Abstract
The proper formation of dendritic arbors is a critical step in neural circuit formation, and as such defects in arborization are associated with a variety of neurodevelopmental disorders. Among the best gene candidates are those encoding cell adhesion molecules, including members of the diverse cadherin superfamily characterized by distinctive, repeated adhesive domains in their extracellular regions. Protocadherins (Pcdhs) make up the largest group within this superfamily, encompassing over 80 genes, including the ∼60 genes of the α-, β-, and γ-Pcdh gene clusters and the non-clustered δ-Pcdh genes. An additional group includes the atypical cadherin genes encoding the giant Fat and Dachsous proteins and the 7-transmembrane cadherins. In this review we highlight the many roles that Pcdhs and atypical cadherins have been demonstrated to play in dendritogenesis, dendrite arborization, and dendritic spine regulation. Together, the published studies we discuss implicate these members of the cadherin superfamily as key regulators of dendrite development and function, and as potential therapeutic targets for future interventions in neurodevelopmental disorders.
Abbreviations
| Pcdh | = | Protocadherin |
| Celsr | = | Cadherin EGF LAG 7-pass G-type receptor 1 |
| EC | = | extracellular cadherin |
| TM | = | transmembrane |
| CNR | = | Cadherin related neuronal receptor |
| PSD | = | Post-synaptic density |
| FAK | = | Focal adhesion kinase |
| PKC | = | Protein kinase C |
| MARCKS | = | Myristoylated alanine-rich C-kinase substrate |
| RNAi | = | RNA interference |
| PYK2 | = | Protein tyrosine kinase 2 |
| Rac1 | = | Ras-related C3 botulinum toxin substrate 1 |
| DSCAM | = | Down syndrome cell adhesion molecule |
| SAC | = | starburst amacrine cell |
| CTCF | = | CCCTC-binding factor |
| Dnmt3b | = | DNA (cytosine-5-)-methyltransferase 3 β |
| RGC | = | Retinal ganglion cell |
| TAF1 | = | Template-activating factor 1 |
| TAO2β | = | Thousand and one amino acid protein kinase 2 β |
| p38 MAPK | = | p38 mitogen-activated protein kinase |
| MEK3 | = | Mitogen-activated protein kinase kinase 3 |
| MEF2 | = | Myocyte enhancer factor 2 |
| FMRP | = | Fragile X mental retardation protein |
| Ds | = | Dachsous |
| PCP | = | planar cell polarity |
| EGF | = | Epidermal growth factor |
| Fjx1 | = | Four jointed box 1 |
| Fj | = | Four jointed |
| fmi | = | Flamingo |
| GPCR | = | G-protein-coupled receptor |
| S2 cells | = | Schneider 2 cells |
| md neuron | = | multiple dendrite neuron |
| da neuron | = | dendritic arborization neuron |
| Gogo | = | Golden Goal |
| LIM domain | = | Lin11, Isl-1 & Mec-3 domain |
| CaMKII | = | Ca2+/calmodulin-dependent protein kinase II |
Introduction
The development of functional neuronal circuitry is a key step in the formation of all nervous systems. Neurons, uniquely among metazoan cell types, interact directly in a point-to-point wiring fashion through synapses, which allows neural circuits to encode and constantly modify vast amounts of information. Each neuron is estimated to make thousands of synapses with partner neurons, and studies in a wide variety of model systems have shown that these connections are formed with a high degree of specificity. Because the synapse is, in many ways, a specialized type of adhesive junction, it is not surprising that many of the molecular cues thus far shown to influence the formation of neural circuits are cell adhesion molecules.
