ABSTRACT
The association between caprine PrP gene polymorphisms and its susceptibility to scrapie has been investigated in current years. As the ORF of the PrP gene is extremely erratic in different breeds of goats, we studied the PrP gene polymorphisms in 80 goats which belong to 11 Pakistani indigenous goat breeds from all provinces of Pakistan. A total of 6 distinct polymorphic sites (one novel) with amino acid substitutions were identified in the PrP gene which includes 126 (A -> G), 304 (G -> T), 379 (A -> G), 414 (C -> T), 428 (A -> G) and 718 (C -> T). The locus c.428 was found highly polymorphic in all breeds as compare to other loci. On the basis of these PrP variants NJ phylogenetic tree was constructed through MEGA6.1 which showed that all goat breeds along with domestic sheep and Mauflon sheep appeared as in one clade and sharing its most recent common ancestors (MRCA) with deer species while Protein analysis has shown that these polymorphisms can lead to varied primary, secondary and tertiary structure of protein. Based on these polymorphic variants, genetic distance, multidimensional scaling plot and principal component analyses revealed the clear picture regarding greater number of substitutions in cattle PrP regions as compared to the small ruminant species. In particular these findings may pinpoint the fundamental control over the scrapie in Capra hircus on genetic basis.
INTRODUCTION
The accumulation of protein aggregates is an amalgamating feature of a large class protein conformational disorders that ultimately target the central nervous system and lead to progressive neurodegeneration e.g. Alzheimer disease and Prion diseases.Citation1 The prion theory forecasts that certain mutations in PrP gene may result in unstable PrP molecules with transitional conformations, thus initiating the disease processCitation2 and are accountable for much of the disparity in pathology and disease transmission. Transmissible spongiform encephalopathies (TSEs), or prion diseases, have cases that arise sporadically (Creutzfeldt-Jakob disease [CJD]) or are inherited (fatal familial insomnia).Citation3 Transmission mechanism of prion diseases across the mammalian species is not as such effective as within speciesCitation4 but being a zoonotic and fatal potential, scrapie prions indicated transmission efficacy comparable to BSE causing CJD.Citation5
Scrapie is a mortal, incurable and transmissible neurodegenerative disease of sheep and goats which occur due to the conversion of normal cell surface glycoprotein (PrPC) into conformationally altered isoform (PrPSc) in the absence of nucleic acids.Citation6 The key difference flanked by the 2 forms is that PrPC is entirely sensitive to proteases, while PrPSc is only degraded in part and its amino-terminal end removed.Citation7 As a natural host of scrapie both in sheep and goats showed analogous incubation period while the clinical manifestation perhaps slightly different. Moreover, the supplementary transmissible spongiform encephalopathies (TSEs) which comprise of bovine spongiform encephalopathy (BSE), Creutzfeldt–Jakob disease (CJD) and transmissible mink encephalopathy (TME), usually portrayed by accretion of an anomalous and disease specific isoform (PrPSc) of an usual host-encoded cellular prion protein (PrPC) in the central nervous system. PrPC is also found in number of peripheral cell types along with glial cells of the CNS but in low level.Citation8,9,10 Scrapie research has centered, for many years on developing more realistic live animal tests to make a diagnosis of infected animals; the identification of the genetic basis; and its probable application in breeding plans. Previously, it has been reported in Pakistan that caprine PrP polymorphisms was present in different breeds of sheep and goats such as in Beetal, Tedy and Pak Angora and it was associated with resistance to some important zoonotic and fatal group of diseases which may cross species barrier.Citation8,11 Monitoring and investigation about the transmission of these agents via blood play vital role and has substantial concern regarding the safety of blood and its products.Citation12
In Pakistan goats are kept mainly for milk and meat purpose and play a critical role in the economy of country as well as farmers (). Presently there are about 58.3 million heads of goats in Pakistan and their population is increasing at rate of 3% per annum.Citation13 As small ruminants are the only animals which are migrating in industrialized and developing countries, different zoonotic diseases spotted in small ruminants are mainly transmitted to human beings as occupational infections which badly affect the breeders and veterinarians. Therefore, the investigation of biological activity of PrPC remained crucial to understand the pathogenesis of prion associated diseases through contaminated food.Citation7,14,15 Though no scrapie case has been documented yet in any of Pakistani goats, identifying the PrP gene polymorphism spectrum in these species may be useful for future breeding plans in terms of scrapie resistance.
TABLE 1. Phenotypic characteristics of goat breeds in Pakistan under study.
