?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
The detection method based on the mathematical expectation (ME) strategy is fast and accuracy for low frequency mutation screening in large samples. Previous studies have found that the 14-bp insertion/deletion (indel) variants of the 3′ untranslated region (3′ UTR) within bovine PRNP gene have been characterized with low frequency (≤5%) in global breeds outside China, which has not been determined in Chinese cattle breeds yet. Therefore, this study aimed to identify the 14-bp indel within PRNP gene in 5 major Chinese indigenous cattle breeds and to evaluate its associations with phenotypic traits. It was the first time to use ME strategy to detect low frequency indel polymorphisms and found that minor allele frequency was 0.038 (Qinchuan), 0.033 (Xianan), 0.013 (Nanyang), 0.003 (Jiaxian), and zero (Ji'an), respectively. Compared to the traditional detection method by which the sample was screened one by one, the reaction time by using the ME method was decreased 62.5%, 64.9%, 77.6%, 88.9% and 66.4%, respectively. In addition, the 14-bp indel was significantly associated with the growth traits in 2 cattle breeds, with the body length of Qinchuan cattle as well as the body weight and waistline of Xianan cattle. Our results have uncovered that the method based on ME strategy is rapid, reliable, and cost-effective for detecting the low frequency mutation as well as our findings provide a potential valuable theoretical basis for the marker-assisted selection (MAS) in beef cattle.
INTRODUCTION
Chinese indigenous cattle breeds are characterized with many potential advantages (e.g. strong disease resistance, good adaptability), however, they also demonstrate several serious shortcomings, e.g., slow growth, serious varieties degradation, so these obvious disadvantages need to be rapidly and accurately improved using marker-assisted selection (MAS). Molecular markers includes the single nucleotide polymorphism (SNP) and the insertion/deletion (indel). Compared with the SNPs, indel variants have the advantage of convenient detection and remarkable effects as well as they also are characterized with low minor allele frequency (<10%) across bovine genome, such as mutations in the bovine ALX4 gene,Citation1 lymphotoxin A geneCitation2 and DGAT1 gene.Citation3 For the minor allele frequency detection, the traditional detection methods by which the sample was screened one by one are very time-consuming and high-cost, therefore, a quick and accurate method is required for application on detecting the low frequency mutation of potential critical candidate genes, which would improve crucial flaws in beef cattle industry using the MAS strategy.
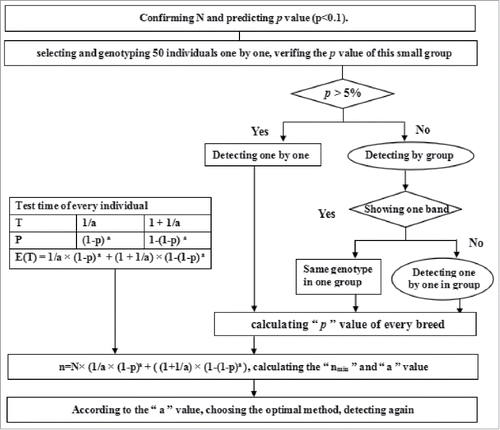
To date, a method based on the theory of mathematical expectation (ME) has been widely applied to detect low frequency events in epidemiology, population gnetics, quantitative genetics, molecular evolution, e.g. the general screening of parasites, tuberculosis, leukemia in large samples.Citation4 In details, principal of the ME method is described as followsCitation4-6: all detected individuals are randomly divided into several mixed groups, and each mixed groups is genotyped; if a mixed group shows the single specific genotype, then all mixed individuals are regarded that possess the same genotype; while if one mixed group demonstrates different genotypes, then the genotype of each individual is required to be identified again. So, this ME method can quickly and accurately detect the genotypes of the individuals, which would save time and decrease expenses.