The establishment of a proper dendritic arbor is a critical component of neuronal circuit formation, and is linked to synaptogenesis as well, both morphologically and in terms of molecular mechanisms.Citation1-3 Dendrites represent the “input” side of the neuron (axons typically represent the “output” side), and as dendritic arbor size and pattern varies widely among distinct neuronal types,Citation4 aberrations from these normal parameters should have major effects on the development of proper circuitry. Consistent with this, disrupted dendrite development, including defects in the dendritic spines onto which most excitatory synapses are made, is associated with autism spectrum disorders and intellectual disabilities, both in available human patient samples and in the brains of transgenic mice engineered to model these disorders.Citation5-8
Over the past decade, many cell adhesion molecules, including members of the cadherin superfamily, have been implicated in developmental brain disorders (see recent reviews refs. 9–11). At the same time, recent publications have demonstrated increasing evidence of major roles for the diverse group of cell adhesion molecules known as the protocadherins (Pcdhs), which represents the largest group within the cadherin superfamily, in the regulation of dendrite arborization and function. For the purposes of this review, we confine our focus to recent work that has demonstrated functions for the clustered (α-, β-, and γ-) Pcdhs, the non-clustered δ-Pcdhs, and the atypical cadherins such as Fat and Flamingo/Celsr, in dendrite development. These molecules all have interesting expression patterns and functional roles in many other aspects of neural development, such as apoptosis, axon outgrowth and targeting, and migration; though we will not discuss these here, information can be found in several other recent reviews (see refs. 12–18).
Protocadherin Classification
The Pcdhs include more than 80 members of the cadherin superfamily: a diverse collection of cell-surface molecules defined by the presence of several ∼110 amino acid extracellular cadherin (EC) motifs.Citation19-21 The canonical members of the superfamily, the “classical” cadherins, are type I transmembrane (TM) proteins that contain 5 EC repeats and a conserved cytoplasmic domain that interacts with the armadillo repeat proteins, β-catenin and p120 ctn, and that mediate calcium-dependent, primarily homophilic, adhesion via the EC domains.Citation22-25 Shintaro Suzuki and colleagues used degenerate PCR to search for additional cadherin molecules, and discovered and named the first protocadherins in the early 1990's.Citation26 The initial molecules to be cloned were structurally quite similar to the classical cadherins, with a single TM domain and 6 or 7 EC repeats, though lacking catenin-binding sites in their cytoplasmic domains.Citation26-29 Such “typical” Pcdhs can be divided into 2 classes: 1) the clustered α-, β-, and γ-Pcdhs, which are encoded by, respectively, the Pcdha, Pcdhb, and Pcdhg gene clusters, together encompassing ∼60 genes in mammals with differing arrangements in other vertebrates; and 2) the non-clustered δ-Pcdhs ().Citation12,15,19,21,30-33 The molecules initially reported by Suzuki and colleagues included both clustered Pcdhs and δ-Pcdhs.
Figure 1. Clustered and Non-Clustered Protocadherins. Schematic domain structures of the clustered (α-, β-, and γ-) Pcdhs, and of the δ1- and δ2- non-clustered Pcdhs. Domains as indicated by legend; double horizontal line represents the plasma membrane. The domain structure of a classical cadherin is shown for comparison. A summary of known dendritic functions appears above each schematic.
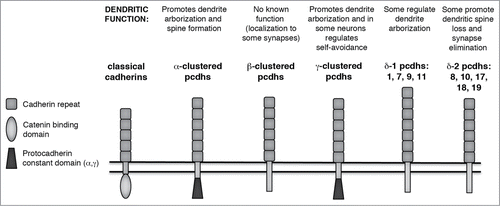
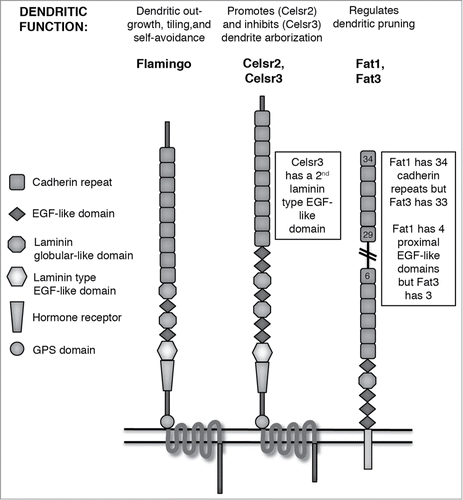
In addition to these Pcdhs, there are also several molecules with larger numbers of EC domains that are referred to in the literature as either Pcdhs or as “atypical cadherins;” because of their many roles in dendrite development, we will include these in our review, but refer to them as atypical cadherins. We discuss 2 groups of these, both of which are found in invertebrates as well as vertebrates: 1) the Fat and Dachsous cadherins, which have 27–34 EC domains and a single TM domain;Citation16 and 2) the 7 TM cadherins, which include Flamingo in Drosophila and its mammalian homologues Celsr1–3 ().Citation17,18
Figure 2. Atypical Cadherins. Schematic domain structures of Flamingo, Celsr, and Fat atypical cadherins. Domains as indicated by legend; double horizontal line represents the plasma membrane. A summary of known dendritic functions appears above each schematic. Because Fat proteins have too many cadherin repeats to easily depict in a reasonably-sized figure, the double-diagonal lines indicate continuation of cadherin repeats between the 6th and 29th.
