ABSTRACT
Cellular Prion Protein (PrPC) is a well-studied protein as the substrate for various progressive untreatable neurodegenerative diseases. Normal functions of PrPC are poorly understood, though recent proteomic and transcriptomic approaches have begun to reveal common themes. We use our compound prp1 and prp2 knockout mutant zebrafish at three days post fertilization to take a transcriptomic approach to investigating potentially conserved PrPC functions during development. Gene ontology analysis shows the biological processes with the largest changes in gene expression include redox processing, transport and cell adhesion. Within these categories several different gene families were prevalent including the solute carrier proteins, cytochrome p450 enzymes and protocadherins. Continuing from previous studies identifying cell adhesion as an important function of PrPC we found that in addition to the protocadherins there was a significant reduction in transcript abundance of both ncam1a and st8sia2. These two genes are involved in the early development of vertebrates. The alterations in cell adhesion transcripts were consistent with past findings in zebrafish and mouse prion protein mutants; however E-cadherin processing after prion protein knockdown failed to reveal any differences compared with wild type in either our double prp1/prp2 mutant fish or after prp1 morpholino knockdown. Our data supports a cross species conserved role for PrPC in the development and maintenance of the central nervous system, particularly by regulating various and important cell adhesion processes.
Introduction
The cellular prion protein (PrPC) is a well-conserved protein across mammals and to a lesser extent across other vertebrates. It has fascinated researchers since its identification as the cause of a variety of neurodegenerative disorders including Creutzfeldt Jakob disease (CJD) in humans, scrapie in sheep, chronic wasting disease (CWD) in cervids and bovine spongiform encephalopathy (BSE) in cattle via a conformational change in PrPC to become scrapie prion protein, or PrPSc [Citation1,Citation2]. Interest is often focussed on the infectious capabilities of the PrPSc conformation to spread disease, including across species, dubbed ‘the protein only hypothesis’[Citation2]. In addition to its ability to misfold into PrPSc, normally folded PrPC has been implicated in the pathology of Alzheimer’s disease by acting as a receptor for soluble amyloid-beta oligomers [Citation3–7]. Despite being subjected to such a large amount of scrutiny, the actual normal physiological functions of PrPC are not well understood, nor how these functions may be affected under disease conditions [Citation8]. Here we perform transcriptomic analysis on wild-type (WT) vs mutant zebrafish, which lack both prp1 and prp2 gene products to identify potential functions of PrPC during early development.
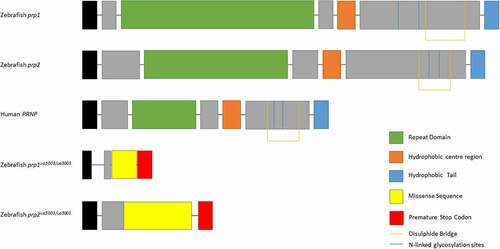
Zebrafish possesses two prion protein genes homologous to mammalian PRNP, prp1 and prp2, due to a whole genome duplication which occurred in the teleost lineage [Citation9,Citation10]. While both prp1 and prp2 are larger than their mammalian counterpart and therefore share little similarity at the amino acid level, all predicted functional domains of PrPC are present in both including an N-terminal signal peptide, a repetitive region, a central hydrophobic domain, a disulphide bridge, two N-linked glycosylation sites and a GPI anchor for attachment to the cell membrane [Citation11] (). This conservation of PrP across evolutionary time indicates that this protein has ancient and important physiological functions.
Figure 1. Prion proteins of zebrafish Prp1, Prp2, human PrPC and Prp1ua5003/ua5003 and Prp2ua5001/ua5001 mutant proteins. Zebrafish prion protein genes are larger however have the same conserved domains as human PRNP: the repeat domain (green), hydrophobic centre region (orange), hydrophobic tail (blue), N-linked glycosylation sites (blue lines) and di-sulphide bridge (orange lines). The prp1ua5003/ua5003 and prp2ua5001/ua5001 alleles have frameshift deletions near the beginning of the coding exon, leading to a missense sequence of amino acids (yellow), pre-mature stop codons (red) and a shortened, nonsense transcript
Transmission of prion diseases to fish by crossing the species barrier has been previously investigated. The difference in size of mammalian to fish PrPs and lower conservation means the chance of transmissibility between species is low but the conserved domains may suggest it is not impossible. Studies have shown that sea bream fed with either scrapie or BSE contaminated brain homogenate results in signs of neurodegeneration and deposits in the brain which reacted to antibodies against sea bream PrP. These deposits developed faster in fish challenged with BSE prions and did not occur in those fed with non-contaminated brain homogenate [Citation12]. While deposits and histological signs of neurodegeneration were observed, there were no clinical symptoms of prion disease and passaging the disease onto additional animals was not reported. Additional in vitro studies using mouse cell culture demonstrated that three different fish PrP proteins, including zebrafish Prp1 and Prp2, did not increase the formation of proteinase K resistant prion conversion [Citation13]. While this supports that it is unlikely fish Prps can misfold into pathogenic species after exposure to mammalian prions, the various nature of different PrPSc strains means it still remains a possibility.
There have been many proposed functions for PrPC including cell adhesion, learning and memory, maintaining circadian rhythm, aspects of the immune response, synaptic function, neuroprotection and more [Citation14,Citation15]. Determining which of these is a direct function of PrPC has proven difficult since animal knockout studies have not shown any obvious overt phenotype in both mice and zebrafish models [Citation16,Citation17]. This is in stark contrast to what can be seen after acute knockdown of PrPC, such as morpholino knockdown of prp1 in zebrafish leading to a lethal phenotype during gastrulation [Citation18]. This phenotype is particularly interesting due to a similar phenotype occurring after knockdown of certain ZIP proteins, from which PrPC may be phylogenetically linked [Citation19]. Discrepancies between chronic stable knockout of PrPC and acute knockdown may suggest robust compensatory mechanisms in mutants allowing for their survival. The lack of overt phenotypes after Prnp gene knockout is surprising as PrPC is evolutionarily well conserved which would suggest an essential function; yet there is little evidence for any particular gene(s) which may be involved in functional redundancy.