Prion protein (PrP) encoded by the PRNP gene plays a critical role in bovine spongiform encephalopathy (BSE) in cattle and buffalo, and scrapie in goat and sheep.Citation7-9 Furthermore, numerous polymorphisms of PRNP gene have been shown to influence the susceptibility of these diseases,Citation10-14 and they significantly decreased the production, milk traits in several animal breeds.Citation15-21 To date, several polymorphisms were reported in the cattle PRNP gene, including a 23-bp indel,Citation22 a 12-bp indel, a 14-bp indel,Citation16 and the variable number of octapeptide repeats in the open reading frame.Citation17 According to the previous reports, the 14-bp indel in 3′ UTR have been characterized with low frequency (≤5%) in breeds outside China. For instance, the minor allele frequency of the 14-bp indel was less than 0.01 in cows and artificial insemination sires of the USA.Citation23 However, the frequency of this indel locus has not been determined in Chinese cattle. Therefore, the 14-bp indel was genotyped in healthy animals of Chinese indigenous cattle breeds.
Qinchuan, Nanyang, Jiaxian and Ji'an cattle are indigenous beef cattle breeds in China,Citation24 whereas Xianan cattle are Chinese first crossbreeding beef breed. However, the frequency of the 14-bp indel polymorphism of PRNP as well as whether this indel affects growth traits in these breeds have not been elucidated until now. Therefore, the aim of the present study was to use ME strategy to determine the 14-bp indel polymorphism in Chinese cattle breeds and to analyze the relationship between this indel and growth traits in healthy cattle. The results may provide the basis for MAS of beef cattle.
MATERIAL AND METHODS
All animal experiments were implemented in accordance with the relevant laws and institutional guidelines and approved by the Institutional Animal Care and Use Committee of Northwest A&F University (NWSUAF).
Animals and Data Collection
A total of 1005 Chinese cattle from 5 cattle breeds, Qinchuan (n = 160), Xianan (n = 214), Jiaxian (n = 199), Nanyang (n = 197), and Ji'an (n = 235) were randomly sampled. These five groups are the main Chinese cattle breeds distributed in the provinces of Shaanxi (Qinchuan), Henan (Xianan, Jiaxian, Nanyang), and Jiangxi (Ji'an). Growth traits and physical parameters in unrelated Nanyang cattle were measured at birth and after 6, 12, 18 and 24 months, which included body weight (kg), body height (cm), body length (cm), heart girth (cm), hucklebone width (cm), average daily weight gain (kg). Records of growth traits in unrelated adult Qinchuan and Jiaxian cattle were also measured at similar intervals, which included body weight (kg), body height (cm), body length (cm), chest girth (cm), chest width (cm), chest depth (cm), cannon circumference (cm), rump length (cm), height at hip cross (cm), hucklebone width (cm), hip width (cm), back height (cm). Meanwhile, records of growth traits in unrelated adult Ji'an and Xianan cattle were at similar intervals, which included body weight (kg), body height (cm), body length (cm), chest girth (cm), hucklebone width (cm), and the average daily weight gain (kg).Citation24,25
Genomic DNA Isolation and DNA Pool Construction
Genomic DNA was isolated from the leukocyte fraction of the blood or the ear tissue following the procedures described by Lan et al.Citation26 and Pan et al.Citation24 The genomic DNA concentration was quantified, and the working solution of each DNA sample was brought to 50 ng/µL.Citation26 In each breed, a total of 50 DNA samples were randomly selected to construct genomic DNA pools, respectively.Citation26 The genomic DNA pools were used as a template for PCR amplification of the 14-bp indel within PRNP gene.
PCR Amplification and Genotyping of the 14-bp Indel of PRNP
Based on bovine PRNP gene sequence (AC_000170.1), a pair of primers (F: 5′-TGGCTTGCACTTTGTGGTAT-3′; R: 5′-CCCACGTCTCCTTAGTACCTT-3′) was designed to amplify DNA pools within region covering the PRNP 3′UTR indel.Citation11,15 The PCR was performed in 25 μL reaction volume containing 1.0 μL genomic DNA (constructed from 10 different individuals), 0.5 μL each primer (forward and reverse primer), 12.5 μL 2× Eco Taq PCR Super mix (+dye) and 10.5 μL ddH2O. Touch Down PCR reaction was carried out as follows: initial denaturation for 4 min at 95°C, followed by 18 cycles of denaturation for 30 s at 94°C, annealing for 30 s at 68°C (with a decrease of 1°C per cycle), and extension for 1000bp/min at 72°C, another 23 cycles of 30 s at 94°C, 30 s at 50°C, and 45 s at 72°C, and a final extension of 10 min at 72°C. The samples were then cooled to 4°C. The PCR products specificity was confirmed by sequencing.Citation11 The 14-bp indel of PRNP was detected in 5 Chinese indigenous cattle breeds by electrophoresis using 3.5% agarose gel stained with ethidium bromide.