Clustered Protocadherins
In mammals, the clustered Pcdh genes are situated in 3 sequential arrays (termed Pcdha, Pcdhb, and Pcdhg) encompassing about 1 MB on human chromosome 5q31 and mouse chromosome 18.Citation30,31,34 Within the Pcdha and Pcdhg clusters, large “variable” exons (14 for Pcdha and 22 for Pcdhg in the mouse) encoding 6 EC domains, a TM domain, and a ∼90 amino acid cytoplasmic domain are each expressed from their own promoters and spliced to 3 small “constant” exons that encode a shared 125 amino acid C-terminal domain (the mouse Pcdhb locus contains 22 variable exons but no constant exons; ). To generate transcripts encoding individual clustered Pcdh isoforms, a given variable exon promoter is “chosen,” through mechanisms that are only now being elucidated,Citation35-37 and transcription through the remaining portion of the cluster proceeds. For the Pcdha and Pcdhg clusters, intervening variable exons are removed when the 5′ variable exon is spliced to the 3 constant exons.Citation38,39 The Pcdhg cluster is further divided into subfamilies based on sequence similarity of the variable exons: Pcdhg A (12 exons), B (7 exons), and C (3 exons) subfamilies, with an additional 2 exons closely homologous to Pcdhg C found at the 3′ end of the Pcdha cluster (αC1 and αC2).Citation30
Based on extensive work from Takeshi Yagi's lab using RT-PCR on single cerebellar Purkinje cells from mice harboring maternal and paternal alleles from distinct mouse strains, most clustered Pcdh variable exons appear to be expressed monoallelically. While both alleles are transcriptionally active in any given neuron, each individual variable exon promoter appears to be active from only one of the 2 alleles. The exceptions to this rule are the ubiquitously-expressed Pcdha C1 and C2 exons, and the related Pcdhg C3, C4, and C5 exons, all of which are biallelically expressed. Each Purkinje cell expresses ∼4 Pcdha isoforms (2 of the monoallelically expressed exons plus the ubiquitous C1 and C2), ∼2 Pcdhb isoforms and ∼7 Pcdhg isoforms (∼4 of the monoallelically expressed exons plus the ubiquitous C3, C4, and C5).Citation13,40-43 Our own studies indicate that γ-Pcdh proteins form cis-multimers that interact in a strictly homophilic manner in trans.Citation44 Because cis-interaction occurs promiscuously, while trans interaction requires isoform matching, the Pcdhg cluster could encode between 104 and 105 distinct adhesive interfaces.Citation44,45 As γ-Pcdhs form complexes with α- and β-Pcdhs, which can also mediate homophilic interactions,Citation46-48 clustered Pcdhs could provide vast combinatorial molecular diversity that likely contributes to dendritic arbor development and function.
