confirmed the findings based on the ADMIXTURE, where, the population formed two separated and differentiated clusters.
Figure 2. Multidimensional Scaling Plot (MDS) based on identity-by-state distance matrix indicating breed differentiation. Each dot corresponds to a different individual. For full definition of breeds see Table .
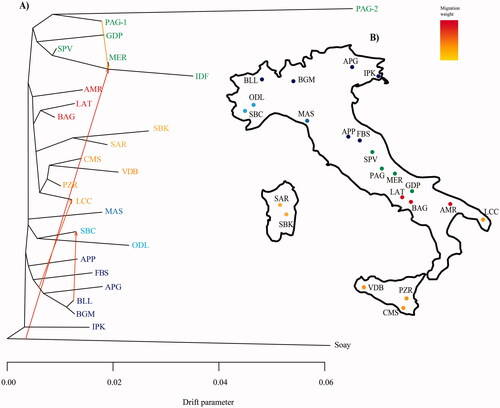
The TREEMIX phylogram shows a clear distribution of clusters according to the geographic origin of breed (Figure ). Indeed, we observe a group including all breeds from Central-Eastern Alps and Central Apennines (IPK, APG, BGM, BLL, APP, FBS), a second group including Western Alps (ODL and SBC), a group including Central Apennines breeds (GDP, SPV, MER, PAG-1, PAG-2) and another group including breeds from Central and Southern Italy (LAT, BAG, AMR), Sicily and Apulia (LCC, CMS, PZR, VDB) and Sardinia (SAR, SBK). In this context, Pagliarola clusters with the Merino-derived Italian breeds (Figure ). This result is not surprising considering the close connection between Pagliarola and some Merino-derived Italian breeds. In fact, up to the 1950s of the last century, the Pagliarola was traditionally reared in the same flocks with Gentile di Puglia, though the management conditions of the two breeds were different (Renieri et al. Citation1984). This topology remained reasonably constant through iterations. The linear method implemented in the OptM (R Package OptM) function indicated a major changing point in the log likelihood at four migration events (Supplementary Materials, Figure S2). A strong migration event is shown between Soay and the node separating Merinizzata and Ile de France breeds. This migration event is probably linked to the Northern European origins of both Ile de France and Soay breeds. On the other hand, the close relationship of Merinizzata with these Northern European breeds is due to its known admixed origin between local Merino-derived breeds and exotic Merino meat breeds including Ile de France (Morbidini et al. Citation1995; Sarti and Panella Citation1999). Another strong migration edge is shown between the base of the crown including Alpagota, Biellese and Bergamasca and the Leccese breeds. This result is in accordance with documented livestock exchange between Southern and Northern regions in order to improve production (Ciani et al. Citation2014). An additional migration event is highlighted between Biellese and Sambucana breeds. In this context, the use of Biellese to improve meat yield in other autochthonous breeds including Sambucana is well known (Bigi and Zanon Citation2008). Finally, another migration event is shown between Pagliarola (PAG-1) and the node including Merinizzata and Ile de France breeds indicating some gene exchange between these breeds. Even if Pagliarola has been considered as one of the local populations of the Apennines (Marchi and Mascheroni Citation1925; Bonadonna Citation1976) from which Appenninica breed has been selected after crosses with meat selected breeds (Sarti et al. Citation2002), neither migration events or drift parameters indicated a genetic relationship between these two breeds.
Figure 3. TREEMIX analysis with the most supported number of migration events with their relative weights (A). Geographic location of the native Italian breeds used in this study. Colours depict the main geographic cluster as highlighted in the TREEMIX analysis. For full definition of breeds see Table .