Studies in zebrafish, from our own lab and others, support a conserved role of zebrafish prion proteins in cell adhesion [Citation18,Citation20]. In addition, proteomic analysis in cell culture has revealed a robust role for mammalian PrPC during epithelial–mesenchymal transition, a cell adhesion event during development, through affecting NCAM1 polysialylation via ST8SIA2 production [Citation21,Citation22]. These studies suggest it plays an important role in the early development of vertebrates and possibly subsequently acts to maintain areas in which it is expressed. Therefore, we have carried out RNA-sequencing analysis on zebrafish larvae to further investigate the role of PrPC during development.
Results
Compound homozygous prp1ua5003;ua5003; prp2ua5001;ua5001 knockout mutant exhibited transcriptomic changes
WT and prp1ua5003/ua5003; prp2ua5001/ua5001 homozygous compound mutant zebrafish larvae underwent RNA-sequencing analysis. Prion compound mutant fish have engineered small deletion mutations near the beginning of the coding sequence leading to frameshifts in each gene, premature stop codons causing truncated proteins and predicted loss of function [Citation17,Citation23] (). Three pools of 50 3dpf WT AB fish and prp1ua5003/ua5003; prp2ua5001/ua5001 compound homozygous mutant fish were collected and sent to Otogenetics for RNA-sequencing (). The age of 3dpf was chosen because it represents a time point, early in development of zebrafish, when the CNS is present, the embryo is available for genetic manipulation and where there is expected to be an overlap in the expression of both prp1 and prp2 [Citation11]. Using a fold change cut-off of log20.5 (i.e. there is either 50% more or 50% less transcript abundance) we found a significant change in the transcript abundance of 1249 genes, with 745 showing an increase in transcript abundance and 504 showing a decrease in transcript abundance in compound mutant prp1ua5003/ua5003; prp2ua5001/ua5001 fish compared to WT ( and ). We have previously shown a decrease in relative transcript abundance of prp1 and prp2 in prp1ua5003/ua5003; prp2ua5001 mutants, predicted to be due to nonsense mediated decay of nonsense mRNA [Citation17], and as expected prp1 and prp2 were among the top genes showing a decrease in transcript abundance.
Table 1. Total number of genes with either a significant increase or decrease in transcript abundance between wild-type and prp1ua5003/ua5003; prp2ua5001/ua5001 homozygous mutant fish with a log2 fold change equal to or greater than 0.5
Figure 2. RNA-Sequencing show 1249 genes with an increase or decrease of log2 fold change of 0.5 between wild-type and compound homozygous prion mutant zebrafish larvae. (A) Scatter graph showing relative FPKM values for wild-type (X-axis) and mutant (Y-axis) genes after RNA-sequencing. (B) Methodology diagram showing RNA-sequencing workflow. Two groups, wild-type and prion mutant (prp1ua5003/ua5003; prp2ua5001/ua5001) homozygous fish, each with three replicates containing a pool of 50 3dpf larvae were processed and sent for RNA-sequencing. Heatmap displays sample of the 25 genes with a biggest differential abundance in FPKM in wild-types compared to mutants. (C) Relative transcript abundance between wild-type and prion mutant fish comparing prp1 and prp2 through both RNA-sequencing and RT-qPCR analysis. D) Relative transcript abundance of ncam1a and st8sia2 through both RNA-sequencing and RT-qPCR analysis. * = P < 0.05
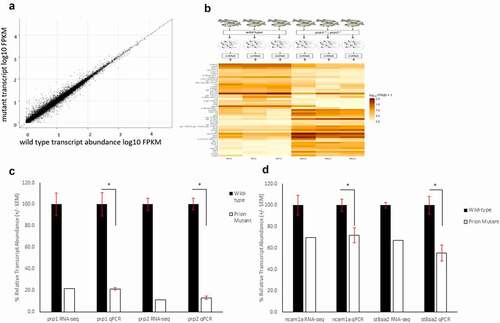
RT-qPCR experiments for both prp1 and prp2 confirmed a significant reduction in transcript abundance for both genes in our mutants of 79% and 87% respectively (). Initial RT-qPCR of select genes (implicated in eye development) does not strongly support validation of the RNA-sequencing results, this could be due the circadian nature of the expression of those genes, and variability due to the low transcript abundance perhaps being difficult to detect through RT-qPCR, though we have yet to prove either explanation (Supplementary Figure 1). On the other hand, changes in transcript abundance for several other genes were verifiable by RT-qPCR ( and described below). For the full results, see the published transcriptome (GEO accession: GSE164423).
Amongst the ten genes showing the largest increase in transcript abundance in mutants compared to WT, five have been linked to proteolytic/hydrolytic processes (cel.1, ela3l, prss59.1, dpp4 and c6ast4). While the proteolytic processing of PrPC itself is becoming increasingly well documented [Citation24–26], its actions in the proteolytic processing of other molecules, whether directly or indirectly is somewhat less appreciated. PrPC is becoming increasingly associated with cell adhesion [Citation18,Citation27,Citation28], proliferation [Citation29,Citation30] and signalling and it is possible it acts in complexes that process other proteins and molecules as part of these events.