The Pcdha genes were the first to be analyzed in the nervous system, with the initial characterization of 8 genes isolated as “CNRs” (Cadherin Related Neuronal Receptors).Citation49 Kohmura et al.Citation49 demonstrated that these α-Pcdh proteins are localized to cortical dendrites and synapses, using fluorescent and immunogold antibody staining, and concentrated in the post-synaptic density (PSD) fraction using biochemistry, though they are clearly also present in some axons.Citation50-52 A similar localization pattern has been observed across several studies of the γ-Pcdhs in forebrain, cerebellar, retinal and spinal cord neurons: These proteins are concentrated in synaptosomes and PSD fractions,Citation53-56 localized at some, but far from all, synapses,Citation53,55-58 and, while also present in axons, observed prominently in dendrites.Citation55,56,58-61 Though the β-Pcdhs have been studied far less than their clustered brethren and no functional analysis has yet been published, it was shown that β16 and β22 demonstrate a similar preferential localization to dendritic and PSD compartments in retinal and cerebellar neurons.Citation62,63
Though the initial analyses of Pcdhg mutant mice by our lab and others focused on roles for these proteins in regulation of neuronal apoptosis and synaptogenesis,Citation53,57,58,64-68 a major role for the γ-Pcdhs in dendrite arborization has recently emerged (). Using cortically-restricted Pcdhg mutant mice, we found that the γ-Pcdhs are essential for the formation of properly complex dendritic arbors in cortical pyramidal neurons.Citation69 The Pcdhg mutant cortex appears to develop normally in terms of neurogenesis, neuronal migration, and establishment of major axonal tracts, but reduced arborization of pyramidal neurons leads to a thinner cortex, primarily due to loss of the cell-sparse, apical dendritic tuft-rich layer I. This is accompanied by aberrantly high activity in a signaling pathway leading from FAK, which is known to be inhibited through binding to the γ-Pcdh constant cytoplasmic domain,Citation70 to PKC and its phosphorylation target MARCKS.Citation69 In Pcdhg cortical mutants, FAK and MARCKS are hyper-phosphorylated and PKC enzymatic activity is increased, all of which is consistent with prior studies of the roles played by these proteins in dendrite arborization.Citation71-76 Confirming that this aberrant signaling pathway is the cause of reduced dendrite arborization, Pcdhg mutant neurons can be rescued by pharmacological inhibition of FAK or PKC, or by transfection with a non-phosphorylatable MARCKS mutant.Citation69
Table 1. Clustered Protocadherin Dendritic Roles
Complementary concurrent work from Qiang Wu's laboratory confirms the importance of γ-Pcdhs in dendrite arborization, and strongly implicates the α-Pcdhs as well.Citation77 Analyzing Pcdha mutant hippocampal neurons both in vivo and in vitro, Suo et al.Citation77 show that mutants exhibit simplified dendritic arbors as well as significant loss of dendritic spine density; the effects, interestingly, are somewhat less severe than those obtained by using RNAi to knockdown γ-Pcdh levels in cultured neurons. As observed previously for Pcdhg,Citation69,70 deletion of the Pcdha cluster leads to aberrantly active (i.e., tyrosine-phosphorylated) FAK and the homologous kinase PYK2/FAK2 in the brain. Dendritic arbor and spine defects similar to those observed in Pcdha mutant neurons are found in wild-type neurons overexpressing PYK2, and mutant neuron defects can be rescued by knocking down PYK2 using RNAi. Using pharmacological inhibitors and constitutively active protein mutants, Suo et al.Citation77 go on to show that the Rho GTPase Rac1 is a positive downstream regulator of dendrite development that is normally inhibited by PYK2. Additionally, lentiviral-mediated Cre-driven Pcdhg mutation in progenitor neurons of the subventricular zone, as reported in Ledderose et al.,Citation78 led to a reduction in dendritic arborization and in dendritic spine density in the olfactory bulb neurons derived from these progenitors. Together with the results of Garrett et al.,Citation69 these data provide strong support for the importance of clustered Pcdhs to the elaboration of complex dendrite arbors and the proper density of postsynaptic spines.Citation70,77,78 This work, building on the initial observation by Chen et al.Citation70 that the α- and γ-Pcdh constant domains bind to, and inhibit, FAK and PYK2, also confirms the importance of this kinase family in linking cell adhesion molecules at the membrane to regulation of multiple downstream signaling pathways that influence cytoskeletal rearrangements necessary for the formation of complex neuronal morphologies.
Though studies using heterologous cell in vitro assays indicate that the γ-Pcdhs can mediate homophilic cell adhesion,Citation44,48 it should be noted that many initially adhesive events at the cell surface of neurons can lead to subsequent repulsion in the proper signaling context. This occurs, for example, in the action of the immunoglobulin superfamily molecule DSCAM, which interacts homophilically to signal repulsion of contacting axon and dendrite branches, mediating a process called self-avoidance that is critical for the function of many neurons whose arbors must spread out to cover an appropriate receptive field.Citation79-82 Mutation of Dscam genes in both Drosophila and mice leads to aberrant self-crossing and fasciculation of dendrites and clumping of cell bodies.Citation83-87 Intriguingly, Lefebvre et al.Citation88 have demonstrated that in at least 2 neuronal subtypes, retinal starburst amacrine cells (SACs) and cerebellar Purkinje cells, loss of the γ-Pcdh proteins leads to an aberrant self-crossing phenotype similar to that observed in Dscam mutants.
