ABSTRACT
Prion-related protein doppel gene (PRND), as an essential member of the mammalian prion gene family, is associated with the scrapie susceptibility as well as phenotype traits, so the genetic variation of the PRND has been highly concerned recently, including the single nucleiotide polymorphism (SNP) and insertion/deletion (indel). Therefore, the objective of present study was to examine the possible indel variants by mathematical expectation (ME) detection method as well as explore its associations with phenotype traits. A novel 20-bp indel was verified in 623 tested individuals representing 4 diversity sheep breeds. The results showed that 3 genotypes were detected and the minor allelic frequency were 0.008 (Lanzhou Fat-Tail sheep, LFTS), 0.084 (Small Tail Han sheep, STHS), 0.021(Tong sheep, TS) and 0.083 (Hu sheep, HS), respectively. Comparing with the traditional method of detecting samples one by one, the reaction times with ME method was decreased by 36.22% (STHS), 37.00% (HS), 68.67% (TS) and 83.33% (LFTS), respectively. Besides, this locus was significantly associated to cannon circumference index (P = 0.012) and trunk index (P = 0.037) in the Hu sheep breed. Notably, it was not concordance with the present result of DNA sequencing (GCTGTCCCTGCAGGGCTTCT) and dbSNPase of NCBI (NC_443194: g.46184887- 46184906delCTGCTGTCCCTGCAGGGCTT). Consequently, it was the first time to detect the new 20-bp indel of sheep PRND gene by ME strategy, which might provide a valuable theoretical basis for marker-assisted selection in sheep genetics and breeding.
INTRODUCTION
The establishment of an association between prion gene (PRNP) polymorphisms and scrapie susceptibility in sheep has enabled the development of breeding programs to increase scrapie resistance in the European Union.Citation1 With the further research on PRNP genotype selection, the polymorphisms of adjacent prion-related doppel gene (PRND), an evolutionarily related paralogue of PRNP, become another key point for animal breeding of resistant varieties. Some studies demonstrated that several polymorphism loci of PRND were associated with scrapie susceptibility, such as G78A (the 26th codon).Citation1 Meanwhile, several polymorphisms of PRND gene had been shown to influence the goat early sex differentiationCitation2 as well as ram fertility,Citation3 especially affecting the differentiation of goat foetal leydig cells.Citation4 Remarkably, at present, there is no study that reveals the relationship of the insertion/deletion (indel) polymorphisms of PRND gene with scrapie susceptibility or growth traits in sheep.
To investigate the association between genetic variation in PRND and growth traits from amounts of samples, molecular markers, including the indel and single nucleotide polymorphism (SNP), are extensive used in the field of animal genetics and breeding nowadays. But the low levels of polymorphism limited application of marker assisted selection (MAS) in sheep breeding. Due to the advantages of convenient detection and remarkable effects, compared with SNP, the indel variants have higher efficiency and wider application. Previous study showed that PRND polymorphic in sheep breeds is poor, for gene sequence of PRND is highly conservative in sheep.Citation3 Therefore, screening samples one by one to detect indel of sheep PRND was time-consuming and high-cost. To detect abundant samples efficiently and accurately, a method based on the theory of mathematical expectation (ME), in our group, has been put and widely applied to detect low frequency mutations.Citation5 Previous study also has demonstrated the feasibility and high efficiency of the ME method in low-frequency detection.Citation5
To detect the possible indel of sheep PRND gene by using ME method, a total of 623 sheep from the 4 domestic sheep breeds (Hu sheep, HS; Small Tail Han sheep, STHS; Lanzhou Fat-Tail sheep, LFTS; Tong sheep, TS) were tested in this study. As excellent sheep varieties of meat and wool, Tong sheep (TS) and Lanzhou Fat-Tail sheep (LFTS) are famous for the large and fat tail, simultaneously, possessing numbers of excellent characteristics, such as crude feed tolerance, higher disease resistance and more resilient than other domestic sheep. While the vast majority of breed sheep is singleton and seasonal oestrus, the Small Tail Han sheep (STHS) and Hu sheep (HS), have a very high reproductive capacity and fast-growth. Particularly, living in warm and moist environment result in some notable flaws in Hu sheep, such as poor resistance of cold, affecting growth and development of sheep. Obviously, to obtain more profiles in sheep breeding, those shortcomings need to be improved and the relevant gene should be detected. Until to now, the relationship between the possible 20-bp indel within PRND of sheep and growth traits in these breeds is unclear. Hence, the objective of this study was to detect the 20-bp indel of sheep PRND gene in 4 Chinese native sheep breeds by using ME method. The result may provide a valuable theoretical basis for MAS in sheep selecting or eliminating while carrying out preventing scrapie project.
