ABSTRACT
The real-time quaking-induced conversion (RT-QuIC) reaction is a sensitive and specific method for detecting prions. However, inhibitory factors present in tissue homogenates can easily interfere with this reaction. To identify the RT-QuIC condition under which low levels of chronic wasting disease (CWD) and bovine spongiform encephalopathy (BSE) prions can be detected in the presence of high concentrations of brain tissue homogenates, reactivities of various recombinant prion proteins (rPrPs) were tested. Among the tested rPrPs, recombinant cervid PrP (rCerPrP) showed a unique reactivity: the reactivity of rCerPrP to CWD and atypical BSE prions was not highly affected by high concentrations of normal brain homogenates. The unique reactivity of rCerPrP disappeared when the N-terminal region (aa 25–93) was truncated. Replacement of aa 23–149 of mouse (Mo) PrP with the corresponding region of CerPrP partially restored the unique reactivity of rCerPrP in RT-QuIC. Replacement of the extreme C-terminal region of MoPrP aa 219–231 to the corresponding region of CerPrP partially conferred the unique reactivity of rCerPrP to rMoPrP, suggesting the involvement of both N- and C-terminal regions. Additionally, rCerN–Mo–CerCPrP, a chimeric PrP comprising CerPrP aa 25–153, MoPrP aa 150–218, and CerPrP aa 223–233, showed an additive effect of the N- and C-terminal regions. These results provide a mechanistic implication for detecting CWD and atypical BSE prions using rCerPrP and are useful for further improvements of RT-QuIC.
Introduction
Prion diseases are fatal neurodegenerative disorders including Creutzfeldt-Jakob disease (CJD), Gerstmann-Straussler-Scheinker syndrome (GSS), and fatal insomnia in humans, scrapie in sheep and goats, chronic wasting disease (CWD) in cervids and bovine spongiform encephalopathy (BSE) [Citation1]. Prions, which are the causative agents of prion diseases, are mainly composed of an abnormal isoform of the prion protein (PrPSc) that are generated from host-encoded cellular isoform of the prion protein (PrPC). Binding of PrPC to PrPSc induces a conformational conversion from the α-helix-rich PrPC to the β-structure-rich PrPSc. Accumulation of PrPSc in the central nervous system is the most characteristic feature of these diseases.
CWD was first identified in 1967 in a group of captive mule deer and was classified as a TSE in 1980. To date, CWD-affected cervids have been found in the US, Canada, South Korea [Citation2,Citation3], and Scandinavian countries [Citation4,Citation5]. Different from other prions, infectious CWD prions have also been detected in secreted body fluids and excretions such as saliva, urine, and faeces [Citation6,Citation7]. Although CWD did not transmit to mice expressing human PrP [Citation8,Citation9], the zoonotic potential of CWD cannot be ignored, as CWD prions are known as experimentally transmissible to several animals including the squirrel monkey [Citation10], and prion properties change during interspecies transmission [Citation11–14].
The classical BSE (C-BSE) was recognized in the United Kingdom in 1986 [Citation15], and since then has spread globally. C-BSE is known as a cause of zoonotic prion diseases as it is transmitted to human via BSE-contaminated products, which has caused the emergence of variant CJD [Citation16]. C-BSE is now under control due to worldwide implementation of control measures such as feed bans. Two atypical BSEs were identified in 2004, and they were classified as H- and L-BSE based on the higher and lower apparent molecular weights of un-glycosylated PrPSc observed in immunoblotting [Citation17]. Atypical BSE cases were mainly disclosed in cattle over the age of 8 years old and are also found worldwide, including in Brazil wherein no C-BSE case has been reported, suggesting that atypical BSEs may be sporadic diseases that occur in aged cattle, similar to sporadic CJD in humans [Citation18]. L-BSE is known to be experimentally transmissible to primates and mice expressing human PrP [Citation19–21], whereas the zoonotic potential of H-BSE has not yet been fully elucidated. Since low levels of prion infectivity and seeding activity of atypical BSE prions were detected in skeletal and intercostalis muscles [Citation22,Citation23] and in tonsillar tissue [Citation24] from cattle affected with atypical BSEs, a potential risk of human transmission via food consumption cannot be ignored.
To disclose the potential presence of CWD and atypical BSEs, highly sensitive and accurate methods are required for detecting low levels of prions in tissue homogenates or body fluids. Bioassays using appropriate gene-modified mice could be sensitive enough to detect low levels of prions [Citation24,Citation25]; however, bioassays require extremely long experimental time periods to detect low levels of infection. Highly sensitive in vitro methods such as protein misfolding cyclic amplification (PMCA) [Citation26] and quaking-induced conversion [Citation27] have been reported to successfully detect the low level of prions. Real-time quaking-induced conversion (RT-QuIC), which detects the amyloid seeding activity of PrPSc, is known as a specific and highly sensitive assay capable of detecting low levels of prions [Citation28–31]. However, the inhibitory factors in tissue homogenates and body fluids can interfere with the reaction [Citation31–33], which then hampers detection of low level prions in the presence of high concentrations of tissue homogenates. In the recent study, we showed that recombinant cervid PrP (rCerPrP) is useful for the detection of atypical BSE prions from tissues of cattle affected with atypical BSEs [Citation22]. In the present study, we show that the RT-QuIC reaction using full-length rCerPrP is less affected by a high concentration of brain tissue homogenates when detecting CWD and atypical BSE prions. We also analysed the region(s) responsible for the unique properties of rCerPrP in the reaction.
Results
Detection of CWD prions in the presence of high concentration of brain tissue homogenates
We analysed the reactivity of six rPrPs to CWD prions in the presence and absence of 0.1% deer NBH (). The endpoints of the reactions using the five rPrPs (rMoPrP, rBvPrP, rHaPrP, rShPrP, and rCerPrP) were from 10−8 to <10−9, in the absence of NBH (, PBS); however, the lag phases were varied with the rPrPs. Among the five rPrPs, the lag phases using rCerPrP were the shortest at each seed dilution. rBoPrP was ineffective in detecting CWD prions, as the endpoint of the reaction was 10−4 (). Interestingly, the reactions of rHaPrP, rMoPrP, and rShPrP were affected by 0.1% deer NBH: the rHaPrP reaction was completely inhibited, and the endpoints of reactions using rMoPrP and rShPrP worsened by 3 and 1 logs, respectively (). Additionally, lag phases of the reactions with each rPrP were significantly prolonged: the lag phases of the reactions using rMoPrP, rBvPrP, and rShPrP were prolonged by 43.5, 27.8, and 29.6 h, respectively, at 10−5 seed dilution in the presence of 0.1% deer NBH (). In contrast, the rCerPrP reaction was not severely affected by 0.1% deer NBH; the endpoint was unchanged, and the lag phase at 10−5 seed dilution was only prolonged by 7.7 h ().