Genetic diversity indices
Genetic diversity indices, which are key parameters in the genetic management of populations, are reported in Table . The results revealed that Pagliarola 1 and Pagliarola 2 tended to display the highest and the lowest variability, respectively, than other Italian breeds. However, we cannot rule out the presence of an ascertainment bias effect due to the use of small samples size in the sub-populations (Bedhiaf-Romdhani et al. Citation2020). Pagliarola 1 showed the highest Ho (0.391 ± 0.182), whereas Pagliarola 2 showed the lowest one (0.274 ± 0.233). Similar outcomes are observed for expected heterozygosity (He = 0.361 ± 0.137; He = 0.254 ± 0.194 for Pagliarola 1 and 2, respectively). The lowest mean values of MAF are recorded for Pagliarola 2 (0.192 ± 0.167) and the highest for Bagnolese (0.287 ± 0.137), while Pagliarola 1 showed an intermediate value (0.276 ± 0.139).
Table 1. Genetic diversity indices for all the analysed breeds.
The effective population size (Ne) estimates for each breed are also reported in Table . The minimum Ne value was found for Pagliarola 2 (Ne = 20), the maximum value was found for Bergamasca (Ne = 130), while Pagliarola 1 showed a small Ne of 48. For the two sub-populations of Pagliarola breed, Ne was smaller (average 34) than for all other breeds (average 94.5). Consistent with our results, Mastrangelo et al. (Citation2017), reported a contemporary effective population size of 25 in the Barbaresca sheep, which, like Pagliarola, is an endangered population that has experienced a dramatic census contraction.
Individual genomic inbreeding have been evaluated using ROH analysis as reported in Figure . The highest mean value of FROH is observed in Pagliarola 2 (0.133), followed by Leccese (0.068) and Valle del Belice (0.067), while the lowest values are observed in Comisana, Bergamasca and Pagliarola 1 (0.008, 0.014 and 0.024, respectively) (Table ). For the Italian sheep breeds, similar low values were previously reported (Mastrangelo et al. Citation2018), in particular for small and endangered breeds, as Altamurana and Barbaresca. The total number of ROH and the total genomic length for each animal of the two Pagliarola sub-populations are shown in Figure . For Pagliarola 1, the majority of the animals clustered near to the origin of coordinates with a total length <100 Mb. For the Pagliarola 2, there were individuals with a larger number of ROH (from 20 to 70) and with the total length of genome in ROH more than 500 Mb. Therefore, Pagliarola 2 showed ROH patterns typically produced by inbreeding or strong population bottlenecks. Generally, in the farming system of local sheep breeds, the farmers use rams born in the flock and the exchange of animals is quite unusual. This leads to an increase of inbreeding within the population due to the uncontrolled mating between genetically related individuals (Mastrangelo et al. Citation2012). Therefore, in these small populations, such as Pagliarola breed, directional mating and the use of selected rams can be considered as encouraging breeding practices to prevent higher increases of inbreeding in order to preserve genetic diversity.
Figure 4. Genomic inbreeding values estimated from runs of homozygosity (FROH) of each breed. For full definition of breeds see Table .
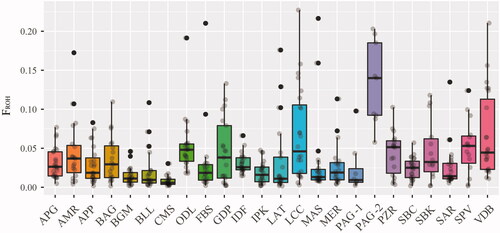
Figure 5. Relationship between the total number of runs of homozygosity (ROH) segments (y-axis) and the total length (Mb) of genome in such ROH (x-axis) for each individual of the two Pagliarola sub-populations: Pagliarola 1 (PAG-1) and Pagliarola 2 (PAG-2).