Turning on a Cre-inducible transgene expressing a single γ-Pcdh isoform can cell-autonomously restore proper self-avoidance in a mutant SAC, but when 2 SACs expressing only this single isoform meet, they aberrantly avoid each other.Citation88 This suggests that, at least in neurons with planar dendritic arbors such as SACs and Purkinje cells, the γ-Pcdhs mediate repulsive interactions in a manner similar to Dscam1 in Drosophila. Recent work from the lab of Le Ma indicates that γ-Pcdh-mediated self-avoidance in mouse Purkinje cell dendrites is genetically independent of a similar self-avoidance role played by Slit/Robo signaling.Citation90 It should be noted that no obvious self-avoidance phenotypes are observed in the non-planar, bushy dendritic arbors of cortical neurons in the absence of the γ-Pcdhs,Citation69 nor are they immediately apparent in other, non-SAC retinal neurons.Citation58 However, the aberrantly clumped axon terminals we previously observed in Pcdhg mutant Ia afferent sensory neurons in the spinal cordCitation89 could be consistent with disrupted self-avoidance, a possibility that remains to be addressed. In any case, it appears that Pcdhg mutant dendrite phenotypes vary among distinct neuronal subtypes. A key question going forward will be to directly assess whether homophilic γ-Pcdh interactions can result in both adhesion or repulsion, as appears to be the case. If so, it is likely that distinct cis-membrane or cytoplasmic signaling partners can modulate the outcome of γ-Pcdh trans-interactions, and these remain to be identified.
Recent studies of mouse mutants in 2 gene regulatory proteins have also indirectly implicated the clustered Pcdhs in dendrite development. Binding sites for the zinc finger transcription factor/insulator protein CTCF are found at many sites within the 3 Pcdh gene clusters, and CTCF positively regulates the expression of these genes in a variety of cell lines.Citation36,91-93 Hirayama et al.Citation94 analyzed a forebrain-restricted conditional CTCF knockout mouse and found reduced cortical and hippocampal neuron dendrite arborization similar to that observed by Garrett et al.Citation69 and Suo et al.,Citation77 as well as disrupted barrel formation in somatosensory cortex. Microarray analysis showed significant downregulation of nearly all Pcdha, Pcdhb, and Pcdhg genes, which is consistent with the demonstrated roles for these proteins in dendrites; it should be noted, however, that 390 genes were significantly altered in the CTCF mutant forebrain, so it is possible that disruption of other genes also contributed to the observed phenotypes.Citation94 In a separate study, it was found that embryos lacking the DNA methyltransferase Dnmt3b exhibit hypomethylation of the clustered Pcdh genes.Citation95 Loss of Dnmt3b cell-autonomously leads to aberrantly constitutive expression of many normally stochastically-expressed Pcdh genes, and disrupted dendrite arborization, including reduced branch number and length as well as increased self-crossing, in Purkinje cells.Citation95 Though, again, it is likely that expression of many other genes are aberrant in the absence of Dnmt3b, these results are consistent with a major role for the clustered Pcdhs in the formation of proper dendritic arbors.
Non-clustered δ-Protocadherins
The largest group of non-clustered Pcdhs is made up of the δ1 (7 EC domains) and δ2 (6 EC domains) families, which are distantly related but exhibit short conserved motifs in their cytoplasmic domains ().Citation96,97 One of the first Pcdhs to be identified 20 y ago was a δ1, Pcdh1 (initially termed Pc42);Citation26 subsequently the δ1 family was expanded to include Pcdh7, Pcdh9, and Pcdh11, while the δ2 family encompasses Pcdh8, Pcdh10, Pcdh17, Pcdh18, and Pcdh19.Citation15 All of the δ-Pcdhs are expressed in the nervous system, with some (e.g., Pcdh10, also known as OL-Pcdh) marking particular subsets of regions and circuits (reviewed in ref. 12). Though several δ-Pcdhs have been reported to exhibit homophilic adhesion (e.g., Pcdh10,Citation98 Pcdh8,Citation99 Pcdh17,Citation100 and Pcdh19Citation101), like the clustered Pcdhs this is generally considered to be modest in comparison to the classical cadherins.Citation12 Several δ-Pcdhs partner in cis with classical cadherins to regulate adhesion: Pcdh8 (also known as paraxial protocadherin in Xenopus and Arcadlin in mammals) antagonizes adhesion by C-cadherinCitation102 and induces the internalization of N-cadherin at synapses in response to increased electrical activity,Citation103 while Pcdh19 can mediate homophilic adhesion only when in a cis-complex with N-cadherin, and this interaction actually precludes homophilic adhesion through N-cadherin itself.Citation104,105 While several of the δ-Pcdhs play interesting roles in axon guidance, and/or have been implicated in various cancers (reviewed in ref. 15), only a few studies have demonstrated roles in dendrite development and/or dendritic spine function thus far ().