MATERIALS AND METHODS
Study Animals and Management Practices
Pakistan is located in South Asia possessing tropical and subtropical environment that is known to be hot (35 + 13°C) and humid (45.38% ± 13.11%) in some areas.Citation13 During summer in Punjab the temperature may go up to 48°C with humidity level (39%). The goats and other livestock animals in Pakistan are kept under similar natural climatic conditions. Adequate feed and water are bequeathed for their maintenance and production. To protect animals from harsh climatic conditions and predators semi closed traditional shelters system were employed where routine vaccination and de-worming were routinely carried out.
Sample Collection
A total of 80 blood samples (3ml from each), having specific phenotypic characteristics were cautiously collected from 11 Pakistani goat breeds as of Jattal (11), Barbari (07), Lebri (09), Tapri (03), Kaghani (05), Khurasani (08), Beetal (11), Bari (03), Lehri (10), Pahari (07) and Damani (06) from all 4 provinces of Pakistan along with Azad Jammu & Kashmir region (AJK) and stored at −20°C till further processing. All the selected goat breeds were identified by an expert and their history about the age; breed and sex were also obtained from the farmers.
Genetic Analysis
Genomic DNA was extracted from EDTA-treated blood buffy coat using a standard DNA isolation protocol as previously described.Citation16 A PCR was standardized for the specific amplification of 876 bp of exon 3 of the PrP gene and for this, a reaction mixture of a total volume of 30 µl was prepared containing nuclease free water, 1x PCR buffer (NH4)2 SO4, 50 mM MgCl2, 10 mM dNTPs mix, 20 pmol each of the forward (5′-1CTTTAAGTGATTTTTACGTGG21-3′) and reverse primers (5′-854TGGCAAAGATTAAGAAGATAATG876-3′), 0.2 unit of Taq DNA Polymerase (Fermentas, Cat #EP0402) and 1µl of DNA as template. The standard amount of PCR reaction mixture, DNA samples, Primers and water was used for amplification of PrP gene. Amplification reactions were performed with a Thermal Cycler (iCycler BioRad, USA) conditions comprised of initial denaturation at 94°C for 05 minutes, followed by denaturation at 94°C for 45 sec, annealing at 60°C for 02 min and extension was at 72°C for 45 sec. Final extension was maintained at 72°C for 10 minutes and storage at 4°C. All amplicons were visualized by staining with ethidium bromide through the electrophoresis of a 05 µl reaction mixture on 2% agarose gels.
Sequence Analysis
Prior to Sanger sequencing, amplicons were cleaned using TIANquick Midi Purification Kit (Tiangen Biotech Beijing Co., Ltd.) while both sense and antisense strands of the amplified DNA were sequenced by using Big Dye terminator cycle sequencing chemistry v 3.1 (Applied Biosystems, USA) and electrophoresis was done by an automated DNA sequencer (ABI Prism 3130xL Genetic Analyzer, Applied Biosystems, USA) from 1st Base Laboratories Singapore. Sequences were analyzed through Codon Code aligner manually followed by the identification of novel SNPs detection.
Bioinformatics Tools
Codon Code aligner software was used to edit and align the PrP sequences while MEGAv.6 softwareCitation17 was used to construct the phylogenetic trees and have insight of phylogeny of Pakistani goat breeds with other mammalian species (). We used homologous PrP sequences ranging from primate members to jawless vertebrate sequences. Moreover, the PrP gene coding sequence of all animals was sequenced and analyzed to identify polymorphisms in selected individuals. Single nucleotide polymorphism distributions were tabulated according to every nucleotide site (). Furthermore, variation profile was summarized in where percentages with respect to being a breed a heterozygous, homozygous, or cumulative variation were calculated. Previously at the protein level, 40 6 amino acid polymorphisms have been described ().
TABLE 2. Polymorphism observed in PrP gene in Pakistani goat breeds of meat and milk potential.
TABLE 3. Level of polymorphism summarized according to percentage observed at each nucleotide position.
TABLE 4. Single nucleotide polymorphisms identified in the PrP region of animals all over the world.
TABLE 5. Comparison between studied PrP sequences of animals all over the world with indeginous genotypes of goat breeds of Pakistan. Entries in bold and underlined format in the reported column were also determined as amino acid polymorphisms in studied breeds. Similarly, one entry placed the 3rd spot in the right most column was the novel polymorphism detected in indigenous goat population.
Genetic Distance
Genetic distances between populations were calculated using Barry and Hartigan evolutionary model called ‘BH87’, asynchronous distance between homologous DNA sequences based on the observed proportions of changes among the 4 bases whereas “Bioconductor” statistical package in R was used to generate genetic distance matrix for selected species.