Figure 1. Reactivity of rPrPs to CWD prions in RT-QuIC in the presence and absence of normal brain homogenates. Representative line graphs for the detection of CWD prions serially diluted with PBS (left column) or 2% brain homogenates in PBS of CWD-negative white-tail deer (normal brain homogenate, NBH) (final concentration of NBH in the reaction mixture was 0.1%) (right column). Detections performed using rMoPrP (red to yellow), rBvPrP (brown), rHaPrP (green), rBoPrP (black), rShPrP (purple), and rCerPrP (blue) are shown. ThT intensities from triplicate wells were plotted against reaction time using different brightness or colours. Dotted lines indicate the thresholds of reaction calculated from the mean ThT fluorescence intensity plus 5 × SD of negative control wells (without brain homogenates). Lag phases (mean ± SD), endpoints, and ratios of the endpoints [Ratio, endpoint (PBS)/endpoint (NBH)] are summarized in the table. *: p < 0.05, **: p < 0.01 by welch t-test
![Figure 1. Reactivity of rPrPs to CWD prions in RT-QuIC in the presence and absence of normal brain homogenates. Representative line graphs for the detection of CWD prions serially diluted with PBS (left column) or 2% brain homogenates in PBS of CWD-negative white-tail deer (normal brain homogenate, NBH) (final concentration of NBH in the reaction mixture was 0.1%) (right column). Detections performed using rMoPrP (red to yellow), rBvPrP (brown), rHaPrP (green), rBoPrP (black), rShPrP (purple), and rCerPrP (blue) are shown. ThT intensities from triplicate wells were plotted against reaction time using different brightness or colours. Dotted lines indicate the thresholds of reaction calculated from the mean ThT fluorescence intensity plus 5 × SD of negative control wells (without brain homogenates). Lag phases (mean ± SD), endpoints, and ratios of the endpoints [Ratio, endpoint (PBS)/endpoint (NBH)] are summarized in the table. *: p < 0.05, **: p < 0.01 by welch t-test](/cms/asset/724cd0f7-f18a-4173-9991-80c66161c434/kprn_a_1858694_f0001_oc.jpg)
Detection of atypical and classical BSE prions in the presence of high concentration of brain tissue homogenates
We analysed the reactivity of six rPrPs to atypical BSE (H- and L-BSE) and classical BSE (C-BSE) in RT-QuIC. Although rBoPrP showed longer lag phases than the other rPrPs, all rPrPs showed similar endpoints for detecting H-BSE prions in the absence of NBH (10−8 or <10−9, ). rBoPrP and rHaPrP seemed to be less effective than the other four rPrPs in detecting L-BSE prions: endpoints of reactions using the former rPrPs (10−6 and 10−7) were lower than the latter rPrPs (10−8 or <10−9). A similar tendency was observed for the detection of C-BSE prions: rBoPrP and rHaPrP (endpoints: 10−4 and 10−5) appeared to be less effective than the four other rPrPs (endpoints: 10−6). Although no difference was observed in the detection endpoints of H-BSE prions among rMoPrP, rBvPrP, rShPrP, and rCerPrP, rCerPrP was able to detect H-BSE prions with shorter lag phases than the other rPrPs (). Interestingly, 0.1% cattle NBH completely interfered with the detection of H-BSE prions with rBvPrP and rHaPrP, detection of L-BSE prions with rMoPrP, rBvPrP, rHaPrP, and rBoPrP, and detection of C-BSE prions with the five rPrPs except for rCerPrP (). Notably, detection of atypical BSE prions with rCerPrP was not severely affected by 0.1% cattle NBH: endpoints of the reaction were unchanged and lag phases were prolonged at some seed dilutions, e.g, at 10−5 to 10−7 for H-BSE, and at 10−4 for L-BSE, but the differences were not that large (<10 h except at 10−7 seed dilution for H-BSE, ). These results suggest that rCerPrP is a good substrate for the detection of atypical BSE as well as CWD prions in the presence of high concentrations of brain tissue homogenates. Moreover, detection of C-BSE prions was affected by 0.1% cattle NBH, which the endpoint of the reaction worsened by 2 logs and the lag phases were significantly prolonged (). Also, 0.1% cattle NBH did not interfere with the detection of atypical BSE prions but interfered with detection of C-BSE prions, suggesting that RT-QuIC using rCerPrP can discriminate atypical BSE from C-BSE prions.
Table 1. Reactivity of rPrPs to atypical and classical BSE prions in RT-QuIC
Involvement of N-terminal region of rCerPrP in its reactivity in the presence of NBH
RT-QuIC reaction is reported to be easily inhibited by high concentrations of tissue homogenates and body fluids [Citation31,Citation33]. In the current study, we found that the reaction of rCerPrP to CWD and atypical BSE prions was not largely affected by NBH. To clarify the mechanism of the unique property of rCerPrP, we analysed the reactivities of rPrPs shown in . The reactions of N-terminal-truncated rCerPrP comprised aa 94–233 (rCerPrP94–233) to CWD, and atypical BSE prions were completely inhibited by NBH (). Furthermore, lag phases for detecting CWD and L-BSE prions using rCerPrP94–233 at the 10−4 seed dilution in PBS containing 0.01% brain homogenates were obviously longer than those using full-length rCerPrP. Replacement of the N-terminal region of rCerPrP (aa 25–153) with the corresponding region of rMoPrP (aa 23–149) (rMoN–CerPrP) had modest influence on the detection endpoints for CWD and atypical BSE prions: the detection endpoints of rMoN–CerPrP for CWD, H-BSE, and L-BSE (10−8, 10−8, and 10−7, respectively, without NBH, ) were slightly lower than those of rCerPrP (10−8, <10−9, and <10−9, respectively, and ). Additionally, the reaction of rMoN–CerPrP to the L-BSE prions was completely inhibited, and reactions of rMoN–CerPrP to CWD and H-BSE were severely affected by 0.1% NBH; the detection endpoints for CWD, H-BSE, and L-BSE prions worsened by 4, 2, and >3 logs, respectively, with significant prolongation of lag phases at each seed dilution (). The reactivities of rMoN–CerPrP were very similar to those of rMoPrP; the reaction of rMoPrP to the L-BSE prions were completely inhibited (), and the reactions of rMoPrP to CWD () and H-BSE prions () were severely affected as observed in the significant prolongation of lag phases in the presence of 0.1% NBH.