Among the ten genes with the biggest decrease in transcript abundance there does not appear to be a consistent biological process linking them. The gene with the biggest reduction in relative transcript abundance is growth hormone releasing hormone (ghrh). Like in mammals Ghrh causes increases in the release of growth hormone during development, particularly in the central nervous system and gut. Secretion of Ghrh is controlled in a circadian manner and has antagonistic effects to somatostatin, with Ghrh promoting short wave sleep while somatostatin promotes deeper REM sleep [Citation31]. Growth hormone levels decrease with age and have been suggested to be involved in the ageing process related to a decrease in physiological functions controlled by the hypothalamus [Citation32]. Interestingly, both somatostatin 1 and somatostatin receptor 5 show a significant increase in transcript abundance in prion mutants (60% and 65% respectively). Through recent collaborations we have shown a disruption in the sleep/wake cycle of prp1ua5003/ua5003; prp2ua5001/ua5001 mutant zebrafish after exposure to amyloid-beta oligomers [Citation7].
Gene ontology analysis of biological processes affected in prp1 and prp2 mutant zebrafish
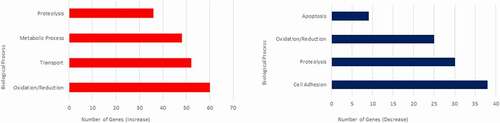
The most populous Biological Process categories of genes altered in zebrafish prion mutants are reported in . Amongst the processes exhibiting a significant increase in transcript abundance, the oxidation/reduction category is represented most often with genes showing a significant increase in transcript abundance (). Gene ontology analysis for genes with a significant decrease in transcript abundance again shows a similar trend to processes previously linked with PrPC [Citation33]. shows the most populated biological process categories with a significant decrease in transcript abundance. Cell adhesion is the largest, with the majority of genes belonging to the protocadherin (pcdh) family showing a significant reduction in transcript abundance, totalling 31 out of the 38 cell adhesion genes. The pcdh genes affected belong to the pcdh2 alpha and gamma sub clusters. Protocadherins are thought to be particularly involved in the cell adhesion of the early central nervous system [Citation34], and this reduction in transcript abundance in our mutant fish may in the future help shed light on some of our previous findings suggesting a delay in neural development after prion protein knockdown [Citation35].
Table 2. Most populated biological process gene ontologies for genes with a log2 fold change of 0.5 or greater
Table 3. Most populated biological process gene ontologies for genes with a log2 fold change of −0.5 or less
Figure 3. Biological Process Gene Ontologies most affected in 3dpf prion mutant (prp1ua5003/ua5003; prp2ua5001/ua5001) zebrafish compared to wild type. Biological processes showing genes with the greatest increase in transcript abundance are shown on the left (red), and genes with the biggest decrease in transcript abundance are shown on the right (blue)
Prion protein is involved in cell adhesion processes in early larval development
Previous work has established a link between PrPC and cell adhesion. Schmitt-Ulms and colleagues used a proteomic and transcriptomic approach in PrPC knockout cells, to show a role for PrPC in the polysialylation of Ncam1 [Citation21]. In zebrafish, Malaga-Trillo and colleagues found a link between Prp1 and cell adhesion including, though not necessarily limited to, effects on the maturation of E-cadherin during embryogenesis [Citation18].
Results from our RNA-sequencing data do not show a significant difference in the transcript abundance of E-cadherin between mutants and WT, though this is not surprising if the role of prion protein is in the maturation of the protein (a proteolytic event) and not of the expression of the gene. In zebrafish, ncam1a is a homologue of NCAM1 and the Ncam1a protein is also polysialylated by St8sia2 [Citation36]. There is a 30% reduction in the transcript abundance of ncam1a and a 33% reduction in the transcript abundance of st8asia2 in our mutant fish compared to WT (). We confirmed this through RT-qPCR, finding a similar reduction in transcript abundance of ncam1a, of approximately 30%, and 50% for st8asia2.
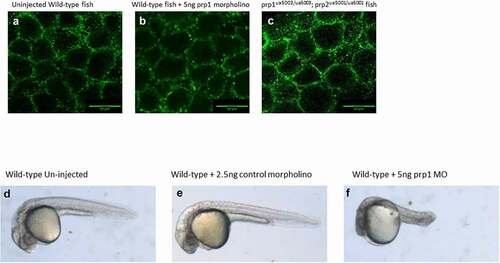
After establishing these changes in ncam1a and st8sia2 transcript abundance we next looked at whether there were changes in the processing of E-cadherin in our prion mutant fish compared to WT. Previous work has established a role of prp1 in regulating E-cadherin processing in zebrafish [Citation18], and our lab has previously shown changes in both E-cadherin and β-catenin localization after morpholino knockdown of prp2 [Citation20]. Zebrafish embryos for both WT, compound mutant fish and prp1 morpholino injected fish were stage-selected for those entering the shield stage of embryogenesis, approximately 6 hpf. We were not able to identify any changes to the processing or localization of E-cadherin either in our prp1ua5003/ua5003; prp2ua5001/ua5001 mutant fish or WT fish injected with 5ng prp1 morpholino (). We kept morpholino injected fish and control injected to fish to see if the morpholino was influencing the fish as they developed. We did not see a significant increase in the number of embryos perishing after 1dpf between the morpholino and control injected embryos (data not shown). By 3dpf morpholino injected fish showed clear signs of necrosis and developmental abnormalities compared to the control injected and un-injected control (). These results would suggest that morpholino knockdown of prp1 was causing an effect compared to the control injected fish. Why this effect is different compared to what has been previously published is not immediately clear. Morpholinos have come under increased scrutiny due to differences seen in morphants compared to mutants; however this could be due to acute knockdown of genes having more impact than stable, chronic knockout [Citation37,Citation38]. We previously discussed at length potential explanations for the disparate results during acute knockdown vs. stable mutation of prion proteins [Citation17] and concluded that results from these morpholino reagents should be interpreted with caution.