MATERIAL AND METHODS
Ethics Statement
All experiments performed in this study were approved by the International Animal Care and Use Committee of the Northwest A&F University (IACUC-NWAFU). Furthermore, the care and use of animals completely complied with local animal welfare laws, guidelines, and policies.
DNA Samples and Data Collection
A total of 623 individuals representing 4 sheep breeds: Hu sheep (HS) (n = 200), Small Tail Han sheep (STHS) (n = 196), Lanzhou Fat-Tail sheep (LFTS) (n = 61) and Tong sheep (TS) (n = 166) were used in this study. These 4 groups are the main Chinese sheep breeds distributed in the provinces of Henan (HS), Gansu (STHS and LFTS), and Shaanxi (TS). All individuals were 2 to 6 years old. Growth traits for all selected unrelated individuals were measured, including body weight (BW), body height (BH), body length (BL), chest circumference (ChC), chest depth (ChD), chest width (ChW), hucklebone width (HuW), hip width (HW), and cannon circumference (CaC); consequently, body length index (BLI), chest circumference index (ChCI), chest width index (ChWI), cannon circumference index (CaCI), hucklebone width index (HuWI) and trunk index (TI) were also calculated on the basis of our reported description.Citation6-8
DNA Isolation and Genomic DNA Pools Construction
DNA samples in this study were extracted from ear tissues (saved in 70 % alcohol at −80°C) and leukocytes of blood (freeze at minus 80°C) by phenol-chloroform method according to our previous reports.Citation9,10 Quality of all the genomic DNA samples were assayed by Nanodrop 1000 (Thermo Scientific, Waltham, MA, USA), diluted to the same standard 50 ng/μL and stored at −20°C for using. Considering work loads and the low frequency of indel in theory, a total of 30 samples were selected randomly to construct a genomic DNA pool for polymerase chain reaction (PCR) to detect potential indel locus in sheep PRND gene.Citation7
Primer Design and PCR Amplification
Based on NCBI SNP database (https://www.ncbi.nlm.nih.gov/snp), one potential indel site was found in sheep PRND gene, so one pair of primers was designed by primer premier Software 5.0 (Premier Biosoft International USA) based on the sheep PRND gene sequence (GenBank No NC_019470.2) (F:5′-TAAAGGCAGAGGCAGAGAAGAA-3′; R: 5′-GAGGTGAAACCGGAATGAGATT-3′). The PCR was performed in 20 μL reaction volume containing 1.0 μL (50 ng) genomic DNA (constructed from 30 different individuals), 0.5 μL each primer (forward and reverse primer), 10 μL 2× Eco Taq PCR Super mix (+dye) and 8 μL ddH2O. Touch Down PCR reaction was performed as follows: initial denaturation for 4 min at 95°C, followed by 18 cycles of denaturation for 30 s at 94°C, annealing for 30 s at 68°C (with a decrease of 1°C per cycle), and extension for 1000 bp/min at 72°C, another 23 cycles of 30 s at 94°C, 30 s at 50°C, and 45 s at 72°C, and a final extension of 10 min at 72°C.Citation6 After cooled to 4°C, the PCR products specificity was confirmed by sequencing. Then, a novel 20-bp indel was detected and the mutation frequency was low. According to previous reports, based on the low frequencies of the 20-bp indel within PRND gene and the sample sizes, we designed the most efficient pooling strategy to detect the all individuals by using ME method.Citation5 Therefore, remaining individuals were genotyped by using ME method.Citation5,10
Statistical Analyses
Genotype and allele frequency were calculated directly. The χ2 test was performed to test whether the polymorphism was in Hardy-Weinberg equilibrium (HWE). Polymorphism information content was calculated by Nei's method implemented in the GDIcall Online Calculator (http://www.msrcall.com/Gdicall.aspx).Citation11 Distribution differences for genotypic and allelic frequencies among/between different breeds were analyzed using the χ2 test implemented in SPSS (Version 18.0) (IBM Corp., Armonk, NY, USA).Citation12 The associations of the 20-bp indel of PRND with several growth traits (e.g. body length (cm)) in different breeds was tested using the analysis of variance (ANOVA) available in SPSS (Version 18.0).Citation13 Meanwhile, the data that did not follow normal distribution and homogeneity of variances were analyzed by the nonparametric (Kruskal-Wallis) test in SPSS (Version 18.0).Citation12
RESULTS
PCR Amplification and Individuals Genotyping by ME Method
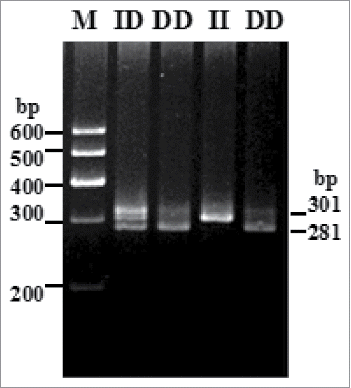
The 20-bp indel of the sheep PRND gene was genotyped by the 3.5% agarose gel electrophoresis. Electrophoretograms revealed the different genotypes of all individuals: one band (301-bp) for genotype II, one band (281-bp) for genotype DD and three bands (301/281-bp) and another homoduplex for genotype ID ().