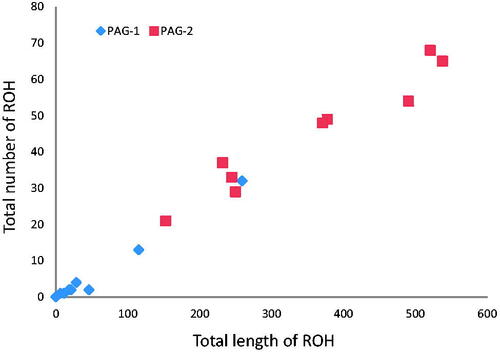
For the two Pagliarola sub-populations, the genetic diversity indices suggest a very different genetic makeup. Indeed, all genetic diversity indices indicate that Pagliarola 2 sub-population could be the result of a strong genetic drift, as underlined by high level of homozygosity (FROH) and a low value of effective population size (Ne) and heterozygosity (Table and Figure ), confirming the results on the genetic structure above reported. One potential explanation for such a trend is a combination of a founder effect facilitated by a small genetic pool from which the Pagliarola 2 sub-population may have originated. Strong differences in terms of genetic drift are not an exception in livestock and have been repeatedly reported also in other species (Lawson Handley et al. Citation2007; Ciani et al. Citation2015; Nicoloso et al. Citation2015; Lenstra et al. Citation2017; Senczuk et al. Citation2020; Moscarelli et al. Citation2021). Moreover, this small genetic background could have been even amplified by possible attempts to keep the subpopulation as ‘pure’ as possible. Unlike Pagliarola 2, Pagliarola 1 showed the highest genetic diversity values when compared with all the other native Italian breeds, likely because of its admixed origin. Such a result is very important to underline that an urgent recovery of this population is needed in order to not lose a marked genetic potential.
FST-outlier analysis
The genome‐wide distribution of pair‐wise FST values was used to identify highly differentiated SNPs between Pagliarola 1 (considered to be the most likely original genetic stock) and the other three Merino-derived Italian breeds (GDP, SPV and MER). Results from the Bayesian approach identified a total of 5 outlier SNPs (Table ) located on OAR01, OAR10 and OAR15. Among the significant markers, two SNPs on OAR01 mapped within know genes (MACF1and GLIS1). The MACF1 gene plays crucial role in biological processes related to feed efficiency in livestock species (Reyer et al. Citation2017; Lam et al. Citation2018). The GLIS1 is reported as a candidate gene for feed efficiency (Cockrum et al. Citation2012) and local adaption (Sweet-Jones et al. Citation2021) in sheep. These results are consistent with the phenotypic characteristics of Pagliarola sheep that have excellent abilities to adapt to harsh environments and with the ability to exploit poor-quality feed compared to the other Merino-derived Italian breeds, for which instead the transhumance was carried out.
Table 2. Results of the BayeScan analysis showing outlier single nucleotide polymorphisms (SNPs) identified between Pagliarola 1 and Merino-derived Italian breeds (Gentile di Puglia, Sopravissana and Merinizzata).
Conclusion
This study has reported, for the first time, genetic diversity and population structure estimates from a genome-wide perspective in the Pagliarola sheep. The genetic structure analyses show that Pagliarola breed own a distinct and unique genetic background composed of two different genetic groups with a common origin but characterised by a different genomic make up. Indeed, Pagliarola 2 showed a marked genomic inbreeding, low values of both heterozygosity and effective population size, suggesting that a strong genetic drift occurred in this population. On the contrary, Pagliarola 1 showed low genomic inbreeding values and high values of heterozygosity.
These results stress the importance of genetic investigations before specific conservation strategies, including a possible reintroduction of the original genetic pool, can be implemented. Indeed, basing on our outcomes we strongly encourage a reintroduction from the original genetic stock of Pagliarola 1 population. Moreover, concerning the genetic relationships with other local breed from the Apennines, although Pagliarola has been considered as one of the ancestral populations from which Appenninica breed derived, we found a closer relationship with Merino-derived breeds. Such a genetic relatedness might be the outcome of the close cultural connection related to transhumance. Indeed, despite Pagliarola breeds was not an active transhumant breed was part of the same herds during the non-transhumant periods. Finally, the BayeScan results demonstrate the importance to preserve the Pagliarola genome as genetic reservoir of alleles particularly adapted to low-input farming system. The information generated in this study strongly suggests that proper conservation measures should be implemented in order to recover the Pagliarola population and its fundamental local products.