Multidimensional Scaling Plot
Multidimensional scaling method convert the structure in the similarity matrix to a simple geometrical picture: the larger the dissimilarity between 2 samples (evaluated through genetic distance profiling), the further apart the points representing the experiments in the picture may be. Furthermore, genetic relationship between groups were examined using an MDS plot of the genetic distance matrix in R.
Principal Component Analysis
Principal component analysis (PCA), a powerful multivariate investigative tool to identify patterns in specifically many dimensions, interrelated data sets and express the data sets by pinpointing their similarities and differences which is based on Eigen analysis. Therefore, PCA used here as a means of constructing an informative graphical representation of the data set by projecting the data onto a lower dimensional space. In the study, control and training data was presented in a 2 dimensions (2D) subspace of the first 2 PCs. The PCs derived by the Eigen analysis of correlation matrix (R = [rij]) was a linear combination of the original p variables (the species) and each PC uncorrelated with the other, meaning these were the new transformed data expressed in terms of the patterns existing in the original data set. The total PCs derived were equal to the number of original variables present in the dataset. The PCs formed were with decreasing order of magnitude of variance of the total variation in the data sets. Thus the first 2 PCs capturing most of the variation in the data set is visualized in a 2D representation. The coefficient of the variables in each of the linear combination, i.e. the PC was defined as loadings. The magnitude of these loadings represented the importance of each variable present. Thus a 2D representation of the loadings of the first 2 PCs were identified as any cluster structure present in the variables (80 PrP homologs), exhibiting the co-occurring pattern of individuals in the control and training data sets.
RESULTS
Observed SNP Profiles
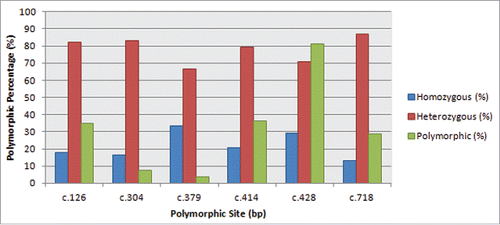
Total six polymorphic sites (at positions 126, 304, 379, 414, 428 and 718) were observed in 80 individuals of 11 breeds which were most notably raised for meat and milk production in Pakistan. In general, c.428 position was found highly polymorphic in all breeds with homozygous G, A and A/G heterozygous genotypes (). In total, 81.25% of individuals were found polymorphic out of them 71.01% individuals were heterozygous for this particular site. Lowest percentage of polymorphism was achieved at c.379 as 3.75%. Similarly, significant percentage of polymorphisms were obtained at c.126, c.304, c.414 and c.718 sites as 35%, 7.5%, 36.25% and 28.75% respectively (). Heterozygous sites were also evaluated in proportion to their relative cumulative polymorphic variation. All individuals showed their preference for being highly heterozygous as compared to be homozygous ().
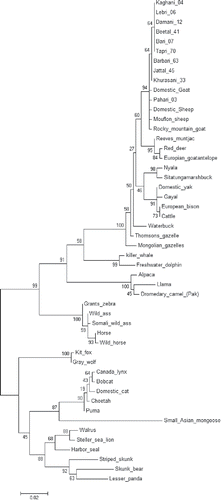
FIGURE 2. NJ Phylogenetic tree constructed using maximum likelihood method between 11 Pakistani goat breeds and 40 other mammalian species. Bootstrap value was kept 1000.
Furthermore, distance analyses (), MDS (), PCA () and phylogenetic tree () was re-constructed with the help of these PrP variables to have an insight of Pakistani goat breeds phylogeny to deepen our understanding in this field of great concern. The results showed that our local goat breeds were in the clade suggesting their genetic relatedness depending on PrP gene. However, deer and antelope were closer relative of Capra hircus on the basis of PrP gene as compared to buffalo, while domestic yak, European bison, gazelles and cattle comes after these species. Similarly, dromedary camel, llama and Bactrian camel from UK, China, Germany, Iran and Pakistan appeared as in a separate clade ().
FIGURE 3. Barry and Hartigan's relative rate test for the comparison of evolutionary distance between PrP homologs in different breeds/species.
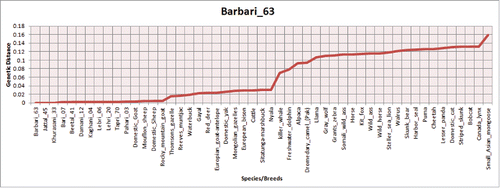
FIGURE 4. Multidimensional Scaling plot of genetic distance between populations (Basques – closed circles, Iberia – open circles, Europe – closed squares, Middle East – cross, Caucasus – closed triangles, North Africa – open triangles). The first 2 axes account for highest genetic variation present in the sample and can be demonstrated that the plot is an accurate representation of the genetic distance matrix. The small ruminant group cluster together on the bottom-left side of the plot, near their bigger ruminant neighbors represented on the upper left place. The canine populations are found near the bottom-right of plot, differentiated from the other 2 ruminant groups.