Table 2. Reactivity of N-terminal truncated rCerPrP in RT-QuIC
Table 3. Reactivity of recombinant chimeric PrP between CerPrP and MoPrP in RT-QuIC
Figure 2. Summary of the reactivity of recombinant chimeric PrPs between CerPrP and MoPrP. rCerPrP, rMoPrP, and their chimeras are indicated with their amino acid differences. Numbers with capital letters are the aa of CerPrP, while those with italics indicate the aa of MoPrP. ‘-’ indicates gaps. A single-letter notation of aa with italics indicate the substituted aa between CerPrP and MoPrP. The RT-QuIC substrate efficiencies were classified as follows: Good, detection endpoints are ≦10−8 and lag phases at 10−4 and 10−5 seed dilutions were <20 h for seeds diluted with PBS; Moderate, detection endpoints were >10−8 or lag phases at 10−4 or 10−5 seed dilutions were ≧20 h. The criteria of the reactivity in the presence of NBH were defined as follows: slightly affected, endpoint ratio was ≧10–2 and prolongation of the lag phase at 10−5 seed dilution (lag phaseNBH − lag phasePBS) was <10 h; moderately affected: endpoint ratio is 10−2 to 10−3 and/or prolongation of the lag phase at 10−5 was from 10–25 h; severely affected, endpoint ratio was <10−3 and/or prolongation of the lag phase at 10−5 seed dilution is >25 h. H, H-BSE; L, L-BSE; W, CWD
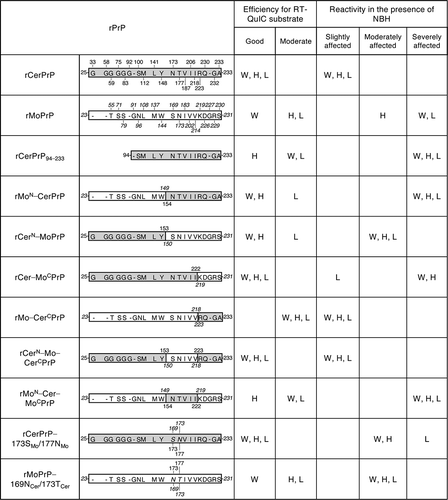
On the contrary, replacing the N-terminal half of MoPrP (aa 23–149) with the corresponding CerPrP (aa 25–153) (rCerN–MoPrP) partially restored reactivity to a level similar to that of rCerPrP. Detection endpoints of CWD, H-, and L-BSE prions using rCerN–MoPrP without NBH (10−8, <10−9, and 10−8, respectively, ) were almost comparable to those using rCerPrP (10−8, <10−9, and <10−9, respectively, and ). In addition, the reactivity of rCerN–MoPrP to CWD, H-, and L-BSE prions was not affected by NBH when endpoints were compared in the absence (PBS) and presence of NBH (). However, the prolongation of lag phases in the detection of CWD, H-, and L-BSE prions using rCerN–MoPrP (11.8, 20.9, and 12.0 h, respectively, at 10−5 seed dilution, ) appeared to be longer than those caused by rCerPrP in the presence of NBH (7.7, 6.5, and 3.0 h, respectively, at 10−5 seed dilution, and ), suggesting that rCerN–MoPrP is more affected by NBH than by rCerPrP. Taken together, these results suggest that the N-terminal region of rCerPrP is necessary for its unique reactivity in RT-QuIC in the presence of NBH.
Involvement of C-terminal region of rCerPrP in its reactivity in the presence of NBH
It was reported that the aa sequence at the C-terminus of PrPC is varied among animal species [Citation34], and aa substitution in this region could alter the efficacy of PrPC conformational conversion [Citation35,Citation36] and transmission kinetics [Citation37]. Thus, we analysed the involvement of the C-terminal region of CerPrP in its reactivity in RT-QuIC. The rCer–MoCPrP, which possesses MoPrP aa 219–231 in the corresponding region of rCerPrP aa 223–233 resulting in 5 amino acid differences, reacted well with CWD, H-, and L-BSE prions (endpoints: <10−9, ) in the absence of NBH if compared with the reactivity of rCerPrP (10−8, <10−9, and <10−9, respectively, and ). Lag phases for the detection of the three prions using rCer–MoCPrP (12.5, 10.6, and 19.7 h, respectively, at 10−5 seed dilution, ) were comparable to those using rCerPrP (10.0, 10.9, and 19.5 h, respectively, and ). However, the detection endpoint of CWD prions using rCer–MoCPrP worsened by >3 log in the presence of 0.1% NBH with prolonged lag phases (). This reactivity is similar to that of rMoPrP to CWD prions (). Although the detection endpoint of H-BSE prions using rCer–MoCPrP worsened only by 1 log in the presence of 0.1% NBH, lag phases for the detection of H-BSE prions using rCer–MoCPrP were significantly prolonged, e.g, lag phases for detecting H-BSE prions using rCerPrP were prolonged by only 6.5 h at 10−5 seed dilution (), but those using rCer–MoCPrP were prolonged by 43.9 h at 10−5 seed dilution in the presence of 0.1% NBH (). On the contrary, rCer–MoCPrP reactivity to L-BSE was not severely affected, and no differences in the detection endpoints with only marginal prolongation of lag phases (around 10 h) at each seed dilutions (). Thus, the reactivity of rCer–MoCPrP to CWD and H-BSE prions in the presence of NBH resembles that of rMoPrP, whereas, its reactivity to L-BSE prions resembles that of rCerPrP. Replacement of MoPrP aa 219–231 with the corresponding CerPrP 223–233 (rMo–CerCPrP) did not affect the detection endpoints of CWD, H-, and L-BSE prions diluted with PBS; <10−9, <10−9, and 10−8, respectively (). However, lag phases in the detection of three prions diluted with PBS using rMo–CerCPrP (about 20.5–26.5 h at 10−4 and 10−5 seed dilutions, ) were longer than those using rCerPrP (less than 19.5 h at 10−4 and 10−5 seed dilutions, and ), but were comparable to those using rMoPrP (14.7–31.2 h at 10−4 and 10−5 seed dilutions, and ), suggesting that rMo–CerCPrP is less efficient than rCerPrP as a substrate. Interestingly, only eleven amino acids in the C-terminal region of CerPrP appeared to confer the unique property of rCerPrP to rMoPrP that made it such that the presence of NBH interferes less with RT-QuIC using rCerPrP. The reactivity of rMoPrP to the three prions was severely interfered in the presence of 0.1% NBH. Detection endpoints worsened by 2–4 logs with a significant prolongation of the lag phases (43.5, 27.0, and >28.8 h for CWD, H-, and L-BSE, respectively, at 10−5 seed dilutions, and ). However, those