The Theory of ME on Low Frequency Mutations
The ME, also known as the mean value, is very important digital feature in the theory of possibility. It represents the overall average value of random variables. The model of ME can be used to analyze abstract methods in practical application.Citation4-6 Given the previous reported data, the frequencies of the 14-bp indel within PRNP were predicted to be low in the tested Chinese cattle breeds.Citation23 Therefore, initial randomly selected 50 animals were genotyped separately (not pooled) to establish the allele frequencies in a population. The experimental results confirmed the low frequency of the 14-bp indel in PRNP gene in the studied Chinese cattle breeds. So, the theory of ME has value of application.Citation4-6
Then, 3 individuals pooled in mixed groups were established by an ME equitation. Subsequently, all animals were divided into pools and were genotyped. If two bands were observed in a pool, the animals were genotyped separately to specifically assign the genotype. In this experiment, the low mutation frequency of 14-bp indel was identified in every cattle breed ().
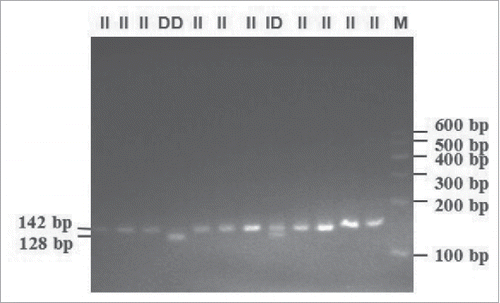
FIGURE 2. Electrophoresis pattern of 14-bp indel in 3′ UTR within bovine PRNP gene (genotyped by 3.5% agarose gel) (II demontrated 142-bp length band; ID demonstrated 142-bp and 128-bp length bands; DD demonstrated 128-bp length band).
Therefore, given the low frequency events, according to the theory of ME, the best method is to use the following equation.Citation4-6:where N is the sample size, a is the sum of a group, p is the probability of the low frequency event (http://www.msrcall.com/DALMcall.aspx).
Using the above equation, we calculated the optimal number of individuals in a mixed group for each breed. We designed the most possible pooling strategy for each cattle breed, and the mixed group was genotyped. The results of individuals were same to the genotype of every individual. Simultaneously, the genotype of every individual was verified by the result obtained from mixed group of 3 individuals.
Statistical Analysis
Genotypic and allelic frequency were calculated directly. The χ2 test was carried out to test whether the polymorphism is in Hardy-Weinberg equilibrium (HWE). Polymorphism information content (PIC) was calculated by Nei's method implemented in the GDIcall Online Calculator (http://www.msrcall.com/Gdicall.aspx).Citation23 Distribution differences for genotypic and allelic frequencies among/between different breeds were analyzed using the χ2 test implemented in SPSS (Version 18.0) (IBM Corp., Armonk, NY, USA).Citation24 The association of the 14-bp indel of PRNP with several growth traits (e.g., body length (cm)) in different breeds was tested using the analysis of variance (ANOVA) available in SPSS (Version 18.0). The data that did not follow normal distribution and homogeneity of variances were analyzed using the nonparametric (Kruskal-Wallis) test in SPSS (Version 18.0).The ANOVA applied the general linear model and the reduced linear model as follows: Yijk = μ + αi + βj + ϵijk, where Yijk is the observation of the reproduction trait (testis weight, etc.) evaluated on the ith level of the fixed factor age (αi) and the jth level of the fixed factor genotype (βj), μ is the overall mean for each trait, and ϵijk is the random error for the ijkth individual.Citation24
RESULTS
Previous reports identified a 14-bp indel in the 3′ UTR of the bovine PRNP gene. The direct 3.5% agarose gel electrophoresis conducted herein detected one band (142-bp) for genotype insertion/insertion (II), one band (128-bp) for genotype deletion/deletion (DD), and 2 bands (142-bp, 128-bp) for genotype insertion/deletion (ID) (). These results were consistent with those previous reports.Citation23
Initially, all individuals were genotyped. The genotypic and allelic frequencies of the 14-bp indel of PRNP were evaluated in the Qinchuan, Xianan, Nanyang, and Jiaxian cattle breeds. The minor allele frequency was 0.038 (Qinchuan), 0.033 (Xianan), 0.013 (Nanyang), 0.003 (Jiaxian), and zero (Ji'an). The effective allele numbers were 1.078 (Qinchuan), 1.067 (Xianan), 1.026 (Nanyang), 1.008 (Jiaxian), and 1.000 (Ji'an). The polymorphism information content values of this indel were low for all analyzed breeds. Moreover, χ2 tests showed that the 14-bp indel in the 3′ UTR of PRNP were in HWE in all analyzed breeds (P > 0.05), except in the Xianan breed.