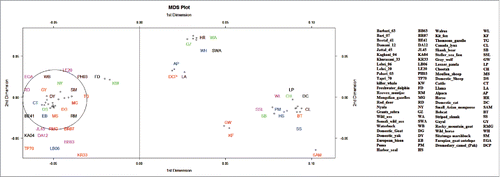
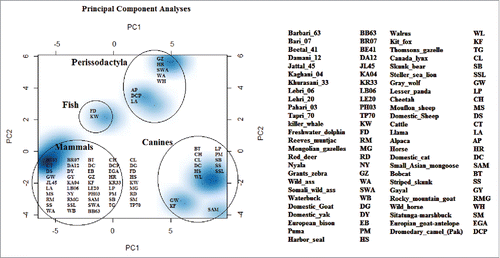
FIGURE 5. Principal component analyses resolve the genetic distance matrix of selected species into PC1, PC2, and PC3 and so on. Out of these, PC1 and PC2 represents large proportion of variation in the dataset and are plotted. PCA results enhanced consistency in our data set as all 3 groups make their spots according to their phylogenetic links. Here PC1 is quoting largest variation in the dataset as compared to PC2. So neighborhood has been revealed between cattle and goat. In that study central nervous system specific human enhancers exhibit 3 distinctive and internally compact clusters exposing the interactive pattern of TFBSs. The control data set of human non-conserved and non-coding elements do not present any vivid cluster structure for the 14 TFBSs that formed internally closely packed clusters in CNS specific enhancers, thus elucidating the significance of clusters in panel A. TFBSs are color coded.
Variations at Protein Sequence Level
Different softwares were used to check out the effect of nucleotide change on primary and secondary structures of protein.Citation11 (Baber et al., 2009;Citation5) The sequence having nucleotide changes at all 6 positions was selected as representative sequence and translated by Expasy Translate tool. As the individuals in this study showed close relatedness with Capra hericus () so the predicted ORF was then aligned with reference prion protein of Capra hircus by using Clustal Omega which resulted in 2 conservative, 2 semi-conservatives and 1 non-conservative variation (Figure S1).
Secondary structure of reference caprine prion protein (Figure S2), as well as altered prion protein was predicted by Phyre2 software which has shown absence of alpha helix at 113-117 and break down of single alpha halix (147-159) of PrPC into 2 (150-152 and 155-159) in major prion protein (Figure S3). These variations may cause change in folding pattern of protein which may convert PrPC to PrPSC.Citation6 This shift could be a major cause of scrapie in goats.
Genetic Distance Evaluation
Distance profiling in biological sciences is an important technique to reveal the level of diversity among individuals of a population/community. Several algorithms were available in form of software packages as well as others have been implemented in Bioconductor. In this study, we used BH87 computational model to generate distance profile against the selected individuals of different breeds/species (). The outcome of this model was a table containing pair wise distance values in it. Diversity pattern, constructed using these computed distance values, is shown in .Citation18
MDS Plot
Multidimensional scaling (MDS) is a tool by which researchers can obtain quantitative estimates of similarity/dissimilarity among groups of items. We provided input in the form of pair wise distance matrix generated for all species mentioned above using BH87 statistical model (). The outcome of MDS, conveyed us the relationships among member sequences. Similar items wherein were seemed to be located proximal to one another, and more diverse items were found proportionately further apart. For example, 3 clusters were uniquely located at variable spots in the MDS diagram (). Member species of each cluster were more related to each other as compared to the individuals of a different cluster as shown in MDS plot. N-1 dimensions (N for number of sequences) were implicitly computed for our total sequence data set. We screened out 1st 2 dimensions only to observe variation pattern in the current data set.Citation19 In this analysis, slightly scattered pattern of large ruminant cluster suggests greater substitution rates in their PrP region as compared to goat and canine species.
2D Loading PCA
Principal components analysis can be used to explore a large data matrix for the direction of largest variation. The whole idea of principal components analysis was to find new directions in the data along with maximal variation. A direction was defined as a linear combination Zk of the data Z by a vector k with weights. So, we employed bioconductor implementation to achieve linear combinations with the help of 2 implicitly calculated Eigen values. These Eigen values provided transformations of our genetic dataset in order to give direction of maximum source variation in the distance profile.