As displayed in , the genotypic and allelic frequencies of 20-bp indel of the sheep PRND gene were evaluated in 4 sheep breeds. The minor allelic frequencies were 0.008 (LFTS), 0.084 (STHS), 0.021(TS) and 0.083 (HS), respectively. Furthermore, the effective allele values were 1.016 (LFTS), 1.182 (STHS), 1.043(TS) and 1.178 (HS), respectively. Results showed that this indel markers were low polymorphic with PIC among studied breeds ranged from 0.0159 to 0.1423. In addition, these loci were not at HWE in all tested breeds (P < 0.05), expect for Lanzhou Fat-Tail sheep.
TABLE 1. Diversity parameters for the novel 20-bp indel of sheep PRND gene.
The low frequency of the 20-bp indel were confirmed in PRND gene in 4 different sheep breeds (). All reaction times of different group in 4 sheep breeds were calculated by using ME method. Combing with minor allelic frequencies (all analyzed breed's minor allelic frequency > 0) and size of each breed, according to the ME method applied equation, the opitical number of individuals in one mixed group (NGn) was 10 (LFTS), 4 (STHS), 7 (TS) and 4 (HS) (). Hence, the predicting reaction times by the formulate of LFTS, STHS, TS and HS were 12, 105, 46 and 107, respectively. The exact reaction times were shown relying on whether there was a single band (281-bp or 301-bp) in a mix group consisted of different sheep: a total of 11, 125, 52 and 126 reaction times were performed in LFTS, STHS, TS and HS. Comparing with the traditional detecting method, by using ME method, the times of PCR were decreased by 36.22% (STHS), 37.00% (HS), 68.67% (TS) and 83.33% (LFTS).
TABLE 2. All reaction times of different group in 4 sheep breeds.
Sequencing of the 20-bp Indel Variants of the Sheep PRND Gene
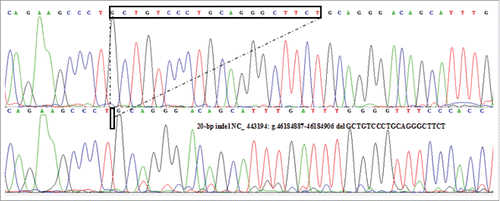
Herein, a novel 20-bp indel in the upstream 2 kb of coding sequence (CDS) of sheep PRND gene was first confirmed. The result of contrasting and analyzing sequence by software (Bioedit UK) showed that the indel sequence was “GCTGTCCCTGCAGGGCTTCT”(), which wasn't consist with the data in NCBI (NC_443194:g.46184887–46184906 del CTGCTGTCCCTGCAGGGCTT).
Association of the Indel Locus and Growth Related Traits of Different Breeds Sheep
Due to all tested sheep were healthy and there are no disease outbreak area in China, our result couldn't reveal this indel loci was relevant to the scrapie. But, the association between the 20-bp indel of PRND gene and the sheep growth traits were investigated in the tested sheep breeds (). Due to the effective number of individuals with ID or DD genotype (mutation genotypes) were less than 5, the data of Lanzhou Fat-Tail sheep and Tong sheep were not included in analysis. The significant relationships were observed between this indel locus and cannon circumference index (P = 0.012), and trunk index (P = 0.037) in the HS breed. This indel locus also appear to have approximately effect on other traits in the HS breed, such as hip width (P = 0.070), and chest circumference index (P = 0.082). Detection and analysis results showed that there was no significantly correlation between the growth traits and this indel of the other tested sheep breed.