The low frequency of 14-bp indel was characterized in PRNP for the 5 Chinese cattle breeds (). Using the ME method, individuals of each breed were assigned by order in groups (the least allowed number in a single group) to mixed groups. Hence, Qinchuan, Xianan, Nanyang, Jiaxian, and Ji'an breeds were divided into 54, 72, 66, 67, and 79 mixed groups (). Dependent on whether there was one single band (142-bp or 128-bp) in the mixed groups from different cattle breeds, the exact detection times were shown. Finally, a total of 90, 100, 81, 70, and 79 reactions times were carried out in Qinchuan, Xianan, Nanyang, Jiaxian, and Ji'an cattle, respectively. Simultaneously, the number of PCR reactions was decreased by 43.7%, 53.3%, 58.9%, 64.8% and 66.4%, respectively, compared to those obtained by the traditional detection methods ().
TABLE 1. Genotypic and allelic frequencies and population indexes for 14-bp indel in the bovine PRNP gene.
TABLE 2. All reaction times of different group in 5 cattle breeds.
In view of the minor allelic frequency of Ji'an cattle breed was 0.000, so, ME was not applied to this breed. Initial randomly selected 50 animals were genotyped separately (not pooled) to establish the allele frequencies in a population. Then, according to the ME evaluation equation, the optimal number of individuals in a single mixed group was 5 (Qinchuan), 6 (Xianan), 8 (Nanyang), and 16 (Jiaxian). Thereby, number of animals to be pooled in mixed groups was established by an ME equitation. Subsequently, all animals were divided into 32, 36, 25, and 13 mixed pools and genotyped, respectively. If two bands were observed in a pool, the animals were genotyped separately to specifically assign the genotype. At last, the exact reaction times were shown. Finally, a total of 60, 75, 44, and 22 reaction times were carried out in Qinchuan, Xianan, Nanyang, and Jiaxian cattle, respectively. Simultaneously, the reaction times were decreased by 62.5%, 64.9%, 77.6%, and 88.9%, respectively, compared to those obtained by the traditional detection method. These results indicated that the ME detection method saves time greatly, which will ultimately reduce the expenses, compared to the traditional methods ().
The associations between the 14-bp indel of the 3′ UTR and the bovine growth traits were evaluated in the tested Chinese cattle breeds (). Because there were less than 5 individuals with ID genotype in Nanyang, Jiaxian, and Ji'an breeds, the respective data were not included in the analysis. The 14-bp indel was significantly correlated to the body length in Qinchuan cattle as well as the body weight and waistline in Xianan cattle; the homozygous insertion genotype was predominant in these breeds, respectively.
TABLE 3. Relationship between the 14-bp indel of PRNP gene and growth traits in Xianan and Qinchuan cattle breeds (LSMa±SE) (P < 0.05).