of rMo–CerCPrP were less affected by 0.1% NBH; their detection endpoints were not changed, but with a slight prolongation of the lag phases was observed (0, 9.3, and 7.6 h, respectively), at 10−5 seed dilutions (). rCerN–Mo–CerCPrP, which is a chimeric PrP composed of CerPrP aa 25–153, MoPrP aa 150–218, and CerPrP aa 223–233, was detected well with CWD, H-, and L-BSE prions without worsening the detection endpoints but had a marginal prolongation of the lag phases, with most of them being less than 10 h long in the presence of NBH (). These results were consistent with the finding that both N- and C-terminal regions of CerPrP are involved in its unique reactivity in RT-QuIC in the presence of NBH. On the contrary, rMoN–Cer–MoCPrP, which has an opposite structure to rCerN–Mo–CerCPrP, showed similar reactivity to rMoPrP, and detection of CWD, H-, and L-BSE prions was severely affected by NBH ().
Effect of CerPrP-specific amino acids in the β2–α2 loop on reactivity in the presence of NBH
Several studies reported that the CerPrP-specific aa Asn173 and Thr177 in the β2–α2 loop (aa 168–178) [Citation38] influence cross-species prion transmission in transgenic mice and affect the reaction of PMCA [Citation39,Citation40]. To clarify the influence of these aa residues on the unique reactivity of rCerPrP in RT-QuIC, we analysed the reactivities of rCerPrP carrying the corresponding MoPrP aa Ser169 and Ans173 (rCerPrP–173SMo/177NMo). Substitution of two CerPrP-specific aa did not affect the detection endpoints of the CWD and atypical BSE prions (10−9 or <10−9, in ), compared with rCerPrP (10−8, <10−9, and <10−9, for CWD, H-, and L-BSE prions, respectively, and ) in the absence of NBH. However, substitutions of the two amino acids affected the reactivity of rCerPrP in the presence of NBH: the detection endpoints worsened by more than >2 log for the CWD and H-BSE prions and by 1 log for L-BSE prions, with >20 h-prolonged lag phases at most seed dilutions (). The substitution of MoPrP Ser169 and Asn173 to the corresponding CerPrP Asn173 and Thr177 (rMoPrP–169NCer/173TCer) reduced the reactivity to H- and L-BSE prions by >1 log in the absence of NBH. The reactivity of rMoPrP to CWD, H-, and L-BSE prions were extremely affected in the presence of NBH as described above; however, detection of three prions using rMoPrP–169NCer/173TCer in the presence of NBH was moderately affected compared to rMoPrP, with the detection endpoints reduced by only 1 log for CWD and L-BSE prions with moderate prolongation of lag phases (8.2–24.5 h at 10−4 and 10−5 seed dilutions, ).
Discussion
rPrPs from Bv, Ha, and human (Hu) have been widely used as substrates to detect the seeding activity of various prions by the RT-QuIC reaction [Citation28–31]. However, few reports are available regarding the utility of CerPrP. The reaction is known to be sensitive to inhibitory factor(s) present in tissue homogenates and body fluids [Citation31–33]. Concentration of PrPSc through immunoprecipitation or iron oxide beads, or lipid removal of through alcohol extraction, have been attempted to reduce the influence of inhibitory factors [Citation32,Citation41,Citation42]; however, a simpler method is still desirable for practical use. In the present study, we demonstrated that rCerPrP reacted with CWD and atypical BSE prions, even in the presence of the highest concentration of NBH (0.1%), in the reaction mixture. Since rCerPrP reactivity was stable and reproducible without the influence of lot differences, we attempted to determine the region(s) responsible for the unique reactivity of rCerPrP. The unique reactivity of rCerPrP disappeared when the N-terminal region (aa 25–93) was truncated. Moreover, replacement of the N-terminal half of MoPrP with the corresponding region of CerPrP partially restored this reactivity, but replacing the N-terminal half of CerPrP with the corresponding region of MoPrP abolished the unique reactivity of rCerPrP (summarized in ). These results indicate that the N-terminal region of the rCerPrP is one of the determinants modulating rCerPrP reactivity in the RT-QuIC reaction in the presence of NBH (). N-terminal truncated rPrPs have been known to act as better substrates for detecting the amyloid seeding activity of PrPSc [Citation30,Citation43]. However, it has been reported that the N-terminal region is one of the essential regions for the binding between PrPC and PrPSc, and PrPSc production [Citation35,Citation44–46]. Deletion of the octapeptide repeat region (aa 51–90 of MoPrP) delayed the accumulation of PrPSc and prolonged the survival of mice inoculated with C-BSE prions [Citation45], suggesting the involvement of the N-terminal region in the efficacy of conformational conversion in certain combinations of PrPC and PrPSc. Amino acid polymorphisms of CerPrP at Gln95 and Gly96 are known to modulate the susceptibility of deer to CWD prions [Citation47,Citation48], also suggesting the influence of the N-terminal region of cervid PrPC on the efficacy of its conformational conversion. There are two possibilities regarding the role of the CerPrP N-terminal region in its unique reactivity in the presence of NBH: first, the presence of the N-terminal region may stabilize intra- or inter-molecular interaction, which enhances the efficacy of amyloid formation. Alternatively, the N-terminal region effectively inhibits the interaction of inhibitory factor(s) in tissue homogenates with rCerPrP; however, the latter is unlikely because the reactivity of rCerPrP to C-BSE was affected in the presence of NBH.