Figure 4. (A–C) There does not appear to be a difference in the maturation or localization of e-cadherin in either 5ng prp1 morpholino injected AB zebrafish or uninjected prp1ua5003/ua5003; prp2ua5001/ua5001 homozygous mutant fish compared to uninjected wild-type controls. (F–E) After 3dpf wild-type larvae injected with prp1 MO (f) show significant signs of necrosis and developmental abnormalities compared to uninjected and control injected larvae (d, e)
KEGG analysis shows decreased transcript abundance in focal adhesion and actin cytoskeleton regulation pathways
The most affected pathway in prion mutants is metabolism, exhibiting both an increase and decrease in relative transcript abundance; however due to the sheer size of this KEGG pathway there was little consistency in processes affected; therefore we focussed our attention on the next most populous pathways.
KEGG analysis shows the two most populated pathways with genes having a decrease in transcript abundance are the focal adhesion kinase (FAK) pathway and actin cytoskeleton regulation pathway. There are two FAK homologues in zebrafish, ptk2ab (fak1a) and ptk2aa (fak1b). While neither shows a significant change in transcript abundance in our zebrafish mutants, the FAK pathway does show several genes with a significant reduction in transcript abundance in close proximity to the FAK genes in the pathway. Genes with direct interactions with FAK showing a significant decrease in transcript abundance include members of the calpain, actinin, talin and integrin families (). There is a significant reduction in transcript abundance in capn2l, tln2a, actn3b and bcar1. All of these have been heavily linked with the regulation of the actin cytoskeleton, affecting cell mobility, division and differentiation [Citation39–43]. There is considerable overlap between genes affected in the FAK pathway and the regulation of the actin cytoskeleton pathway:raf1b, actn3b, bcar1, capn2l, itga9, pak6b, pik3r2, rac1b show a significant decrease in transcript abundance in both.
Figure 5. Snapshot of the Focal Adhesion Kinase KEGG pathway. Gene products highlighted in red show those with a significant decrease in transcript abundance, with the specific gene italicized underneath
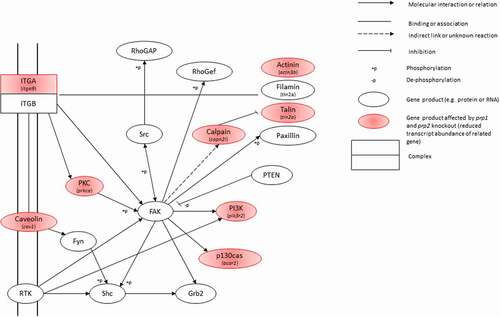
Taken alongside the large number of protocadherin family members also showing a reduction in transcript abundance (), as well as ncam1a and st8sia2 this suggests that prp1 and prp2 help regulate the processes of cell adhesion and differentiation during early development.
Discussion:
Conserved roles of PrPC across species
Despite numerous animal knockout models, there is yet to be a clear and obvious phenotype attributed to the loss of PrPC. This could be due to the age at which the animals were being observed, with evidence both from our lab and others that PrPC may be important in the early development of organisms. Acute transient knockdown of Shadoo (‘shadow of prion protein’) in PrPC knockout mice led to embryonic lethality [Citation44], though this effect was not seen in a combined knockout model of Shadoo and PrPC [Citation45]. Morpholino knockdown of prp1 in zebrafish led to arrest during gastrulation attributed to deficits in cell adhesion [Citation18,Citation46] and our own analysis of prp1 and prp2 in zebrafish suggests further, if non-essential, roles in early development [Citation17,Citation20,Citation35]. This does not account for the relatively diverse and high expression levels of prion protein after development and throughout adulthood suggesting its function may be pleotropic. In the current study, we focus on changes to the transcriptome of zebrafish larvae in our prp1ua5003/ua5003; prp2ua5001/ua5001 mutant fish during early development and identify changes in the transcript abundance of several gene families and related biological processes and further focus on cell adhesion.
Previous transcriptomic and proteomic approaches to investigate changes after the loss of PrPC in mice or mammalian cells have found changes in a consistent set of biological processes including cell adhesion, apoptosis, proteolysis, protection against ROS, the immune system and aspects of the cell cycle [Citation27,Citation33,Citation47,Citation48]. Here, our own transcriptomic approach and comparison of the biological process gene ontologies finds a similar group of processes affected. It is worth noting that we did not see great similarity at the individual gene level with that of other studies. This is likely due to the age of the animals in question. Our zebrafish were 3 days post fertilization (dpf) and would have undergone gastrulation. Similar studies have used either younger zebrafish morphants [Citation49], or E6.7 and E7.5 mice [Citation33] which would not have begun or completed gastrulation. This difference in the relative ages and developmental stages, as well as a different species, may account for this lack of gene expression similarity. We also used whole zebrafish larvae, as opposed to specifically the brain or cell culture, as we were interested in a role of PrPC across the development of the entire organism. There was still a large overlap in the categories of biological process affected overall.