TABLE 3. Relationship between the 20-bp indel locus of PRND gene and growth related traits in Hu sheep (LSMa ± SE) (P < 0.05).
DISCUSSION
Herein, this novel 20-bp indel in the upstream 2 kb of CDS of sheep PRND gene was first found, and this indel sequence was not concordance to the available indel region of PRND gene in NCBI datebase (rs590381771). The difference of breeds (tested breed was Texel in the NCBI) and environment may contribute to this sequence variety.
Simultaneously, the present study was the first report of using ME method to detect the novel 20-bp indel of sheep PRND gene as well as finding significant effect on growth traits in 4 Chinese indigenous sheep breeds (LFTS, STHS, TS and HS). In present study, confirmed the low frequency of this indel by randomly detecting 50 individuals one by one, we established the most efficient pooling strategy for each breed basing on the ME method. Finally, a total of 11, 125, 52 and 126 reaction times were performed in LFTS, STHS, TS and HS. Obviously, comparing with the one by one detecting method, the real times of ME method PCR were decreased a lot. Doubtless, the successful application of ME method in our study was also consistent with previous study.Citation6
Furthermore, our result also found that the 20-bp indel of sheep PRND gene wasn't at HWE in all tested breeds (P < 0.05) expect for Lanzhou Fat-Tail sheep, which might be associated with the cultural background of different sheep breed. Due to migration or crossbreeding, the distribution of STHS, TS and HS was extensive in the Chinese territory. Hence, the migration might be a factor of HWE deviation. In particular, the detected Lanzhou Fat-Tail sheep was in HWE, which might since the small tested sample (n = 61) of this breed. Additionally, owing to the regulatory SNPs possibly affecting gene expression, thus the associations between the indel loci and sheep growth traits were also analyzed.Citation14 In Hu sheep, individuals with genotype DD tends to have bigger hip width than others. Hence, we can select ewe with genotype DD, for big hip width benefit female delivery. Additionally, in Hu sheep, individuals with genotype ID exhibited bigger cannon circumference index (CaCI) and trunk index (TI) than others with genotype II, predicting that binding sites of transcription factor might existed in the indel loci. Analogy to the adjacent PRNP, when the 20-bp indel within sheep PRND was inserted or deleted, the expression of other growth related genes would be influenced by bonding with miRNA.Citation15 Meanwhile, previous study shown several gene, which affecting male reproduction, also influence animal growth, such as SPAG17 gene which has an impact on animal skeletal malformations and bone abnormalities.Citation16 As a member of male reproduction related gene,Citation2,3 PRND be conjectured to play an vital role in sheep growth which support our result. Moreover, our result showed that the PIC values of this indel were low for all studied breeds, which was consistent with the previous conclusion of PRND sequence conserved highly.
Because the all detected sheep were uninfected, the present study can't provide the dependable results about the association of this indel and scrapie susceptibility. However, in sheep, susceptibility to scrapie is strongly associated with polymorphisms of PRNP.Citation17-20 As an evolutionarily related paralogue of PRNP, the PRND gene polymorphisms also might be closely related to scrapie. Furthermore, this novel indel situated in the upstream 2 kb of CDS of PRND, nearby the prion-related protein (testis-specific) gene (PRNT). Previous study have identified the polymorphism of PRNT in sheep, but the similarity function of gene and relationship between polymorphism of PRNT and mutations of PRND still need to be further researched.Citation21
Overall, the detection based on ME method, for low-frequency mutation of the 20-bp indel of sheep PRND, was accessible and efficient. In addition, the detected 20-bp indel within PRND was first confirmed in animal (especially in sheep). Furthermore, this 20-bp indel significantly affected growth traits suggesting that it might be a potential useful DNA marker for the selection of high quality individuals in MAS for sheep breeding.