Ethical approval
All procedures involving animal sample collection followed the recommendation of directive 2010/63/EU. Sampling was carried out by trained veterinarians within the frame of vaccination campaigns, hence no permission from the animal research ethics committee was necessary. Veterinarians adhered to standard procedures and relevant national guidelines to ensure appropriate animal care.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available upon request from the corresponding author.
Additional information
Funding
References
- Ajmone-Marsan P. 2010. A global view of livestock biodiversity and conservation-GLOBALDIV. Anim Genet. 41(Suppl 1):1–5.
- Ajmone-Marsan P, Colli L, Han JL, Achilli A, Lancioni H, Joost S, Crepaldi P, Pilla F, Stella A, Taberlet P, et al. 2014. The characterization of goat genetic diversity: towards a genomic approach. Small Rumin Res. 121(1):58–72.
- Alexander DH, Lange K. 2011. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinf. 12(1):1–6.
- Anagnostou P, Capocasa M, Dominici V, Montinaro F, Coia V, Destro-Bisol G. 2017. Evaluating mtDNA patterns of genetic isolation using a re-sampling procedure: a case study on Italian populations. Ann Hum Biol. 44(2):140–148.
- Anzalone DA, Palazzese L, Iuso D, Martino G, Loi P. 2018. Freeze-dried spermatozoa: an alternative biobanking option for endangered species. Anim Reprod Sci. 190:85–93.
- Barbato M, Orozco-terWengel P, Tapio M, Bruford MW. 2015. SNeP: a tool to estimate trends in recent effective population size trajectories using genome-wide SNP data. Front Genet. 6:109.
- Bedhiaf-Romdhani S, Baazaoui I, Ciani E, Mastrangelo S, Sassi MB. 2020. Genetic structure of Tunisian sheep breeds as inferred from genome-wide SNP markers. Small Rumin Res. 191:106192.
- Beynon SE, Slavov GT, Farré M, Sunduimijid B, Waddams K, Davies B, Haresign W, Kijas J, MacLeod IM, Newbold CJ, et al. 2015. Population structure and history of the Welsh sheep breeds determined by whole genome genotyping. BMC Genet. 16(1):65.
- Bigi D, Zanon A. 2008. Atlante delle razze autoctone. Bov Equini Ovicaprini Suini Allev Ital Edagricole.
- Biscarini F, Nicolazzi EL, Stella A, Boettcher PJ, Gandini G. 2015. Challenges and opportunities in genetic improvement of local livestock breeds. Front Genet. 6:33.
- Bittante G. 2011. Italian animal genetic resources in the Domestic Animal Diversity Information System of FAO. Ital J Anim Sci. 10(2):e29.
- Bonadonna T. 1976. Etnologia zootecnica. Torino: UTET.
- Brisighelli F, Blanco-Verea A, Boschi I, Garagnani P, Pascali VL, Carracedo A, Capelli C, Salas A. 2012. Patterns of Y-STR variation in Italy. Forensic Sci Int Genet. 6(6):834–839.
- Ceccobelli S, Lorenzo PD, Panella F, Lasagna E, Sarti FM. 2016. Morphological and genetic characterisation of Pagliarola breed and its genetic relationships with other three indigenous Italian sheep breeds. Ital J Anim Sci. 15(1):47–54.
- Ciani E, Ciampolini R, D’Andrea M, Castellana E, Cecchi F, Incoronato C, d’Angelo F, Albenzio M, Pilla F, Matassino D, et al. 2013. Analysis of genetic variability within and among Italian sheep breeds reveals population stratification and suggests the presence of a phylogeographic gradient. Small Rumin Res. 112(1-3):21–27.
- Ciani E, Crepaldi P, Nicoloso L, Lasagna E, Sarti FM, Moioli B, Napolitano F, Carta A, Usai G, D'Andrea M, et al. 2014. Genome-wide analysis of Italian sheep diversity reveals a strong geographic pattern and cryptic relationships between breeds. Anim Genet. 45(2):256–266.