Prp1 and prp2 regulation of cell adhesion genes during development
One of the more dramatic phenotypes involving prion protein is the gastrulation arrest reported by some scientists in early zebrafish embryos caused by morpholino knockdown of prp1 leading to disruption of the localization of E-cadherin [Citation18]; however we have been unable to replicate this ourselves (). This may be due to a difference in concentration of morpholino. We have previously shown that higher morpholino doses still cause phenotypes in our prp1 mutant zebrafish, which suggests that the morpholinos have non-specific effects. As such we elected to use lower morpholino doses which did not result in phenotypes in our mutants [Citation17]. As the gastrulation arrest associated with E-cadherin had robust controls demonstrating rescue of the phenotype those results are unlikely to be due to off-target effects at the concentration used [Citation18]. As previous work has also described the effects of PrPC on cell adhesion, particularly the polysialylation of NCAM1 by ST8SIA2 as a requirement for cells to undergo epithelial to mesenchymal transition [Citation21] and through direct interaction with NCAM1 for neuronal differentiation [Citation30], we investigated whether there were similar changes in expression of the zebrafish ncam1a and st8sia2 and further identified significant decreases in transcript abundance of protocadherins. Transcript abundance of ncam1a was significantly reduced in our prion mutants, as was the transcript abundance of st8sia2.
Cell adhesion is the largest gene ontology category with a significant decrease in transcript abundance in our mutants. There are 38 genes associated with the cell adhesion process affected at the chosen log2 fold change cut-off, 31 of which belong to the protocadherin family. Protocadherins are the largest subfamily of cadherin cell adhesion molecules and are primarily expressed within the central nervous system where they are important for its early and continued development [Citation34]. Outside of the chosen fold change cut-off used in the gene ontology analysis are further protocadherins, including members of the pcdh1 alpha and gamma clusters, and two non-clustered delta protocadherins, pcdh19 and pcdh10b. All of these show a reduction in transcript abundance in our prp1ua5003/ua5003; prp2ua5001/ua5001 compound mutants compared to WT.
The age of the zebrafish used for RNA-sequencing was determined by our previous work on prp1 morphants and prp2 mutants while trying to capture a time where both genes are expected to be expressed [Citation17,Citation35]. Combined with the prp1 morpholino data in , these results may suggest a role of prp1 and prp2 in the expression and regulation of protocadherins and other cell adhesion genes such as ncam1a in development of the CNS. Furthermore, genes affected in the FAK pathway would suggest that these processes may be affected through controlling the migration and differentiation of cells which would also support the gastrulation phenotype seen by others [Citation18].
Prion protein mutant fish show decrease in focal adhesion and actin regulation transcript abundance
Aside from the metabolism KEGG pathway, KEGG analysis shows that the two most affected pathways with a decrease in gene transcript abundance are the focal adhesion kinase pathway and the regulation of actin cytoskeleton pathway. There are 11 genes affected in the FAK pathway and 13 genes affected in the regulation of actin cytoskeleton pathway; between the two there is an overlap of 7 genes. Combined, this suggests the involvement of prion protein in not only cell adhesion processes but also processes which regulate cell motility and differentiation. In addition to cell motility the FAK pathway is heavily involved in angiogenesis [Citation50] which previous transcriptomic studies have shown to be a biological process affected in developing PrPC knockout mice [Citation33].
Neuroprotection and roles in immune function
Further, of particular interest is the decreased relative transcript abundance of pcdh19, a non-clustered protocadherin which has been shown to be one of the highest genetic risk factors relating to epilepsy [Citation51]. Mice lacking PrPC have been shown to be at an increased risk of seizures [Citation52] and we have also shown this in our prp1−/- and prp2−/- knockout zebrafish [Citation17,Citation53]. This adds to the increasing amount of data showing PrPC plays a neuroprotective role in vertebrates; this may explain why many phenotypes now becoming apparent occur only after stress is put on the animal.
Finally, Ncam1 has been shown to be expressed in cells involved in the innate immune system including natural killer (NK) cells [Citation54], which also express PrPC. The expression of PrPC in immune system cells and tissues is an understudied area of research but there is evidence to suggest it is involved in immune quiescence [Citation55]. This coincides with its higher expression levels in tissues where inflammation could be severely damaging, such as the CNS and testes. Regulation of Ncam1 by PrPC may therefore be a method in which immune suppression is enacted in these tissues to prevent further damage under stress; however more work is required to properly establish this.
Concluding remarks
To conclude, here we present a transcriptome analysis comparing WT zebrafish and our prp1ua5003/ua5003; prp2ua5001/ua5001 mutant zebrafish early in development (3dpf). We find significant changes in transcript abundance of genes in several different biological process gene ontology categories including cell adhesion, proteolysis and oxidation/reduction processes. Importantly, while there is not much overlap at the individual gene level compared to similar studies done in mice our results do overlap considerably at the categorical level. This implies an important, cross-species conserved role of PrPC in the early development of organisms.
The data support past conclusions that PrPC participates in cell adhesion pathways. Further, the data implicate a shared role for PrPC in regulating NCAM1 and its adhesion functions via ST8SIA2; this shared function of PrPC between mammals and fish is consistent with this being part of an ancient role for PrPC early in its evolution [Citation19].
Materials and methods
Animal ethics, zebrafish fish lines and husbandry
Zebrafish were raised, maintained and bred following Animal Care and Use Committee: Biosciences procedures at the University of Alberta following guidelines set by the Canadian Council of Animal Care. Fish were kept at the University of Alberta fish facility at 28 ͦC under a 14:10 hour light/dark cycle as previously described [Citation56]. The AB strain of zebrafish was used as WT fish as controls for experiments, as well as the background for the prp1ua5003/ua5003; prp2ua5001/ua5001 compound homozygous mutants which we previously generated in our lab [Citation17,Citation23].