PrP gene along with its homologs was the targeted genomic regions of the selected species. For this purpose, previously calculated genetic distance profile using BH87 computational model (), was used to generate initially 2 linear combinations such as PC1 and PC2 residing in the as vertical and horizontal axis. In total, 70 homologous genes from different organisms were included in our data set. According to PCA, it revealed that highest variation may be due to the presence of top most cluster which were resident very far from 2 other clusters. In a group, the members were less variable with respect to each other.Citation20
Final identification of the cluster structures in the binding site preferences of the training and control data set was achieved with the application of Principal Component Analysis (PCA). The first 2 PCs explained maximum of the total variation present in the patterned correlated data set. The 2D loading plot of the first 2 PCs derived from PCA on control data exposed a scenario of interactive pattern of prions across the species in the training data set. The 2D loading plot exhibits clarity of 3 distinct sets of closely packed prions, i.e., 3 cluster configurations of species. The cluster configurations presented coincide with the cluster configuration perceived in RX = [r2ij]. The species which co-occurring with the PrPs packed in cluster-3 were also merged in it.
Phylogenetic Tree Reconstruction
A NJ phylogenetic tree was reconstructed using PrP sequences of 50 individuals including 10 sample sequences. Rest of other homologs was retrieved from GenBank. The rectangular view showed that all our local goat breeds placed themselves in one clade indicating their genetic relatedness based on PrP gene. In the tree diagram (), our breeds can be observed as the closest relative of Capra hircus, then domestic yak, European bison, gazelles and cattle comes after these species. Killer whale, dolphin appeared in a separate clade. Wild ass, horse and zebra came as close relative in another separate clade. Similarly, alpaca, llama and Bactrian camel from Pakistan appeared as in a separate clade. In the last clade fox, wolf, canada lynx, bobcat, cheetah, puma, mongoose, walrus, sea lions, seal, skunk bear and lesser panda appeared.
As far as the first clade was concerned, regular trend of all goats were observed in which all breeds showed close relatedness with each other. Another important thing was also observed that domestic sheep placed itself into goat clade. Barbari-58, Khurasani-36, Lehri-43, Kaghani-03, Beetal-26 and Mouflon sheep place themselves closer to other mammals.
DISCUSSION
Ovine and caprine management has endured huge economic losses due to scrapie epidemics in Europe. In the majority of countries, the prevalence of natural scrapie in goats remained underrated as compared to sheep. Studies to produce information on PrP gene polymorphisms and their distribution in goat breeds domesticated in diverse countries have become progressively more important, especially after the discovery of BSE or prions in goats in the 1990s.Citation21 Surely, identification of candidate PrP gene polymorphic alleles that may confer susceptibility or resistance to goat scrapie, was a beginning step in understanding the likelihood of any selection program. Modern gene sequence technologies have now paved the path for breeders to make sheep and goats resistant against distressing disorders including scrapie by helping them in determining the allele frequencies, which is a prerequisite to devise an appropriate breeding plan for reasons of animal welfare as well as human food safety and food security.
This study provides new evidence in Pakistani goats for an association of PrP gene polymorphisms with resistance to classical scrapie. In contrast there is no indication for gene variants studied in an association with increased susceptibility to classical scrapie, such as the one found for codon 136 valine polymorphism of European ovine PrP.Citation22 Of the 6 PrP coding region polymorphisms that were observed in investigated herds (G102W, S127G, N135S, H143R, K231R and S240P), one N135S was described for the first time in the Pakistani goat population. In conformity with previous association studies, we found that the goat PrP gene was highly variable in all the 11 breeds studied with majority of animals showing H143R polymorphism. Among caprine PrP polymorphisms I142M, H143R, N146S/D, R154H, R211Q and Q222K which has been reported both in EuropeanCitation23 and Asian sheep breedsCitation24 in association to scrapie resistance, only H143R was detected for the studied goats.Citation13 This polymorphism has also been identified in other Pakistani goat breeds like Pak Angora, Teddy and BeetalCitation8 along with G102WCitation25 and K231RCitation26 in European caprine (). Moreover, F141L and R154H that were risk factors for typical scrapie and has been reported in some EuropeanCitation27,28,29,30 and AsiaticCitation31 breeds, were not detected in the populations of Pakistani studied goats. Our genetic analysis also revealed an association of S127G with a decreased probability to develop clinical scrapie in agreement to European breeds.Citation22 It appears that domestic goat populations maintain a number of PrP polymorphisms as might be expected from balancing selection and the majority of these polymorphisms were associated with a degree of protection against prions compared to the wild type. We were interested to see if a goat population from a region which had never any confirmed scrapie outbreaks would show equally diverse PrP genetics as the UK or other scrapie affected countries (). A comprehensive list of already reported polymorphisms as well as published from Pakistani biologists was generated. Out of 6 variants, 5 named G102W, S127G, H143R, K231R and S240P were in correspondence with the variant list from all over the world. Here we reported a novel genotype N135S identified in Pakistani goats would of great interest for future correspondence. In general, c.428 position was observed as highly polymorphic in all breeds with homozygous G, A and A/G heterozygous genotypes (). With respect to that site, approximately 81.25% of individuals were found polymorphic (). Out of them, 71.01% individuals were heterozygous for this particular site. Lowest percentage of polymorphism was achieved at c.379 as 3.75%. Similarly, significant percentage of polymorphisms were obtained at c.126, c.304, c.414 and c.718 sites as 35%, 7.5%, 36.25% and 28.75% respectively (). Whereas, heterozygous sites were evaluated as proportion to the cumulative polymorphic variation. Surprisingly, all species had adopted the heterozygous trend as compared to be homozygous () which might had made them less susceptible to scrapie.