- Ciani E, Lasagna E, D’Andrea M, Alloggio I, Marroni F, Ceccobelli S, Delgado Bermejo JV, Sarti FM, Kijas J, Lenstra JA, et al. 2015. Merino and Merino-derived sheep breeds: a genome-wide intercontinental study. Genet Sel Evol. 47(1):64.
- Cockrum RR, Pickering NK, Anderson RM, Hyndman DL, Bixley MJ, Dodds KG, Stobart RH, McEwan JC, Cammack KM. 2012. Identification of single nucleotide polymorphisms associated with feed efficiency in rams. In: Conference: Western section American society of Animal Science [place unknown]; p. 79.
- Fabrizio P. 1928. La pecora del Molise [Sheep of Molise]. Italia Agricola. 1965(6):283–295.
- Fitak RR. 2021. optM: an R package to optimize the number of migration edges using threshold models. (in preparation)
- Foll M, Gaggiotti O. 2008. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics. 180(2):977–993.
- Gaouar SB, Lafri M, Djaout A, El-Bouyahiaoui R, Bouri A, Bouchatal A, Maftah A, Ciani E, Da Silva AB. 2017. Genome-wide analysis highlights genetic dilution in Algerian sheep. Heredity. 118 (3):293–301.
- Hoffmann I. 2010. Climate change and the characterization, breeding and conservation of animal genetic resources. Anim Genet. 41(s1):32–46.
- Kijas JW, Lenstra JA, Hayes B, Boitard S, Neto LRP, Cristobal MS, Servin B, McCulloch R, Whan V, Gietzen K, et al. 2012. Genome-Wide Analysis of the world’s sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol. 10(2):e1001258.
- Kijas JW, Townley D, Dalrymple BP, Heaton MP, Maddox JF, McGrath A, Wilson P, Ingersoll RG, McCulloch R, McWilliam S, et al. 2009. A Genome Wide Survey of SNP variation reveals the genetic structure of sheep breeds. PLOS One. 4(3):e4668.
- Lam S, Miglior F, Fonseca P, Seymour D, Asselstine V, Brito L, Schenkel F, Cánovas A. 2018. Identification of variants associated with divergent feed efficiency groups using multiple RNA-sequencing datasets from dairy and beef cattle.In: Proc World Congr Genet Appl to Livest Prod. Vol. Electronic. Auckland, New Zealand; p. 773.
- Lawson Handley L-J, Byrne K, Santucci F, Townsend S, Taylor M, Bruford MW, Hewitt GM. 2007. Genetic structure of European sheep breeds. Heredity. 99(6):620–631.
- Lenstra JA, Tigchelaar J, Biebach I, Hallsson JH, Kantanen J, Nielsen VH, Pompanon F, Naderi S, Rezaei H-R, Saether N, et al. 2017. Microsatellite diversity of the Nordic type of goats in relation to breed conservation: how relevant is pure ancestry? J Anim Breed Genet. 134(1):78–84.
- Marchi E, Mascheroni E. 1925. Zootecnia speciale. Torino: UTET.
- Mastrangelo S, Ciani E, Sardina MT, Sottile G, Pilla F, Portolano B. 2018. Runs of homozygosity reveal genome-wide autozygosity in Italian sheep breeds. Anim Genet. 49(1):71–81.
- Mastrangelo S, Di Gerlando R, Tolone M, Tortorici L, Sardina MT, Portolano B. 2014. Genome wide linkage disequilibrium and genetic structure in Sicilian dairy sheep breeds. BMC Genet. 15(1):108.
- Mastrangelo S, Portolano B, Di Gerlando R, Ciampolini R, Tolone M, Sardina MT. 2017. Genome-wide analysis in endangered populations: a case study in Barbaresca sheep. Animal. 11(7):1107–1116.
- Mastrangelo S, Sardina MT, Riggio V, Portolano B. 2012. Study of polymorphisms in the promoter region of ovine β-lactoglobulin gene and phylogenetic analysis among the Valle del Belice breed and other sheep breeds considered as ancestors. Mol Biol Rep. 39(1):745–751.