RNA-sequencing analysis of WT and prp1ua5003/ua5003; prp2ua5001/ua5001 mutant larvae
AB WT and prp1ua5003/ua5003; prp2ua500/ua5001 fish (ZFIN ID: ZDB-ALT-181,113-1 and ZDB-ALT-130,724-2) were bred and raised to 3dpf. 50 larvae were taken to form three replicates of each group, WT and mutant, totalling six different samples. Each pool of 50 larvae was homogenized in TRIzol (Invitrogen/ThermoFisher Scientific catalogue no. 15,596,026) with a rotor stator homogenizer (VWR catalogue no. 47,747–370, Radnor, PA) and shipped to Otogenetics (Atlanta, GA) for Illumina PE100-125 and HiSeq2500 sequencing and DNAnexus Platform standard RNAseq analysis at a depth of greater than 41 million reads. Read alignments and annotation were done using the TopHat and Bowtie pipelines and initial quantification analysis of differential gene expression was done using Cufflinks [Citation57,Citation58]. Upon receipt of results it was found that two of the three prp1ua5003/ua5003; prp2ua5001samples might have been contaminated with WT transcripts. We took a conservative approach and filtered these samples out of the analysis. The integrity of the remaining prp1ua5003/ua5003; prp2ua5001/ua5001 sample was rigorously screened to ensure it lacked WT transcript by assessing SNPs that were consistently present in mutant vs WT samples. Further analysis was performed using the R Programming Language (Version 4.0.0) packages CummRbund and ggplot2 [Citation59–61].
RT-qPCR detection of selected genes of interest
Experiments were performed in compliance with the MIQE guidelines (Minimum Information for Publication of Quantitative Real-Time PCR Experiments [Citation62]). RNA samples for all genes were extracted from either 3dpf WT AB or compound homozygous prp1ua5003/ua5003; prp2ua5001/ua5001 mutant zebrafish. RNA extraction was done from pools of 15–20 larvae previously stored in RNAlater (Ambion/ThermoFisher Scientific, catalogue no. AM7021) and processed using the RNeasy Kit (Qiagen catalogue #74,104, Toronto, ON, Canada) following the manufacturers protocol. Homogenization of larvae was done in RLT buffer with a rotor stator homogenizer as stated above. RNA concentration was quantified using a Nanodrop 2000 spectrophotometer (Thermo Scientific). RNA integrity was confirmed using an Agilent RNA 6000 NanoChip and Agilent 2100 Bioanalyser for numbers of at least 7/10. cDNA was generated using a qScript Supermix kit (Quanta BioSciences catalogue #95,048–100, Beverly, MA, USA) and qPCR carried out as described previously [Citation17]. Three technical replicates were used for each biological replicate and transcript abundance was normalized to β-actin. Statistical analysis for relative fold change in transcript abundance was done using RQ values. Primers used for the genes were as follows: prp1 forward: 5ʹ-ATCCGGCACTTATTGAGCAG-3ʹ, prp1 reverse: 5ʹ-CACTTCGGAGATGCTGTGTC-3ʹ, prp2 forward: 5ʹ-CCAACTCTGCAGCTAGTACA-3ʹ, prp2 reverse: 5ʹ-CAGTGTCGCCGTCATTATCA-3ʹ, st8sia2 forward: 5ʹ- GACCAACCATGTCCAGATCAAAC-3ʹ, st8sia2 reverse: 5ʹ- TGGATCTCATCACAAAAGCGAGTA-3ʹ, ncam1a forward: 5ʹ-GTAGCTGGAAAAAGGCCCCT-3ʹ, ncam1a reverse: 5ʹ-AACAGTGGCAGCTACCTGTC −3ʹ. All primers were validated before use.
For the RT-qPCR primers used for genes related to eye development see supplementary Table 1.
Morpholino injections in zebrafish embryos
An antisense prp1 morpholino oligonucleotide (MO) was purchased from Gene Tools, LLC (Philomath, OR) and has been previously described by us and others [Citation18,Citation35] (ZFIN ID: ZDB-MRPHLNO-100,423-6), a standard negative control morpholino was also acquired and used in experiments (5ʹ-CCTCTTACCTCAGTTACAATTTATA-3ʹ). Injection solutions consisted of 1.0 μl KCl, 1.0 μl 0.25% dextran red, MO specific volume resulting in a 5ng/μl concentration for prp1-MO or 2.5ng/μl for the standard MO and the volume finalized to 10 μl with nuclease free water. Embryos identified to be at the 1–2 cell stage were mounted on an agarose plate and injected with 1nl of injection solution with the volume previously calibrated using an ocular micrometre, injecting into mineral oil. Larvae at 3dpf were imaged using a Leica M164 dissecting microscope with a Leica DFC 400 camera.
E-cadherin immunohistochemistry
Embryos identified at the shield stage of development (approximately 6 hpf) were manually dechorionated and fixed in 4% paraformaldehyde and processed for antibody staining. An anti-mouse E-cadherin antibody (BD Biosciences, 610,181) at a 1:5000 dilution was used, and embryos were imaged using a Zeiss LSM 700 scanning confocal microscope and Zen 2010 software (Carl Zeiss Imaging). Images were analysed with ImageJ.
Gene ontology, KEGG pathway and statistical analysis
Genes identified to have either a log2 fold change of 0.5 or greater (increase in transcript abundance) or −0.5 or lower (decrease in transcript abundance) were selected for gene ontology and KEGG pathway analysis using DAVID version 6.8 [Citation63,Citation64]. Additional statistical analysis and visualization was carried out using the tidyverse group of R packages [Citation65] and Microsoft Excel.
Supplemental Material
Download Zip (96.6 KB)Acknowledgments
Operating funds were from Alberta Prion Research Institute–Alberta Innovates BioSolutions and the Alzheimer Society of Alberta and the Northwest Territories. PLAL was supported by Studentships from Alzheimer Society of Canada and Alberta Innovates Health Solutions. We appreciate the technical assistance of Richard Kanyo.