ABBREVIATIONS
| BH | = | body height |
| BL | = | body length |
| BLI | = | body length index |
| bp | = | base pair |
| BW | = | body weight |
| CaC | = | cannon circumference |
| CaCI | = | cannon circumference index |
| CDS | = | coding sequence |
| ChC | = | chest circumference |
| ChCI | = | chest circumference index |
| ChD | = | chest depth |
| ChW | = | chest width |
| ChWI | = | chest width index |
| DD | = | deletion/deletion |
| HS | = | Hu sheep |
| HuW | = | hucklebone width |
| HuWI | = | hucklebone width index |
| HW | = | hip width |
| HWE | = | Hardy-Weinberg equilibrium |
| Ho | = | homozygosity |
| He | = | heterozygosity |
| ID | = | insertion/deletion |
| II | = | insertion/insertion |
| Indel | = | insertion/deletion |
| LFTS | = | Lanzhou Fat-Tail sheep |
| MAS | = | marker-assisted selection |
| ME | = | mathematical expectation |
| Ne | = | effective allele numbers |
| PCR | = | polymerase chain reaction |
| PIC | = | Polymorphism information content |
| PRNP | = | prion gene |
| PRND | = | prion-related doppel gene |
| PRNT | = | prion-related protein (testis-specific) gene |
| SNP | = | single nucleotide polymorphism |
| STHS | = | Small Tail Han sheep |
| TD-PCR | = | touch-down PCR |
| TI | = | trunk index |
| TS | = | Tong sheep |
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
No conflict of interest exits in the submission of this manuscript, and manuscript is approved by all authors for publication.
FUNDING
This work was funded by the National Natural Science Foundation of China (N0.31662624). We greatly thanked the staffs of TS elite reservation farm, Baishui county, Shannxi Province, Ruilin Sci-Tech Cluture and Breeding Limit Company Yongjing county, Gansu Province, and shanshan agriculture and animal husbandry Sci-tech company Mengjin county, Henan Province for collecting samples.
REFERENCES
- Mesquita P, Batista M, Marques MR, Santos IC, Pimenta J, Silva Pereira M, Carolino I, Santos Silva F, Oliveira Sousa MC, Gama LT, et al. Prion-liked Doppel gene polymorphisms and scrapie susceptibility in portuguese sheep breeds. Anim Genet 2010; 41(3):311-4; PMID:19968641; http://dx.doi.org/10.1111/j.1365-2052.2009.01992.x
- Kocer A, Gallozzi M, Renault L, Tilly G, Pinheiro I, Le Provost F, Pailhoux E, Vilotte JL. Goat PRND expression pattern suggests its involvement in early sex differentiation. Dev Dyn 2007; 236:836-42; PMID:17226816; http://dx.doi.org/10.1002/dvdy.21066
- Pereira RM, Mesquita P, Batista M, Baptista MC, Barbas JP, Pimenta J, Santos IC, Marques MR, Vasques MI, Silva Pereira M, et al. Doppel gene polymorphisms in Portuguese sheep breeds: Insights on ram fertility. Anim Reprod Sci 2009; 114:157-66; PMID:19028030; http://dx.doi.org/10.1016/j.anireprosci.2008.10.003
- Allais-Bonnet A, Castille J, Pannetier M, Passet B, Elzaïat M, André M, Montazer-Torbati F, Moazami-Goudarzi K, Vilotte JL, Pailhoux E. A specific role for PRND in goat foetal Leydig cells is suggested by prion family gene expression during gonad development in goats and mice. FEBS Open Bio 2016; 6(1):4-15; PMID:27047737; http://dx.doi.org/10.1002/2211-5463.12002
- Yang Q, Zhang SH, Liu LL, Cao XK, Lei CZ, Qi XL, Lin FP, Qu WD, Qi XS, Liu JM, et al. Application of mathematical expectation (ME) strategy for detecting low frequency mutations: An example for evaluating 14 bp insertion/deletion (indel) within the bovine PRNP gene. Prion 2016; 10:409-19; PMID:27580010; http://dx.doi.org/10.1080/19336896.2016.1211593
- Jia WC, Wu XF, Li XC, Xia T, Lei CZ, Chen H, Pan CY, Lan XY. Novel genetic variants associated with mRNA expression of signal transducer and activator of transcription 3 (STAT3) gene significantly affected goat growth traits. Small Ruminant Res 2015; 129:25-36; http://dx.doi.org/10.1016/j.smallrumres.2015.05.014
- Lan XY, Zhao HY, Li ZJ, Zhou R, Pan CY, Lei CZ, Chen H. Exploring the novel genetic variant of PITX1 gene and its effect on milk performance in dairy goats. J Integr Agr 2013; 12(1):118-26; http://dx.doi.org/10.1016/S2095-3119(13)60212-9