Table 2. Nonclustered Protocadherin Dendritic Roles
Pcdh7 is a δ1-Pcdh also known as NF-protocadherin in Xenopus, where it is expressed in retinal ganglion cells (RGCs) and their targets in the optic tectum. Expression of a dominant negative Pcdh7 lacking the extracellular domain in the retina led to significantly reduced RGC dendrite arborization, though it should be noted that the disruption of axonogenesis was far more severe.Citation106 Expression of a dominant-negative template-activating factor (TAF1), which despite being predominantly known as a nuclear protein is also found at the cell membrane where it binds to the C-terminus of Pcdh7, produced an exact phenocopy, suggesting that Pcdh7 acts through TAF1 to promote axonogenesis and dendrite arborization in RGCs.Citation106 Intriguingly, a meta-analysis of genome-wide association studies for human epilepsies recently implicated Pcdh7 as a potential risk factor for these disorders.Citation107
The rat ortholog of the δ2-family member Pcdh8 was initially identified as Arcadlin in a screen for mRNAs rapidly induced in hippocampus following seizures.Citation99 The protein is found in cortical and hippocampal dendrites and co-localizes with the excitatory postsynaptic marker PSD-95 as well as with N-cadherin, but is also detected in axons and growth cones.Citation99,103 Pcdh8 can be co-immunoprecipitated with N-cadherin and blocks its homophilic adhesion in an in vitro assay. Homophilic interaction of Pcdh8 activates the kinase TAO2β, which in turn phosphorylates and activates p38 MAPK via another kinase, MEK3; this leads to the internalization of the Pcdh8/N-cadherin cis-complex, which limits dendritic spine density in hippocampal neurons. Consistent with this, Pcdh8 knockout neurons exhibit significantly increased spine density, but only in the presence of N-cadherin.Citation103 Though effects on dendrite arborization were not reported, these data demonstrate a role for Pcdh8 in the regulation of dendritic spine density and synaptogenesis, and demonstrate an important neuronal function for the observed cis-interactions between δ2-Pcdhs and classical cadherins.
Similarly, a prominent role at dendritic spines has been reported recently for the δ2 protein Pcdh10, also known as OL-Pcdh.Citation108 The activity-regulated transcription factor MEF2 (myocyte enhancer factor 2) had previously been shown to promote synapse elimination through a mechanism involving the translation-regulating RNA binding protein FMRP (fragile X mental retardation protein), encoded by a gene disrupted in the most common inherited cause of intellectual disability and autism, fragile X syndrome. Comparison of the known MEF2 target genes with the known FMRP-binding mRNAs identified Pcdh10, deletion of which has also been associated with autism.Citation109 Using cultured cortical neurons, Tsai et al.Citation108 found that shRNA knockdown of Pcdh10 prevented synapse elimination induced by introduction of constitutively-active MEF2. Active MEF2 leads to the ubiquitination (by the E3 ligase murine double minute 2) and proteosomal degradation of the spine protein PSD-95, and this mechanism depends on Pcdh10, which could be co-immunoprecipitated with both ubiquitinated PSD-95 and proteosomal components.Citation108 Together with the results of Yasuda et al.,Citation103 Tsai et al.Citation108 suggests that multiple δ2-Pcdhs can regulate synapse density and dendritic spine number, likely through promoting synapse elimination.