Disclosure statement
The authors have no competing interests.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Kovács GG, Trabattoni G, Hainfellner JA, et al. Mutations of the prion protein gene: phenotypic spectrum. J. Neurol. 2002;249:1567–1582.
- Prusiner S. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144.
- Um JW, Nygaard HB, Heiss JK, et al. Alzheimer amyloid-β oligomer bound to postsynaptic prion protein activates Fyn to impair neurons. Nat. Neurosci. 2012;15:1227–1235.
- Laurén J, Gimbel DA, Nygaard HB, et al. Cellular prion protein mediates impairment of synaptic plasticity by amyloid-β oligomers. Nature. 2009;457:1128–1132.
- Kostylev MA, Kaufman AC, Nygaard HB, et al. Prion-protein-interacting amyloid-β oligomers of high molecular weight are tightly correlated with memory impairment in multiple Alzheimer mouse models. J. Biol. Chem. 2015;290:17415–17438.
- Larson M, Sherman MA, Amar F, et al. The complex PrP(c)-Fyn couples human oligomeric Aβ with pathological tau changes in Alzheimer’s disease. J Neurosci. 2012;32:16857–71a.
- Özcan GG, Lim S, Leighton PLA, et al. Sleep is bi-directionally modified by amyloid beta oligomers. eLife. 2020;9. bioRxiv 610014. DOI:https://doi.org/10.1101/610014.
- Leighton PLA, Ted Allison W, Zerr I. Protein misfolding in prion and prion-like diseases: reconsidering a required role for protein loss-of-function. J Alzheimers Dis. 2016;54:3–29.
- Postlethwait JH, Woods IG, Ngo-Hazelett P,et al. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 2000;10:1890–1902.
- Postlethwait JH, Yan Y-L, Gates MA, et al. Vertebrate genome evolution and the zebrafish gene map. Nat. Genet. 1998;18:345–349.
- Cotto E, André M, Forgue J, et al. Molecular characterization, phylogenetic relationships, and developmental expression patterns of prion genes in zebrafish (Danio rerio). FEBS J. 2005;272:500–513.
- Salta E, Panagiotidis C, Teliousis K, et al. Evaluation of the possible transmission of BSE and scrapie to gilthead sea bream (Sparus aurata). PLoS One. 2009;4:e6175.
- Salta E, Kanata E, Ouzounis C, et al. Assessing proteinase K resistance of fish prion proteins in a scrapie-infected mouse neuroblastoma cell line. Viruses. 2014;6:4398–4421.
- Castle AR, Gill AC. Physiological functions of the cellular prion protein. Front Mol Biosci. 2017;4:19. doi: https://doi.org/10.3389/fmolb.2017.00019.
- Wulf M-A, Senatore A, Aguzzi A. The biological function of the cellular prion protein: an update. BMC Biol. 2017;15:34.
- Steele AD, Lindquist S, Aguzzi A. The prion protein knockout mouse: a phenotype under challenge. Prion. 2007;1:83–93.
- Leighton PLA, Kanyo R, Neil GJ, et al. Prion gene paralogs are dispensable for early zebrafish development and have nonadditive roles in seizure susceptibility. J Biol Chem. 2018;293:12576–12592.
- Málaga-Trillo E, Solis GP, Schrock Y, et al. Regulation of embryonic cell adhesion by the prion protein. PLoS Biol. 2009;7:0576–0590.
- Schmitt-Ulms G, Ehsani S, Watts JC, et al. Evolutionary descent of prion genes from the ZIP family of metal Ion transporters. PLoS One. 2009;4:e7208.
- Huc-Brandt S, Hieu N, Imberdis T, et al. Zebrafish prion protein PrP2 controls collective migration process during lateral line sensory system development. PLoS One. 2014;9:1–22.
- Mehrabian M, Brethour D, Wang H, et al. The prion protein controls polysialylation of neural cell adhesion molecule 1 during cellular morphogenesis. PLoS One. 2015;10:1–23.
- Mehrabian M, Hildebrandt H, Schmitt-Ulms G. NCAM1 polysialylation: the prion protein’s elusive reason for being? ASN Neuro. 2016;8:175909141667907.
- Fleisch VC, Leighton PLA, Wang H, et al. Targeted mutation of the gene encoding prion protein in zebrafish reveals a conserved role in neuron excitability. Neurobiol. Dis. 2013;55:11–25.
- Liang J, Kong Q. α-Cleavage of cellular prion protein. Prion. 2012;6:453–460.
- Mcdonald AJ, Dibble JP, Evans EGB, et al. A new paradigm for enzymatic control of -cleavage and -cleavage of the prion protein*. J Biolog Chem. 2013;289(2):803–813. https://doi.org/https://doi.org/10.1074/jbc.M113.502351.
- Lewis V, Johanssen VA, Crouch PJ, et al. Prion protein ‘gamma-cleavage’: characterizing a novel endoproteolytic processing event. Cell Mol Life Sci. 2016;73:667–683.
- Mehrabian M, Ehsani S, Schmitt-Ulms G. An emerging role of the cellular prion protein as a modulator of a morphogenetic program underlying epithelial-to-mesenchymal transition. Front Cell Dev Biol. 2014;2:53.
- Rousset M, Leturque A, Thenet S. The nucleo-junctional interplay of the cellular prion protein: a new partner in cancer-related signaling pathways? Prion. 2016;10:143–152.
- Richardson DD, Tol S, Valle-Encinas E, et al. The prion protein inhibits monocytic cell migration by stimulating β1 integrin adhesion and uropod formation. J. Cell Sci. 2015;128:3018–3029.
- Prodromidou K, Papastefanaki F, Sklaviadis T, et al. Functional cross-talk between the cellular prion protein and the neural cell adhesion molecule NCAM is critical for neuronal differentiation of neural stem/precursor cells. Stem Cells. 2014;32:1674–1687.