- Lan XY, Pan CY, Chen H, Zhang CL, Li JY, Zhao M, Lei CZ, Zhang AL, Zhang LZ. An AluI PCR-RFLP detecting a silent allele at the goat POU1F1 locus and its association with production traits. Small Ruminant Res 2007; 73(1):8-12; http://dx.doi.org/10.1016/j.smallrumres.2006.10.009
- Zhang SH, Sun K, Bian YN, Zhao Q, Wang Z, Ji CN, Li CT. Developmental validation of an X-Insertion/Deletion polymorphism panel and application in HAN population of China. Sci Rep 2015; 5:18336; PMID:26655948; http://dx.doi.org/10.1038/srep18336
- Zhang XY, Wu XF, Jia WC, Pan CY, Li XC, Lei CZ, Chen H, Lan XY. Novel nucleotide variations, haplotypes structure and associations with growth related traits of goat at Motif-Binding factor (ATBF1) gene. Asian-Australas J Anim Sci 2015; 28(10):1394-406; PMID:26323396; http://dx.doi.org/10.5713/ajas.14.0860
- Czarnik U, Grzybowski G, Zabolewicz T, Strychalski J, Kaminski S. Deletion/insertion polymorphism of the prion protein gene (PRNP) in Polish red cattle, Polish White-backed cattle and European bison (Bison bonasus L., 1758). Genetika 2009; 45(4):519-525; PMID:19507705
- Pan CY, Wu CY, Jia WC, Xu Y, Hu SR, Lei CZ, Lan XY, Chen H. A critical functional missense mutation (H173R) in the bovine PROP1 gene significantly affects growth traits in cattle. Gene 2013; 531(2):398-402; PMID:24029076; http://dx.doi.org/10.1016/j.gene.2013.09.002
- Li Y, Wang K, Jiang YZ, Dai CF, Zou QY, Zheng J. 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) inhibits human ovarian cancer cell proliferation. Cell Oncol (Dordr) 2014; 37(6):429-37; PMID:25404385; http://dx.doi.org/10.1007/s13402-014-0206-4
- Lan XY, Zhao HY, Li ZJ, Li AM, Lei CZ, Chen H, Pan CY. A novel 28-bp insertion-deletion polymorphism within goat PRNP gene and its association with production traits in Chinese native breeds. Genome 2012; 55(7):547-52; PMID:22794197; http://dx.doi.org/10.1139/g2012-040
- Hou JX, An XP, Song YX, Gao TY, Lei YN, Cao BY. Two mutations in the caprine MTHFR 3′ UTR regulated by microRNAs are associated with milk production traits. PLoS One 2015; 10(7):e0133015; PMID:26186555; http://dx.doi.org/10.1371/journal.pone.0133015
- Teves ME, Sundaresan G, Cohen DJ, Hyzy SL, Kajan I, Maczis M, Zhang Z, Costanzo RM, Zweit J, Schwartz Z, et al. Spag17 deficiency results in skeletal malformations and bone abnormalities. PLoS One 2015; 10(5):e0125936; PMID:26017218; http://dx.doi.org/10.1371/journal.pone.0125936
- Papasavva-Stylianou P, Kleanthous M, Toumazos P, Mavrikiou P, Loucaides P. Novel polymorphisms at codons 146 and 151 in the prion protein gene of Cyprus goats, and their association with natural scrapie. Vet J 2007; 173(2):459-62; http://dx.doi.org/10.1016/j.tvjl.2005.09.013
- González L, Martin S, Hawkins SA, Goldmann W, Jeffrey M, Sisó S. Pathogenesis of natural goat scrapie: modulation by host PRNP genotype and effect of co-existent conditions. Vet Res 2010; 41(4):48; PMID:20374697; http://dx.doi.org/10.1051/vetres/2010020
- Guan F, Pan L, Li J, Tang H, Zhu C, Shi G. Polymorphisms of the prion protein gene and their effects on litter size and risk evaluation for scrapie in Chinese Hu sheep. Virus Genes 2011; 43(1):147-52; PMID:21556743; http://dx.doi.org/10.1007/s11262-011-0609-5
- Goldmann W, Ryan K, Stewart P, Parnham D, Xicohtencatl R, Fernandez N, Saunders G, Windl O, González Bossers A, et al. Caprine prion gene polymorphisms are associated with decreased incidence of classical scrapie in goat herds in the United Kingdom. Vet Res 2011; 42(1):110; PMID:22040234; http://dx.doi.org/10.1186/1297-9716-42-110
- Mesquita P, Garcia V, Marques MR, Santos Silva F, Oliveira Sousa MC, Carolino I, Pimenta J, Fontes CM, Horta AE, Prates JA, et al. The prion-related protein (testis-specific) gene (PRNT) is highly polymorphic in Portuguese sheep. Anim Genet 2016; 47(1):128-32; PMID:26538093; http://dx.doi.org/10.1111/age.12380