Figure 3. Regions responsible for the unique reactivity of rCerPrP in RT-QuIC. The three-dimensional structure of CerPrP (PDB ID: 4YXH) was drawn using open-source PyMOL. The regions responsible for the unique reactivity of rCerPrP, N- and C-terminal regions and the β2–α2 loop are shown using blue, yellow, and red circles, respectively. The aa of rCerPrP in the β2–α2 loop and C-terminus are shown in the single-letter notation. Hydrophobic aa differed between rCerN–Mo–CerCPrP and rCerPrP–173SMo/177NMo and regions in which rCerPrP are replaced with (aa 153–222) are shown around the structure. The side chains of aa with aa numbers except for three aa at C-terminal end were drawn with sticks and the aa residues were coloured indicated with magenta
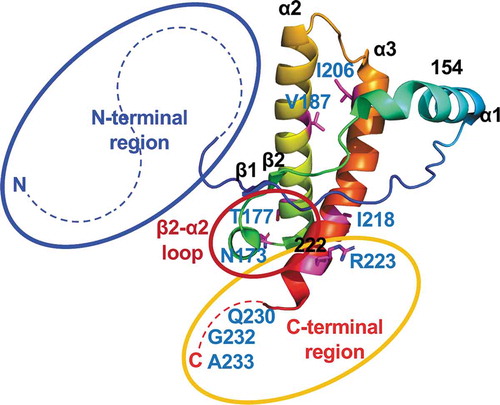
Although rMo–CerCPrP was not a good substrate, i.e., the lag phases for detecting CWD and atypical BSE prions were >20 h at 10−4 and 10−5 seed dilutions (), the C-terminal region of rCerPrP is also involved in the unique reactivity of rCerPrP, and the reactivity of rMo–CerCPrP in RT-QuIC was less affected with NBH than that of rMoPrP ( and ). However, the unique reactivity of rCerPrP in the presence of NBH disappeared due to N-terminal truncation even in the presence of the C-terminal region of CerPrP (rCerPrP94–233) ( and ), suggesting that a cooperative effect of the C-terminal region with the N-terminal region. Indeed, rCerN–Mo–CerCPrP showed better reactivity than rCer–MoCPrP when used as a substrate and was less affected in the presence of NBH compared to rCer–MoCPrP, suggesting an additive effect of the N- and C-terminal regions of CerPrP (). The NMR structure of rHuPrP and rMoPrP revealed the intramolecular interaction between the C-terminal region (aa 219–226) and the N-terminal flexible region [Citation49] or the C-terminal region (aa 215–223) and the β2–α2 loop (aa 164–174) [Citation34]. These intramolecular interactions could be destabilized by substituting Gln217Mo to Arg, which is a mutation associated with GSS [Citation50], and HuPrP Glu219 to Lys, which is a protective polymorphism against sporadic CJD [Citation51]. Thus, intra- or inter-molecular interactions between the N- and C-terminal regions of rCerPrP may be strong enough to overcome the influence of possible inhibitor(s) in NBH and to promote the conformational conversion of rCerPrP.
Since rCerN–Mo–CerCPrP possessed five amino acids from MoPrP including amino acid substitutions at aa 173 and aa 177, we expected that the reactivity to CWD and to atypical BSE prions of rCerPrP–173SMo/177NMo, which possesses only two amino acid differences from CerPrP, would be closer to that of rCerPrP compared to that of rCerN–Mo–CerCPrP. The detection endpoints of rCerPrP–173SMo/177NMo for detecting CWD and atypical BSE prions were comparable to that of rCerN–Mo–CerCPrP when the seeds were diluted with PBS. However, the reactivity of rCerPrP–173SMo/177NMo was more susceptible to the inhibitory effect of NBH than to that of rCerN–Mo–CerCPrP even though the N- and C-terminal regions of rCerPrP–173SMo/177NMo were composed of CerPrP (). One possible explanation for this may be an incompatibility of amino acids in the aa 154–222 region of CerPrP, which contains five aa differences between Cer and MoPrP. Indeed, aa 173 and 177 of rCerPrP–173SMo/177NMo were substituted by Ser169 and Asn173 of MoPrP, respectively, which are located within the β2–α2 loop (aa 168–178 of CerPrP) [Citation38], whereas the remaining three aa are Val187 (α2-helix: aa 176–197 of CerPrP), Ile206, and Ile218 (α3-helix: aa 202–226 of CerPrP) (). An amino acid substitution at Ser170 of HuPrP with the corresponding Asn from BvPrP, which in turn corresponds to aa 173 of CerPrP, increased the conversion of HuPrPC into the protease-resistant PrP (PrP-res) by CWD prions during protein PMCA analysis [Citation36]. Rabbit (Rb) PrP is known to be difficult to convert into PrP-res; however, substitution at Ile202 of RbPrP using the corresponding Val from MoPrP, which corresponds to aa 206 of CerPrP, increased the conversion of RbPrPC into PrP-res by RML prions during PMCA [Citation35]. Additionally, substitution of Ile215 of HuPrP with the Val from BvPrP, which corresponds to aa 218 of CerPrP, worsened the conversion efficiency of HuPrPC by the RML prions [Citation36]. The β2–α2 loop is known to interact with the α3-helix through hydrophobic interactions and a disulphide bridge, but single amino acid substitutions within these regions decreased hydrophobic interactions [Citation52]. Therefore, heterologous amino acid combinations in the β2–α2 loop and α3-helix may influence PrP stability and affect the conversion efficiency of rPrP during RT-QuIC.
In the current study, we showed that full-length rCerPrP as a substrate is useful for detecting CWD and atypical BSE prions in tissues with low level of prions, since the reaction of rCerPrP was not highly affected by high NBH concentrations. Additionally, we found that at least the N- and C-terminal regions of CerPrP are involved in the unique reactivity of the rCerPrP. These results will be useful for optimizing artificial rPrP for RT-QuIC reactions.