- Steiger A, Guldner J, Hemmeter U, et al. Effects of growth hormone- releasing hormone and somatostatin on sleep eeg and nocturnal hormone secretion in male controls. Neuroendocrinology. 1992;56:566–573.
- Kim K, Choe HK. Role of hypothalamus in aging and its underlying cellular mechanisms. Mech Ageing Dev. 2019;177:74–79.
- Khalifé M, Young R, Passet B, et al. Transcriptomic analysis brings new insight into the biological role of the prion protein during mouse embryogenesis. PLoS One. 2011;6:e23253.
- Hayashi S, Takeichi M. Emerging roles of protocadherins: from self-avoidance to enhancement of motility. J Cell Sci. 2015;128:1455–1464.
- Kaiser DM, Acharya M, Leighton PLA, et al. Amyloid beta precursor protein and prion protein have a conserved interaction affecting cell adhesion and CNS development. PLoS One. 2012;7:e51305.
- Rieger S, Volkmann K, Köster RW. Polysialyltransferase expression is linked to neuronal migration in the developing and adult zebrafish. Dev Dyn. 2008;237:276–285.
- Rossi A, Kontarakis Z, Gerri C, et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–233.
- Place ES, Smith JC. Zebrafish atoh8 mutants do not recapitulate morpholino phenotypes. PLoS One. 2017;12:1–12.
- Camacho Leal MDP, Costamagna A, Tassone B, et al. Conditional ablation of p130Cas/BCAR1 adaptor protein impairs epidermal homeostasis by altering cell adhesion and differentiation 06 Biological Sciences 0601 Biochemistry and Cell Biology. Cell Commun Signal. 2018;16:1–13.
- Gupta V, Discenza M, Guyon JR, et al. α‐Actinin‐2 deficiency results in sarcomeric defects in zebrafish that cannot be rescued by α‐actinin‐3 revealing functional differences between sarcomeric isoforms. FASEB J. 2012;26:1892–1908.
- Thomas-Jinu S, Gordon PM, Fielding T, et al. Non-nuclear pool of splicing factor SFPQ regulates axonal transcripts required for normal motor development. Neuron. 2017;94:322–336.e5.
- Wu Q, Zhang J, Koh W, et al. Talin1 is required for cardiac Z-disk stabilization and endothelial integrity in zebrafish. FASEB J. 2015;29:4989–5005.
- Lepage SE, Bruce AEE. Characterization and comparative expression of zebrafish calpain system genes during early development. Dev Dyn. 2008;237:819–829.
- Young R, Passet B, Vilotte M, et al. The prion or the related Shadoo protein is required for early mouse embryogenesis. FEBS Lett. 2009;583:3296–3300.
- Daude N, Wohlgemuth S, Brown R, et al. Knockout of the prion protein (PrP)-like Sprn gene does not produce embryonic lethality in combination with PrPC-deficiency. Proc. Natl. Acad. Sci. 2012;109:9035–9040.
- Sempou E, Biasini E, Pinzón-Olejua A, et al. Activation of zebrafish Src family kinases by the prion protein is an amyloid-β-sensitive signal that prevents the endocytosis and degradation of E-cadherin/β-catenin complexes in vivo. Mol Neurodegener. 2016;11:18.
- Mehrabian M, Brethour D, MacIsaac S, et al. CRISPR-Cas9-based knockout of the prion protein and its effect on the proteome. PLoS One. 2014;9:e114594.
- Mehrabian M, Brethour D, Williams D, et al. Prion protein deficiency causes diverse proteome shifts in cell models that escape detection in brain tissue. PLoS One. 2016;11:e0156779.
- Nourizadeh-Lillabadi R, Seilø Torgersen J, Vestrheim O, et al. Early embryonic gene expression profiling of zebrafish prion protein (PrP2) morphants. PLoS One. 2010;5:e13573.
- Zhao X, Guan JL. Focal adhesion kinase and its signaling pathways in cell migration and angiogenesis. Adv Drug Deliv Rev. 2011;63:610–615.
- Cooper SR, Jontes JD, Sotomayor M. Structural determinants of adhesion by protocadherin-19 and implications for its role in epilepsy. Elife. 2016;5:1–22.
- Carulla P, Llorens F, Matamoros-Angles A, et al. Involvement of PrP C in kainate-induced excitotoxicity in several mouse strains. Sci. Rep. 2015;5. DOI:https://doi.org/10.1038/srep11971.
- Kanyo R, Leighton PLA, Neil GJ, et al. Amyloid-β precursor protein mutant zebrafish exhibit seizure susceptibility that depends on prion protein. Exp Neurol. 2020;328:113283.
- Abel AM, Yang C, Thakar MS, et al. Natural killer cells: development, maturation, and clinical utilization. Front Immunol. 2018;9:1–23.
- Bakkebø MK, Mouillet-Richard S, Espenes A, et al. The cellular prion protein: a player in immunological quiescence. Front Immunol. 2015;6. DOI:https://doi.org/10.3389/fimmu.2015.00450.
- Westerfield M. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio). Eugene: University of Oregon Press; 2000.
- Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578.
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111.
- R Core Team. R: a language and environment for statistical computing. 2020.
- Goff L, Trapnell C, Kelley D. cummeRbund: analysis, exploration, manipulation, and visualization of Cufflinks high-throughput sequencing data. R package version. 2014;2(0):1–45.
- Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag; 2016. doi:https://doi.org/10.1007/978-3-319-24277-4.
- Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622.
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13.
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
- Wickham H, Averick M, Bryan J, et al. Welcome to the {tidyverse}. J. Open Source Softw. 2019;4:1686.