- Milanesi M, Capomaccio S, Vajana E, Bomba L, Garcia JF, Ajmone-Marsan P, Colli L. 2017. BITE: an R package for biodiversity analyses. bioRxiv.:181610.
- Morbidini L, Panella F, Pollidori P, Sarti DM, Sarti FM. 1995. Verifica delle caratteristiche riportate nello standard della razza Merinizzata Italiana da carne, [A check of traits carried in the standard of Italian Merinos breed [sheep]]. Allev Ovini E Caprini Italy.
- Moscarelli A, Sardina MT, Cassandro M, Ciani E, Pilla F, Senczuk G, Portolano B, Mastrangelo S. 2021. Genome-wide assessment of diversity and differentiation between original and modern Brown cattle populations. Anim Genet. 52(1):21–31.
- Nicoloso L, Bomba L, Colli L, Negrini R, Milanesi M, Mazza R, Sechi T, Frattini S, Talenti A, Coizet B, et al. 2015. Genetic diversity of Italian goat breeds assessed with a medium-density SNP chip. Genet Sel Evol. 47:62.
- Pickrell J, Pritchard J. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8(11):1002967.
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81(3):559–575.
- Renieri C, Silvestrelli M, Casciotti D, Ronchi B. (Perugia U (Italy) I di PA). 1984. La popolazione ovina Pagliarola: studio demografico, [The ‘Pagliarola’ sheep population. Demographic study]. Vergaro Italy. 9.
- Reyer H, Shirali M, Ponsuksili S, Murani E, Varley PF, Jensen J, Wimmers K. 2017. Exploring the genetics of feed efficiency and feeding behaviour traits in a pig line highly selected for performance characteristics. Mol Genet Genomics. 292(5):1001–1011.
- Rochus CM, Tortereau F, Plisson-Petit F, Restoux G, Moreno-Romieux C, Tosser-Klopp G, Servin B. 2018. Revealing the selection history of adaptive loci using genome-wide scans for selection: an example from domestic sheep. BMC Genomics. 19(1):71.
- Sarti DM. 2002. L’Appenninica. Passato – presente – futuro [ovini], [The Appenninica. Past – present – future [sheep]]. Allev Ovini E Caprini Italy. 19(3):4.
- Sarti DM, Panella F, Pauselli M, Sarti FM, Lasagna E. 2002. L’allevamento degli ovini, [Sheep farming]. Il Divulg Agenzia Territ Sostenibilita Aliment Agro-Ambient Ed Energ Ed Bologna Italy.
- Sarti FM, Panella F. 1999. Study of the most suitable traits to use in linear morphological evaluation of Merinizzata Italiana sheep. Proceeding 13th National Congress ASPA Piacenza Italy. p. 206–208.
- Senczuk G, Mastrangelo S, Ajmone-Marsan P, Becskei Z, Colangelo P, Colli Ferretti L, Karsli T, Lancioni H, Lasagna E, Marletta D, et al. 2021. On the origin and diversification of Podolian cattle breeds: testing scenarios of European colonization using genome-wide SNP data . Genet Sel Evol. 53(1):48.
- Senczuk G, Mastrangelo S, Ciani E, Battaglini L, Cendron F, Ciampolini R, Crepaldi P, Mantovani R, Bongioni G, Pagnacco G, et al. 2020. The genetic heritage of Alpine local cattle breeds using genomic SNP data. Genet Sel Evol. 52(1):40.
- Sweet-Jones J, Yurchenko AA, Igoshin AV, Yudin NS, Swain MT, Larkin DM. 2021. Resequencing and signatures of selection scan in two Siberian native sheep breeds point to candidate genetic variants for adaptation and economically important traits. Anim Genet. 52(1):126–131.
- Wickham H. 2011. ggplot2. WIREs Comp Stat. 3(2):180–185.