DISCUSSION
This study is the first report of the 14-bp indel in the 3′ UTR of PRNP gene in 5 Chinese indigenous cattle breeds: Qinchuan, Xianan, Nanyang, Jiaxian, and Ji'an. Given the low frequency of the 14-bp indel within the 3′ UTR in cows and artificial insemination sires of the USA (0.006 and zero, respectively),Citation23 in Vietnamese native cattle (0.18) and Laos native populations of Asian origin (0.22)Citation27 (), we hypothesized that the frequency of the 14-bp indel of the 3′ UTR in PRNP gene may be low in Chinese cattle breeds. We used an ME strategy to detect the allele frequency in all individuals. Simultaneously, we verified experimentally that the frequency of the 14-bp indel in the 3′ UTR was low in 5 cattle breeds. The results of our study were consistent with previous results.Citation10,28 However, they did not corroborate the results presented by Shimogiri,Citation10 who reported the frequency in Japanese Brown of more than 0.10.Citation27 Those discrepancies might be due to the small sample size (n = 64). Overall, the 14-bp indel within the PRNP gene was characterized by low frequency in main Chinese cattle breeds.
Furthermore, our results confirmed that the 14-bp indel in the 3′ UTR of PRNP gene was at HWE in populations of 5 Chinese indigenous cattle breeds except Xianan (P > 0.05), which consisted with the cultural background of different cattle breeds. Xianan cattle are the first crossbreeding beef cattle in China. Hence, crossbreeding might also be a cause of HWE deviation. According to the previous reported data, the 14-bp indel of the 3′ UTR in PRNP was at HWE in most populations of breeds outside China, with the exception of the Japanese Brown,Citation29 artificial insemination sires in USA,Citation23 Japanese Short horn,Citation29 and Kuchinoshima populationsCitation29 (P > 0.05), which might due to the small sample size of these breeds. There were no significant differences in the distribution of genotypic and allelic frequencies for the 14-bp indel among the Qinchuan, Xianan, Nanyang, Jiaxian, and Ji'an breeds (χ2 test, P = 1.000). Moreover, we analyzed the reported data for foreign breeds and obtained the results similar to those reported herein for the 5 Chinese native breeds. Thus, we verified that the distribution of genotypic and allelic frequencies in the 14-bp indel of PRNP showed no significant differences among the different cattle breeds.
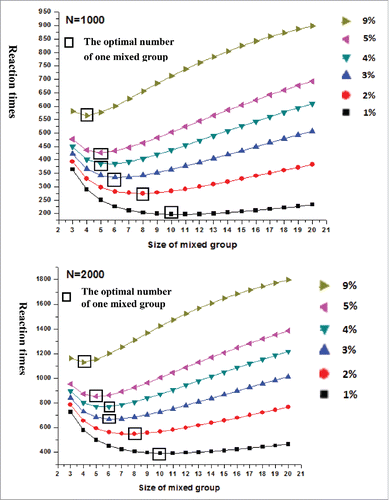
The low frequency of the 14-bp indel in the 3′ UTR of PRNP in Chinese cattle breeds obtained experimentally confirmed that the ME method can be applied to detect low frequency mutations.Citation4-6 This method could be used to calculate the optimal reaction time from the low frequency mutation and sample size. Using the GDIcall Online Calculator (http://www.msrcall.com/Gdicall.aspx), we designed the best possible pooling strategy for 5 cattle breeds, excluding Ji'an cattle breed which mutation frequency was zero. We found that the optimal number of individuals in a group was 5, 6, 8, and 16 in Qinchuan, Xianan, and Nanyang, Jiaxian, respectively, if all reaction time was least (). Moreover, we calculated other methods for division of individuals into groups with different low frequency mutations and sample size by using this equation and identified the best method among them (). Concurrently, using the results of the previous reports on low frequency mutations in breeds outside China, we proposed a better method, which was based on a new method we put forward (http://www.msrcall.com/Gdicall.aspx).Citation12 Thus, for the experiments with U.S. Sires,Citation30 each group should comprise 4 individuals, and any effect that may alter the accuracy of the experiment could be eliminated if the concentration of DNA in all individuals is the same. In other breeds, the ME method was also convenient for quick and accurate detection of the low frequency mutations.
FIGURE 3. Reaction times about different sizes of one mixed group (different mutation frequency(1%–9%) when N = 1000 and N = 2000).