This important PrP sequences with important variation profiles from scrapie resistant individuals were further exploited to discover its evolutionary mechanism. In distance plot, MDS, PCA and phylogenetic analysis, these species/breeds had shown their origin were of great interest. Analyzing all sequences with respect to multiple angles generated their origin association with other species/breeds and on the basis of this gene deer and antelope species were clustered with Capra hircus. Through sequence analyses we came up with an important finding that killer whale has the lowest rate of variation while mongoose seemed to possess the largest rate of substitutions (). Similarly, MDS and PCA () diagrams elucidated that all canine sequences had observed the highest level of variations described by PC1 (representing 75% of variation alone).
ABBREVIATIONS
| MRCA | = | Most Recent Common Ancestors |
| TSEs | = | Transmissible Spongiform Encephalopathies |
| CJD | = | Creutzfeldt-Jakob disease |
| TME | = | Transmissible mink encephalopathy |
| GPI | = | Glycosyl-phosphatidyl inositol |
| CDS | = | Coding DNA sequence |
| ORF | = | Open Reading Frame |
| MDS | = | Multidimensional scaling |
| EDTA | = | Ethylenediamine tetra-acetic acid |
| SNPs | = | Single Nucleotide Polymorphism |
| PrPc | = | Cellular prion protein |
| PrPsc | = | Scrapie prion protein |
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
We have no conflicts of interest to disclose; all authors approved the manuscript and this submission.
KPRN_A_1178422_Supplementary_Material.zip
Download Zip (135.3 KB)Funding
This work was supported by the Natural Science Foundation of China (Project No. 31472166).
REFERENCES
- Aguzzi A, Lakkaraju AK. Cell Biology of Prions and Prionoids: A Status Report. Trends in cell biology. (2016); 26(1):40–51; http://dx.doi.org/10.1016/j.tcb.2015.08.007
- Huang Z, Gabriel JM, Baldwin MA, Fletterick RJ, Prusiner SB, Cohen FE. Proposed three-dimensional structure for the cellular prion protein. Proc Natl Acad Sci 1994; 91(15):7139–43.
- Colby DW, Prusiner SB. Prions. Cold Spring Harb. Perspect Biol 2011; 3:a006833; PMID:21421910
- Beringue V, Vilotte JL, Laude H. Prion agent diver- sity and species barrier. Vet Res 2008; 39(4):1–30; PMID:18519020
- Cassard H, Torres JM, Lacroux C, Douet JY, Bene- stad SL, Lantier F, Reine F. Evidence for zoonotic potential of ovine scrapie prions. Nat Commun 2014; 5:5821; PMID:25510416
- Wang J, Wang X, Gao X, Vortmeyer AO. Prion diseases and their prpsc-based molecular diag- nostics. J Neurol Neurosci 2002; 4(6):56.