Materials and methods
Brain materials
Brain tissue from six CWD-affected deer was pooled and used as a source of CWD prions. Two brains of unaffected white-tail deer in USA were pooled and used as negative control [Citation25]. Each tissue was homogenized in phosphate-buffered saline (PBS) at a concentration of 20% and was frozen at – 80°C until use. Brain homogenate of H-BSE-affected cattle [Citation53] was kindly provided by Dr Iwamaru Y, National Institute of Animal Health, Japan. Ten-percent of brain homogenates from C-BSE [Citation54] and L-BSE [Citation55]-affected cattle were prepared with PBS and stored at – 80°C.
Construction of the expression system of recombinant PrPs
Expression plasmids for the full-length recombinant mouse and hamster PrPs (rMoPrP, rHamPrP; amino acids (aa) 23–231) were kindly provided by Dr Atarashi R, Miyazaki University, Japan. An expression plasmid for the full-length bank vole rPrP (rBvPrP; aa 23–230) was kindly provided by Dr Caughey B, National Institute of Health, USA. Genes encoding for full-length bovine PrP (BoPrP; aa 25–242), CerPrP (aa 25–233, genotype: G96M132S225Q226 [Citation56]) and sheep (ARQ) PrP (ShPrP) (aa 25–233) were amplified from the corresponding genome DNA (Supplementary Table 1). The gene fragment encoding N-terminal truncated rCerPrP (aa 94–233) was amplified from that of the full-length rCerPrP using primers listed in Supplementary Table 2. Each gene fragment was subcloned using the Zero Blunt TOPO PCR Cloning Kit (Invitrogen, USA). Nucleotide sequences were determined using BigDye v3.1 (Applied Biosystems, USA) and ABI-3130 Avant sequencer (Applied Biosystems). Each gene fragment with the correct nucleotide sequence was inserted into the NdeI and BamHI sites of pET11a (Novagen, USA).
Genes encoding the N-terminal replaced chimera of rCerPrP and rMoPrP (rMoN–CerPrP, rCerN–MoPrP) were generated using assembly PCR [Citation57] with primer sets described in the Supplementary Table 2. Genes encoding the C-terminal replaced chimeras (rCer–MoCPrP, rMo–CerCPrP) were generated through two consecutive PCRs with primers in Supplementary Table 2. Genes encoding rCerPrP and rMoPrP with two amino acid substitutions (rCerPrP–173SMo/177NMo, rMoPrP–169NCer/173TCer) were generated by assembly PCR using primer sets listed in Supplementary Table 2. Each PrP gene was inserted into pET11a for the expression and purification of rPrPs.
Expression and purification of rPrPs
Expression and purification of rPrPs were performed as described elsewhere [Citation58] with minor modifications. Briefly, BL21(DE3)pLysS cells (Invitrogen) transformed with the expression plasmid were pre-cultured in 5 mL L-Broth medium overnight at 37°C, and were further cultured in 200 mL MagicMedia E. coli expression medium (Invitrogen) for 30 h at 37°C. Bacterial cells were lysed using CelLytic B cell lysis reagent (Sigma, USA) with lysozyme (Sigma) and benzonase (Millipore, USA) for 30 min at room temperature (r.t.). Inclusion bodies were collected by centrifugation at 14,400 × g for 10 min at 4°C. The resulting pellets were washed three times using the 10% CelLytic B cell lysis reagent and stored at −80°C until use. Pellets were thawed and solubilized in denaturing buffer (100 mM sodium phosphate [pH 8.0], 10 mM tris(hydoroxymethyl)aminomethane [tris], 6 M guanidine hydrochloride [GdnHCl]) by rotating for 2 h at r.t. After centrifugation at 5,170 × g for 30 min at 4°C, the supernatant was mixed with Ni-NTA superflow resin (Qiagen, Germany), equilibrated with the denaturing buffer on a rotator for 1 h at r.t., and the Ni-NTA superflow resin was loaded onto the XK16 column (GE Healthcare, UK). The rPrP was refolded using a linear gradient of 6 to 0 M GdnHCl in refolding buffer (100 mM sodium phosphate [pH 8.0], 10 mM tris) using the ÄKTAexplore 10S system (GE Healthcare) and eluted by a linear gradient of 0 to 500 mM imidazole in 10 mM tris (pH 5.8). The fractions were pooled and dialysed against ultra-pure water. After filtration, rPrP concentration was determined by measuring absorbance at 280 nm.
RT-QuIC reaction
RT-QuIC with rMoPrP, rBvPrP, rBoPrP, rShPrP and rCerPrP was performed using reaction conditions described elsewhere [Citation22]. The reaction using rHaPrP was performed with 60 μg/ml substrate and 350 mM NaCl without SDS. Brain homogenates (20%) from uninfected deer or cattle (hereafter referred to as normal brain homogenate [NBH]) were diluted to 2% with PBS and were used as the negative control or diluent for the seed of the corresponding species. Then, 2% BHs of prion-infected animals were prepared from stock solutions (10% or 20%) (Supplementary Figure 1) and were serially diluted with PBS or species-matched 2% NBH, and 5 μl of each dilution was added into the three wells as seed. Final concentrations of brain tissue homogenates of the seed in the reaction mixture were from 10−4 [0.01%] to 10−9 [0.0000001%] after diluting with PBS. When the seeds were diluted with 2% NBH, the final concentration of the NBH in the reaction was 0.1%. Reactions were performed using the Infinite F200 microplate reader (TECAN, Switzerland) at 37°C, or at 42°C for rBvPrP, using the same cycles as described elsewhere [Citation22].
Data analysis
Thresholds of the reactions were calculated as mean thioflavin T (ThT) fluorescence intensity plus 5 × SD from the negative control wells without seed. The reactions were considered positive when the ThT fluorescence intensity exceeded the threshold [Citation59], with the following exception: if ThT fluorescence intensity temporarily exceeded the threshold within the first 1 h, the data from the first 1 h were excluded from calculations. If the oscillated waveforms were continuously observed throughout the reaction, it was considered as negative even the intensity exceeded the threshold. If oscillated waveforms were observed prior to the appearance of the typical ThT fluorescence curve observed from rPrP fibrils, the reaction was judged as positive once the typical fluorescence curve exceeded the threshold. The endpoint of the reaction was determined as the highest seed dilution that gave a positive reaction in three independent experiments with three replicates. If positive reactions were observed at 10−9 dilution, the endpoint was set as <10−9. The lag phase (h) was defined as the time required for the fluorescence intensity to exceed the threshold.