Due to the low number of heterozygous animals (n < 5) in Jiaxian, Nanyang, and Ji'an cattle breeds, we only analyzed the association of the 14-bp indel with growth traits of Qinchuan and Xianan cattle breeds. The results of the association analysis suggested that the 14-bp indel in the 3′ UTR might affect body length of Qinchuan cattle and body weight and waistline of Xianan cattle. It could be concluded that the 14-bp indel in the 3′ UTR or the adjacent area might contain the binding sites of transcriptions factors of the genes affecting the growth traits. When the 14-bp indel in the 3′ UTR of PRNP was inserted or deleted, the structure and function of genes related to the growth traits would be affected by bonding with miRNA.Citation31
In summary, an economic ME method was presented to quickly and accurately detect low frequency mutation, such as the 14-bp indel in the 3′ UTR of the PRNP gene, which would save time and reduce expenses. Besides, the detected 14-bp indel significantly affect growth traits, which might be a potential useful DNA marker for MAS in cattle.
ABBREVIATIONS
| BSE | = | bovine spongiform encephalopathy |
| PrP | = | Prion protein (PrP) |
| PRNP | = | prion protein gene |
| 3′ UTR | = | 3′-untranslated region |
| Indel | = | insertion/deletion |
| SNPs | = | single nucleotide polymorphisms |
| SPSS | = | statistical product and service solutions |
| GLM | = | general linear mode |
| bp | = | base pair |
| HWE | = | Hardy-Weinberg equilibrium |
| Ho | = | homozygosity |
| He | = | heterozygosity |
| Ne | = | effective allele numbers |
| PIC | = | Polymorphism information content |
| ME | = | mathematical expectation |
| PCR | = | polymerase chain reaction |
| ANOVA | = | analysis of variance |
| II | = | insertion/insertion |
| ID | = | insertion/deletion |
| DD | = | deletion/deletion |
| MAS | = | marker-assisted selection |
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
We confirm that this manuscript has not been published in whole or in part and is not being considered for publication elsewhere. The authors have no conflicts of interest to declare.
Funding
This work was supported by the National Natural Science Foundation of China (NO.31672400), the National Project of Scientific Innovation Experiment for Undergraduate of Northwest A&F University (No. 201510712097), Science and Technology Overall Project of Shaanxi Province (No. 2014KTZB02-02-02-02), Program of National Beef Cattle and Yak Industrial Technology System (No. CARS-38), Project of Biological Species Capacity Building and Industrialization from National Development and Reform Commission (No. 2014) and Project of Major Sci-Tech of Henan Province (No. 141100110200).
REFERENCES
- Brenig B, Schmeitz E, Hardt M, Scheuermann P, Freick M. A 20 bp Duplication in Exon 2 of the Aristaless-Like Homeobox 4 gene (ALX4) is the candidate causative mutation for tibial hemimelia syndrome in galloway cattle. PLoS One 2015; 10(6):e0129208; PMID:26076463; http://dx.doi.org/10.1371/journal.pone.0129208
- Behl JD, Mishra P, Verma NK, Niranjan SK, Dangi PS, Sharma R, Behl R. Nucleotide polymorphisms in the bovine lymphotoxin A gene and their distribution among Bos indicus zebu cattle breeds. Gene 2016; 579(1):82-94; PMID:26724420; http://dx.doi.org/10.1016/j.gene.2015.12.049
- Rosse Ida C, Steinberg Rda S, Coimbra RS, Peixoto MG, Verneque RS, Machado MA, Fonseca CG, Carvalho MR. Novel SNPs and INDEL polymorphisms in the 3′UTR of DGAT1 gene: in silico analyses and a possible association. Mol Biol Rep 2014; 41(7):4555-63; PMID:24676595; http://dx.doi.org/10.1007/s11033-014-3326-z