- Westergard L, Heather M, Christensen David A, Harris . The cellular prion protein (PrPC): Its physi- ological function and role in disease. Biochimicaet- BiophysicaActa 2007; 1772:629–644
- Babar ME, Farid A, Benkel BF, Ahmad J, Sajid IA, Imran M, Hussain T, Nadeem A. Genetic variability at seven codons of the prion protein gene in nine Pakistani sheep breeds. J Genet 2008; 87(2):187–90; PMID:18776650; http://dx.doi.org/10.1007/s12041-008-0029-z
- Ford MJ, Burton LJ, Morris RJ, Hall SM. Selective expression of prion protein in peripheral tissues of the adult mouse. Neuroscience 2002; 113:177–92; PMID:12123696
- Moser M, Colello RJ, Pott U, Oesch B. Developmental expression of the prion protein gene in glial cells. Neuron 1995; 14:509–17; PMID:7695897; http://dx.doi.org/10.1016/0896-6273(95)90307-0
- Babar ME, Farid A, Benkel BF, Ahmad J, Nadeem A, Imran M. Frequencies of PrP genotypes and their implication for breeding against scrapie susceptibil- ity in nine Pakistani sheep breeds. Mol Biol Rep 2009; 36(3):561–5; PMID:18297414; http://dx.doi. org/10.1007/s11033-008-9214-7
- Mabbott, Neil Andrew. “Prospects for safe and effective vaccines against prion diseases.” Expert review of vaccines 14.1 (2015): 1–4. DOI: 10.1586/14760584.2015.965691
- Hussain A, Babar ME, Imran M, Haq IU, Javed MM. Detection of four novel polymorphisms in PrP gene of Pakistani sheep (Damani and Hashtnagri) and goats (Kamori and Local Hairy) breeds. Virol J 2011; 8(1):1.
- Harris DA, Peters PJ, Taraboulos A, Lingappa VR, DeArmond SJ, Prusiner SB. Cell biology of prions, in: SB Prusiner (Ed.), Prion Biology and Diseases. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY 2004; 483–544
- Hussein, Mansour F., and Saud I. Al-Mufarrej. “Prion Diseases: A Review.” Scientific Journal of King Faisal University (Basic and Applied Sciences) 5.2 (2004): 1425
- Maryam, J. Babar ME, Nadeem A, Hussain T. “Genetic variants in interferon gamma (IFN-γ) gene are associated with resistance against ticks in Bos taurus and Bos indicus."Molecular biology reports 39.4 (2012): 4565–4570
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA6: Molecular evolutionary genetics analysis using maximum likelihood, evolu- tionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28:2731–9; PMID:21546353; http://dx.doi.org/10.1093/molbev/msr121
- Barry, Daniel, and J. A. Hartigan. “Statistical analysis of hominoid molecular evolution.” Statistical Science (1987): 191–207. http://dx.doi.org/10.1214/ss/1177013353
- Cox TF, Cox MA. Multidimensional scaling. Boca Raton, FL: CRC Press; 2000.
- Venables, WN, “Ripley," BD. (15197). Modern Applied Statistics with S—Plus. 2nd edition. New York: Springer-Verlag; 2002
- Foster JD, Parnham DW, Hunter N, Bruce M. Distribution of the prion protein in sheep terminally affected with BSE following experimental oral transmission. J Gen Virol 2001; 82(10):2319–26; PMID:11562525
- Goldmann W, Ryan K, Stewart P, Parnham D, Xicohtencatl R, Fernandez N, Foster J. Caprine prion gene polymorphisms are associated with decreased incidence of classical scrapie in goat herds in the United Kingdom. Vet Res 2011; 42(1):110; PMID:22040234
- Holko I, Novackova A, Holkova T, Kmet V. PrP genotyping of sheep breeds in Slovakia. Vet Rec 2005; 157(20):628; PMID:16284332
- Barillet F, Mariat D, Amigues Y, Faugeras R, Caillat H, Moazami-Goudarzi K, Perrin-Chauvineau C. Identification of seven haplotypes of the caprine PrP gene at codons 127, 142, 154, 211, 222, and 240 in French Alpine and Saanen breeds and their association with classical scrapie. J Gen Virol 2009; 90(3):769–776; PMID:19218225
- Goldmann W, Chong A, Foster J, Hope J, Hunter N. The shortest known prion protein gene allele occurs in goats, has only three octapeptide repeats and is non-pathogenic. J Gen Virol 1998; 79:3173–6; PMID:9880037; http://dx.doi.org/10.1099/0022-1317-79-12-3173
- Zhang L, Li N, Fan B, Fang M, Xu W. PRNP polymor- phisms in Chinese ovine, caprine and bovine breeds. Anim Genet 2004; 35:457–61; PMID:15566469; http:// dx.doi.org/10.1111/j.1365-2052.2004.01204.x
- Dawson, M. Hoinville LJ, Hosie BD, Hunter N, “Guidance on the use of PrP genotyping as an aid to the control of clinical scrapie.” Veterinary Record 142.23 (1998): 623–625
- Buschmann, Anne, Lühken G, Schultz J, Erhardt G, Groschup MH. “Neuronal accumulation of abnormal prion protein in sheep carrying a scrapie-resistant genotyp(PrPARR/ARR).” Journal of General Virology 85.9 (2004): 2727–2733.