Disclosure of potential conflicts of interest
The authors declare that there are no conflict of interest.
Supplemental Material
Download MS Word (239.7 KB)Acknowledgments
We thank Dr Atarashi R, Miyazaki University, Japan and Ms Matsuo A, Nagasaki University, for kind instruction about the methods for RT-QuIC. We thank Dr Iwamaru Y, the National Institute of Animal Health, Japan for providing the brain homogenate of H-BSE affected cattle. We also thank Zensho Co., Ltd, for the BSL3 facility.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–13383.
- Haley NJ , Hoover EA . Chronic wasting disease of cervids: current knowledge and future perspectives. Annu Rev Anim Biosci. 2015;3:305–325.
- Williams ES , Miller MW . Chronic wasting disease in deer and elk in North America. Rev Sci Off Int Epiz. 2002;21(2):305–316.
- Osterholm MT , Anderson CJ , Zabel MD , et al. Chronic wasting disease in cervids: implications for prion transmission to humans and other animal species. mBio. 2019 Jul 23;10(4):e01091-19.
- Vikoren T , Vage J , Madslien KI , et al. First detection of chronic wasting disease in a wild red deer (Cervus elaphus) in Europe. J Wildl Dis. 2019 Oct;55(4):970–972.
- Mathiason CK , Powers JG , Dahmes SJ , et al. Infectious prions in the saliva and blood of deer with chronic wasting disease. Science. 2006 Oct 6;314(5796):133–136.
- Tamgüney G , Miller MW , Wolfe LL , et al. Asymptomatic deer excrete infectious prions in faeces. Nature. 2009 Sep 24;461(7263):529–532.
- Kong Q , Huang S , Zou W , et al. Chronic wasting disease of elk: transmissibility to humans examined by transgenic mouse models. J Neurosci. 2005 Aug 31;25(35):7944–7949.
- Tamgüney G , Giles K , Bouzamondo-Bernstein E , et al. Transmission of elk and deer prions to transgenic mice. J Virol. 2006 Sep;80(18):9104–9114.
- Race B , Meade-White KD , Miller MW , et al. Susceptibilities of nonhuman primates to chronic wasting disease. Emerg Infect Dis. 2009 Sep;15(9):1366–1376.
- Baron T , Vulin J , Biacabe AG , et al. Emergence of classical BSE strain properties during serial passages of H-BSE in wild-type mice. PLoS One. 2011;6(1):e15839.
- Capobianco R , Casalone C , Suardi S , et al. Conversion of the BASE prion strain into the BSE strain: the origin of BSE? PLoS Pathog. 2007 3; Mar(3): e31.
- Huor A , Espinosa JC , Vidal E , et al. The emergence of classical BSE from atypical/Nor98 scrapie. Proc Natl Acad Sci U S A. 2019;116(52):26853–26862.
- Simmons MM , Chaplin MJ , Konold T , et al. L-BSE experimentally transmitted to sheep presents as a unique disease phenotype. Vet Res. 2016 Nov 8;47(1):112.
- Wells GA , Scott AC , Johnson CT , et al. A novel progressive spongiform encephalopathy in cattle. Vet Rec. 1987 Oct 31;121(18):419–420.
- Will RG , Ironside JW , Zeidler M , et al. A new variant of Creutzfeldt-Jakob disease in the UK. Lancet. 1996 Apr 6;347(9006):921–925.
- Jacobs JG , Langeveld JP , Biacabe AG , et al. Molecular discrimination of atypical bovine spongiform encephalopathy strains from a geographical region spanning a wide area in Europe. J Clin Microbiol. 2007 Jun;45(6):1821–1829.
- Houston F , Andréoletti O . Animal prion diseases: the risks to human health. Brain Pathol. 2019 Mar;29(2):248–262.
- Comoy EE , Casalone C , Lescoutra-Etchegaray N , et al. Atypical BSE (BASE) transmitted from asymptomatic aging cattle to a primate. PLoS One. 2008;3(8):e3017.
- Mestre-Frances N , Nicot S , Rouland S , et al. Oral transmission of L-type bovine spongiform encephalopathy in primate model. Emerg Infect Dis. 2012 Jan;18(1):142–145.
- Béringue V , Herzog L , Reine F , et al. Transmission of atypical bovine prions to mice transgenic for human prion protein. Emerg Infect Dis. 2008 Dec;14(12):1898–1901.
- Sawada K , Suzuki A , Yamasaki T , et al. Estimation of prion infectivity in tissues of cattle infected with atypical BSE by real time-quaking induced conversion assay. J Vet Med Sci. 2019 Jun 6;81(6):846–850.
- Suardi S , Vimercati C , Casalone C , et al. Infectivity in skeletal muscle of cattle with atypical bovine spongiform encephalopathy. PLoS One. 2012;7(2):e31449.
- Balkema-Buschmann A , Eiden M , Hoffmann C , et al. BSE infectivity in the absence of detectable PrP(Sc) accumulation in the tongue and nasal mucosa of terminally diseased cattle. J Gen Virol. 2011 Feb;92(Pt 2):467–476.
- McNulty E , Nalls AV , Mellentine S , et al. Comparison of conventional, amplification and bio-assay detection methods for a chronic wasting disease inoculum pool. PLoS One. 2019;14(5):e0216621.
- Saborio GP , Permanne B , Soto C . Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature. 2001;411:810–813.
- Orru CD , Wilham JM , Hughson AG , et al. Human variant Creutzfeldt-Jakob disease and sheep scrapie PrP(res) detection using seeded conversion of recombinant prion protein. Protein Eng Des Sel. 2009 Aug;22(8):515–521.
- Atarashi R , Satoh K , Sano K , et al. Ultrasensitive human prion detection in cerebrospinal fluid by real-time quaking-induced conversion. Nat Med. 2011 Feb;17(2):175–178.
- Orru CD , Groveman BR , Raymond LD , et al. Bank vole prion protein as an apparently universal substrate for RT-QuIC-based detection and discrimination of prion strains. PLoS Pathog. 2015 Jun;11(6):e1004983.