- Zhang YE, Liu GY, Ji AB, Sun JP. The application of the mathematical expectation in the disease census. J Math Med 2003; 16(1):83-4 (in Chinese)
- Guo LJ, Zhang Y. As application examples of the mathematical expectation. The public science and technology 2006; 7:169. ( in Chinese)
- Stephen C. Newman. Biostatistical Methods in Epidemiology; 1rd ed. New York; John Wiley & Sons Publishers; 2001
- Prusiner SB. Prions. Proc Natl Acad Sci USA 1998; 95(23):13363-83; PMID:9811807; http://dx.doi.org/10.1073/pnas.95.23.13363
- Maheshwari A, Fischer M, Gambetti P, Parker A, Ram A, Soto C, Concha-Marambio L, Cohen Y, Belay ED, Maddox RA, et al. Recent US case of variant Creutzfeldt-Jakob disease-global implications. Emerg Infect Dis 2015; 21(5):750-9; PMID:25897712; http://dx.doi.org/10.3201/eid2105.142017
- Sigurdson CJ, Miller MW. Other animal prion diseases. Br Med Bull 2003; 66:199-212
- Kashkevich K, HumeNanyang A, Ziegler U, Groschup MH, Nicken P, Leeb T, Fischer C, Becker CM, Schiebel K. Functional relevance of DNA polymorphisms within the promoter region of the prion protein gene and their association to BSE infection. FASEB J 2007; 21(7):1547-155; PMID:17255470; http://dx.doi.org/10.1096/fj.06-7522com
- Nakamura I, Xue G, Sakudo A, Saeki K, Matsumoto Y, Ikuta K, Onodera T. Novel single nucleotide polymorphisms in the specific protein 1 binding site of the bovine PRNP promoter in Japanese Black cattle: impairment of its promoter activity. Intervirology 2007; 50(3):190-6; PMID:17283444; http://dx.doi.org/10.1159/000099217
- Xue G, Sakudo A, Kim C, Onodera T. Coordinate regulation of bovine prion protein gene promoter activity by two Sp1 binding site polymorphisms. Biochem Biophys Res Commun 2008; 372(4):530-5; PMID:18505676; http://dx.doi.org/10.1016/j.bbrc.2008.05.085
- Sander P, Hamann H, Drögemüller C, Kashkevich K, Leeb T. Bovine prion protein (PRNP): promoter polymorphisms modulated PRNP expression and may be responsible for differences in bovine spongiform encephalopathy susceptibility. J Biol Chem 2005; 280(45):37408-14; PMID:16141216; http://dx.doi.org/10.1074/jbc.M506361200
- Msalya G, Shimogiri T, Nishitani K, Okamoto S, Kawabe K, Minesawa M, Maeda Y. Indels within promoter and intron 1 of bovine prion protein gene modulate the gene expression levels in the medulla oblongata of two Japanese cattle breeds. Anim Genet 2009a; 41(2):218-21http://dx.doi.org/10.1111/j.1365-2052.2009.01983.x
- Brandsma JH, Janss LL, Visscher AH. Association between PrP genotypes and litter size and 135 days weight in texel sheep. Livest Prod Sci 2004; 85:59-64; http://dx.doi.org/10.1016/S0301-6226(03)00116-7
- Lan XY, Zhao HY, Li ZJ, Li AM, Lei CZ, Chen H, Pan CY. A novel 28-bp insertion–deletion polymorphism within goat PRNP gene and its association with production traits in Chinese native breeds. Genome 2012a; 55(7):547-52; http://dx.doi.org/10.1139/g2012-040
- Lan XY, Zhao HY, Wu CY, Hu SR, Pan CY, Lei CZ, Chen H. Analysis of genetic variability at codon 42 within caprine prion protein gene in relation to production traits in Chinese domestic breeds. Mol Biol Rep 2012b; 39(4):4981-8; http://dx.doi.org/10.1007/s11033-011-1294-0
- Alexander BM, Stobart RH, Russel WC, O'Rourke KI, Lewis GS, Logan JR, Duncan JV, Moss GE. The incidence of genotypes at codon 171 of the prion protein gene (PRNP) in five breeds of sheep and production traits of ewes associated with those genotypes. J Anim Sci 2005; 83:455-9; PMID:15644519