- Goldmann W, Baylis M, Chihota C, Stevenson E, Hunter N. Frequencies of PrP gene haplotypes in British sheep flocks and the implications for breeding programmes. J Appl Microbiol. 2005;98(6):1294–302. PMID: 15916643.
- Saunders GC, Cawthraw S, Mountjoy SJ, Hope J, Windl O. PrP genotypes of atypical scrapie cases in Great Britain. J Gen Virol. 2006 Nov;87(Pt 11):3141–9. PMID:17030846.
- Lan, Z. Wang ZL, Liu Y, Zhang X."Prion protein gene (PRNP) polymorphisms in Xinjiang local sheep breeds in China.” Archives of virology 151.10 (2006): 2095–2101
- Vaccari G, Panayiotidis CH, Acin C, Peletto S, Bar- illet F, Acutis PL, Bossers A, Langeveld J, van Keu- len L, Sclaviadis T, et al. State-of-the-art review of goat TSE in the European Union, with special emphasis on PRNP genetics and epidemiology. Vet Res 2009; 40:48; PMID:19505422; http://dx.doi.org/10.1051/vetres/2009031
- Billinis C, Panagioidis HC, Psychas V, Argyroudis S, Nicolaou A, Leontides S, Papadopoulos O, Skla- viadis T. Prion protein gene polymorphisms in natu- ral goat scrapie. J Gen Virol 2002; 83:713–21; PMID:11842266; http://dx.doi.org/10.1099/0022-1317-83-3-713
- Agrimi U, Conte M, Morelli L, Di Bari MA, Di Guardo G, Ligios C, Antonucci G, Aufiero GM, Pozzato N, Mutinelli F, et al. Animal transmissible spongiform encephalopathies and genetics. Vet Res Commun 2003; 27:31–8; PMID:14535366; http:// dx.doi.org/10.1023/B:VERC.0000014115.18327.26
- Papasavva-Stylianou P, Kleanthous M, Toumazos P, Mavrikiou P, Loucaides P. Novel polymorphisms at codons 146 and 151 in the prion protein gene of Cyprus goats, and their association with natural scrapie. Vet J 2007; 173:459–62; PMID:16314132; http://dx.doi.org/10.1016/j.tvjl.2005.09.013
- Goldmann W, Martin T, Foster J, Hughes S, Smith G, Hughes K, Dawson M, Hunter N. Novel polymorphisms in the caprine PrP gene: a codon 142 mutation associated with scrapie incu- bation period. J Gen Virol 1996; 77:2885–91; PMID:8922485; http://dx.doi.org/10.1099/0022-1317-77-11-2885
- Acutis PL, Colussi S, Santagada G, Laurenza C, Maniaci MG, Riina MV, Peletto S, Goldmann W, Bossers A, Caramelli M, et al. Genetic variability of the PRNP gene in goat breeds from Northern and Southern Italy. J Appl Microbiol 2008; 104:1782–9; PMID:18217941; http://dx.doi.org/10.1111/j.1365-2672.2007.03703.x
- Acutis PL, Bossers A, Priem J, Riina MV, Peletto S, Mazza M, Casalone C, Forloni G, Ru G, Caramelli M. Identification of prion protein gene polymor- phisms in goats from Italian scrapie outbreaks. J Gen Virol 2006; 87, 1029–33; PMID:16528054; http://dx.doi.org/10.1099/vir.0.81440-0
- Wopfner F, Weidenhofer G, Schneider R, Von Brunn A, Gilch S, Schwarz TF, Werner T, Schatzl M. Analysis of 27 mammalian and 9 avian PrPs reveals high conservation of flexible regions of the prion protein. J Mol Biol 1999; 289:1163–78; PMID:10373359; http:// dx.doi.org/10.1006/jmbi.1999.2831
- Zhou RY, Li XL, Li LH, Wang HY, Lu JG. Poly- morphism of the PRNP gene in the main breeds of indigenous Chinese goats. Arch Virol 2008; 153:979–82; PMID:18369524; http://dx.doi.org/10.1007/s00705-008-0074-1
- Vaccari G, Di Bari MA, Morelli L, Nonno R, Chiappini B, Antonucci G, Marcon S, Esposito E, Fazzi P, Palaz- zini N, et al. Identification of an allelic variant of the goat PrP gene associated with resistance to scrapie. J Gen Virol 2006; 87:1395–402; PMID:16603543; http:// dx.doi.org/10.1099/vir.0.81485-0
- Kurosaki Y, Ishiguro N, Horiuchi M, Shinagawa M. Polymorphisms of caprine PrP gene detected in Japan. J Vet Med Sci 2004; 67:321–3; PMID:15805738; http:// dx.doi.org/10.1292/jvms.67.321