- Peden AH , McGuire LI , Appleford NEJ , et al. Sensitive and specific detection of sporadic Creutzfeldt-Jakob disease brain prion protein using real-time quaking-induced conversion. J Gen Virol. 2012 Feb;93(Pt 2):438–449.
- Wilham JM , Orru CD , Bessen RA , et al. Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog. 2010 Dec 2;6(12):e1001217.
- Hoover CE , Davenport KA , Henderson DM , et al. Endogenous brain lipids inhibit prion amyloid formation in vitro. J Virol. 2017 May 1;91(9):e02162-16.
- Orru CD , Wilham JM , Raymond LD , et al. Prion disease blood test using immunoprecipitation and improved quaking-induced conversion. mBio. 2011;2(3):e00078–11.
- Billeter M , Riek R , Wider G , et al. Prion protein NMR structure and species barrier for prion diseases. Proc Natl Acad Sci U S A. 1997;94:7281–7285.
- Erana H , Fernandez-Borges N , Elezgarai SR , et al. In vitro approach to identify key amino acids in low susceptibility of rabbit prion protein to misfolding. J Virol. 2017 Dec 15;91(24):e01543-17.
- Kurt TD , Aguilar-Calvo P , Jiang L , et al. Asparagine and glutamine ladders promote cross-species prion conversion. J Biol Chem. 2017 Nov 17;292(46):19076–19086.
- Watts JC , Giles K , Serban A , et al. Modulation of Creutzfeldt-Jakob disease prion propagation by the A224V mutation. Ann Neurol. 2015 Oct;78(4):540–553.
- Gossert AD , Bonjour S , Lysek DA , et al. Prion protein NMR structures of elk and of mouse/elk hybrids. Proc Natl Acad Sci U S A. 2005 Jan 18;102(3):646–650.
- Harrathi C , Fernandez-Borges N , Erana H , et al. Insights into the bidirectional properties of the sheep-deer prion transmission barrier. Mol Neurobiol. 2019 Aug;56(8):5287–5303.
- Sigurdson CJ , Nilsson KP , Hornemann S , et al. A molecular switch controls interspecies prion disease transmission in mice. J Clin Invest. 2010 Jul;120(7):2590–2599.
- Denkers ND , Henderson DM , Mathiason CK , et al. Enhanced prion detection in biological samples by magnetic particle extraction and real-time quaking-induced conversion. J Gen Virol. 2016 Aug;97(8):2023–2029.
- Orru CD , Wilham JM , Vascellari S , et al. New generation QuIC assays for prion seeding activity. Prion. 2012 Apr-Jun;6(2):147–152.
- Orru CD , Hughson AG , Groveman BR , et al. Factors that improve RT-QuIC detection of prion seeding activity. Viruses. 2016 May 23;8(5):140.
- Abalos GC , Cruite JT , Bellon A , et al. Identifying key components of the PrPC-PrPSc replicative interface. J Biol Chem. 2008 Dec 5;283(49):34021–34028.
- Hara H , Miyata H , Das NR , et al. Prion protein devoid of the octapeptide repeat region delays bovine spongiform encephalopathy pathogenesis in mice. J Virol. 2018;92(1):e01368–17.
- Lawson VA , Priola SA , Wehrly K , et al. N-terminal truncation of prion protein affects both formation and conformation of abnormal protease-resistant prion protein generated in vitro. J Biol Chem. 2001 Sep 21;276(38):35265–35271.
- Duque Velasquez C , Kim C , Herbst A , et al. Deer prion proteins modulate the emergence and adaptation of chronic wasting disease strains. J Virol. 2015 Dec;89(24):12362–12373.
- Johnson CJ , Duque Velasquez C , Vanderllo JP , et al. Prion protein polymorphisms affect chronic wasting disease progression. PLoS One. 2011;6(3):e17450.
- Zahn R , Liu A , Luhrs T , et al. NMR solution structure of the human prion protein. Proc Natl Acad Sci U S A. 2000;97(1):145–150.
- Liemann S , Glockshuber R . Influence of amino acid substitutions related to inherited human prion diseases on the thermodynamic stability of the cellular prion protein. Biochemistry. 1999;38:3258–3267.
- Biljan I , Giachin G , Ilc G , et al. Structural basis for the protective effect of the human prion protein carrying the dominant-negative E219K polymorphism. Biochem J. 2012 Sep 1;446(2):243–251.
- Lee J , Chang I , Yu W . Atomic insights into the effects of pathological mutants through the disruption of hydrophobic core in the prion protein. Sci Rep. 2019 Dec 16;9(1):19144.
- Okada H , Iwamaru Y , Imamura M , et al. Experimental H-type bovine spongiform encephalopathy characterized by plaques and glial- and stellate-type prion protein deposits. Vet Res. 2011 Jun;23(42):79.
- Shindoh R , Kim CL , Song CH , et al. The region approximately between amino acids 81 and 137 of proteinase K-resistant PrPSc is critical for the infectivity of the Chandler prion strain. J Virol. 2009 Apr;83(8):3852–3860.
- Hagiwara K , Yamakawa Y , Sato Y , et al. Accumulation of mono-glycosylated form-rich, plaque-forming PrPSc in the second atypical bovine spongiform encephalopathy case in Japan. Jpn J Infect Dis. 2007;60:305–308.
- Moreno JA, Telling GC . Molecular mechanisms of chronic wasting disease prion propagation. Cold Spring Harb Perspect Med. 2018 Jun 1;8(6):a024448.
- Suzuki A , Yamasaki T , Hasebe R , et al. Enhancement of binding avidity by bivalent binding enables PrPSc-specific detection by anti-PrP monoclonal antibody 132. PLoS One. 2019;14(6):e0217944.
- Atarashi R , Moore RA , Sim VL , et al. Ultrasensitive detection of scrapie prion protein using seeded conversion of recombinant prion protein. Nat Methods. 2007 Aug;4(8):645–650.
- Henderson DM , Davenport KA , Haley NJ , et al. Quantitative assessment of prion infectivity in tissues and body fluids by real-time quaking-induced conversion. J Gen Virol. 2015 Jan;96(Pt 1):210–219.