- De Vries F, Borchers H, Hamann H, Drogemuller C, Reinecke S, Lupping W, Distl O. Associations between the prion protein genotype and performance traits of meat breeds of sheep. Vet Rec 2004; 155:140-3; PMID:15338706; http://dx.doi.org/10.1136/vr.155.5.140
- Ioannides IM, Mavrogenis AP, Papachristoforou C. Analysis of PrP genotypes in relation to reproductive and production traits in Chios sheep. Livest Sci 2009; 122(2–3):296-301; http://dx.doi.org/10.1016/j.livsci.2008.09.022
- Walawski K, Czarnik U. Prion octapeptide-repeat polymorphism in Polish Black-and-White cattle. J Appl Genet 2003; 44(2):191-5; PMID:12773797
- Zhu XY, Feng FY, Xue SY, Hou T, Liu HR. Bovine spongiform encephalopathy associated insertion/deletion polymorphisms of the prion protein gene in the four beef cattle breeds from North China. Genome 2011; 54(10):805-11; PMID:21923635; http://dx.doi.org/10.1139/g11-043
- Czarnik U, Grzybowski G, Zabolewicz T, Strychalski J, Kaminski S. Deletion/insertion polymorphism of the prion protein gene (PRNP) in Polish red cattle, Polish White-backed cattle and European bison (Bison bonasus L., 1758). Genetika 2009; 45(4):519-25; PMID:19507705
- Pan CY, Wu CY, Jia WC, Xu Y, Hu SR, Lei CZ, Lan XY, Chen H. A critical functional missense mutation (H173R) in the bovine PROP1 gene significantly affects growth traits in cattle. Gene 2013; 531(1):398-402; PMID:24029076; http://dx.doi.org/10.1016/j.gene.2013.09.002
- Zhang S, Dang Y, Zhang Q, Qin Q, Lei C, Chen H, Lan X. Tetra-primer amplification refractory mutation system PCR (T-ARMS-PCR) rapidly identified a critical missense mutation (P236T) of bovine ACADVL gene significantly affecting growth traits. Gene 2015; 559(2):184-8; PMID:25620159; http://dx.doi.org/10.1016/j.gene.2015.01.043
- Lan XY, Pan CY, Chen H, Zhang CL, Li JY, Zhao M, Lei CZ, Zhang AL, Zhang L. An AluI PCR-RFLP detecting a silent allele at the goat POU1F1 locus and its association with production traits. Small Ruminant Res 2007; 73(1):8-12; http://dx.doi.org/10.1016/j.smallrumres.2006.10.009
- Shimogiri T, Msalya G, Myint SL, Okamoto S, Kawabe K, Tanaka K, Mannen H, Minezawa M, Namikawa T, Amano T, et al. Allele distributions and frequencies of the six prion protein gene (PRNP) polymorphisms in Asian native cattle, Japanese breeds, and mythun (Bos frontalis). Biochem Genet 2010; 48(9–10):829-39; PMID:20623331; http://dx.doi.org/10.1007/s10528-010-9364-x
- Uchida L, Heriyanto A, Thongchai C, Hanh TT, Horiuchi M, Ishihara K, Tamura Y, Muramatsu Y. Genetic diversity in the prion protein gene (prnp) of domestic cattle and water buffaloes in Vietnam, Indonesia and Thailand. J Vet Med Sci 2014; 76(7):1001-8; PMID:24705506; http://dx.doi.org/10.1292/jvms.13-0642
- Sander P, Hamann H, Pfeiffer I, Wemheuer W, Brenig B, Groschup MH, Ziegler U, Distl O, Leeb T. Analysis of sequence variability of bovine prion protein gene (PRNP) in GermaNanyang cattle breeds. Neurogenetics 2004; 5(1):19-25; PMID:14727152; http://dx.doi.org/10.1007/s10048-003-0171-y
- Seabury CM, Womack JE, Piedrahita J, Derr JN. Comparative PRNP genotyping of US cattle sires for potential association with BSE. Mamm Genome 2004; 15(10):828-33; PMID:15520885; http://dx.doi.org/10.1007/s00335-004-2400-6
- Hou J, An X, Song Y, Gao T, Lei Y, Cao B. Two mutations in the caprine MTHFR 3′ UTR regulated by microRNAs are associated with milk production traits. PLoS One 2015; 10(7):e0133015; PMID:26186555; http://dx.doi.org/10.1371/journal.pone.0133015