Fat and Dachsous Atypical Cadherins
The Fat and Dachsous (Ds) atypical cadherins were initially described in Drosophila, where they interact heterophilically to play important roles in tissue growth and patterning through the Hippo pathway, and in planar cell polarity (PCP), the process by which cells are directionally organized along the plane of a cell sheet, perpendicular to each cell's apical-basal axis; mammals have 4 homologous Fat genes (fat1–4; fat4 is considered to be the ortholog of Drosophila fat) and 2 Dachsous genes (Ds1 and 2).Citation16 These genes encode very large type I transmembrane proteins, with Fat1–4 having 34 cadherin repeats and Ds1–2 having 27; Fat proteins also exhibit varying numbers of EGF-like and laminin-like domains (). Relatively little is known about the roles of Fat and Ds proteins in nervous system development, outside of the fact that the genes are expressed there (). The intracellular domain of Fat1 interacts with Homer scaffolding proteins, suggesting a possible synaptic role.Citation110 This possibility has yet to be addressed due to the perinatal lethality of fat1 mutant mice, which present with kidney defects and partially-penetrant holoprosencephaly and anophthalmia;Citation111 fat1/fat4 double mutants also exhibit a high incidence of exencephaly.Citation112 Fat2 is expressed by cerebellar Purkinje cells,Citation113 while Fat3 protein localizes to both axons and dendrites throughout the nervous system.Citation114
Table 3. Atypical Cadherin Dendritic Roles
One study has provided evidence that Fat3 is critical for the formation of dendritic arbors by amacrine cells of the retina.Citation115 Mature amacrine cells, situated in the inner nuclear layer or ganglion cell layer of the retina, exhibit a unipolar morphology, with a dendritic arbor projecting to distinct sublaminae within the synaptic inner plexiform layer. During development, however, their arbors are initially multipolar, with neurites pruned as the amacrine cells migrate into the inner nuclear layer. Deans et al.Citation115 show that in fat3 mutant retina, this pruning process is defective, and some amacrine cells form 2 dendritic arbors: a proper one within the inner plexiform layer, and an aberrant one facing the opposite direction within the inner nuclear layer, forming a supernumerary plexiform layer. Though it is not known whether Fat3 interaction with Ds1 or 2 is important for this role, it was shown that Fjx1, the mammalian homolog of Drosophila 4-jointed, a Golgi-kinase that can modulate Fat-Ds binding,Citation16 interacts genetically with fat3. Fat3 heterozygotes that also carry a homozygous mutation in Fjx1 exhibit a mild phenotype, consistent with the fact that Drosophila Fj typically promotes Fat signaling.Citation115 Consistent with this is the prior demonstration that Fjx1 can negatively regulate dendrite extension and branching in hippocampal neurons.Citation116 This suggests a possible role for Ft/Ds signaling in the regulation of dendrite arborization in a variety of neuronal types, which remains to be shown directly.
7TM Atypical Cadherins
Along with the clustered Pcdhs, the 7TM atypical cadherins, which include Flamingo in Drosophila and its mammalian homologues Celsr1–3 (),Citation17,18 play perhaps the best-characterized roles in axon and dendrite development (). Most of these were first uncovered for Drosophila flamingo (fmi), which like ft and ds is also known for roles in PCP. Subsequent analyses of rodent Celsr genes have shown that many of these functions in neurite development are evolutionarily conserved. The structure of the 7TM cadherins sets them apart from other members of the family: They each contain 7 transmembrane domains similar to G-protein-coupled receptors (GPCRs) of the secretin receptor family, a hormone receptor domain, 2 laminin-G-like domains, varying numbers of EGF-like domains, and 9 cadherin repeats situated at the N-terminus ().Citation18 Cell aggregation assays using Drosophila S2 cells indicate that Fmi/Celsr proteins engage in homophilic adhesion that is dependent on the presence of the cadherin repeats.Citation117-119 While it is unclear whether Fmi or the Celsrs act as true GPCRs, some mutant phenotypes (see below) can be rescued by constructs lacking much of the extracellular region (including the cadherin repeats),Citation120 and Celsr cadherin repeat binding can trigger intracellular Ca2+ release,Citation121 suggesting that these proteins are capable of mediating signaling as well as adhesion. While we focus here only on dendritic functions, we note that both Fmi and Celsr1–3 have been shown to play critical roles in axon outgrowth and guidance in a number of studies of multiple neuronal types (e.g. in refs. 122–126; reviewed in ref. 18).
Fmi (a.k.a. starry night) was initially cloned by Chae et al.Citation118 and Usui et al.,Citation117 in studies that identified a role in PCP of the wing in collaboration with frizzled, the receptor for Wg/Wnts. Concurrently, it was isolated in a mutagenesis screen for genes affecting dendritic patterning in md (multiple dendrite) sensory neurons, in which fmi mutant neurons exhibited excessive growth of dorsal dendritic arbors.Citation127 A follow-up studyCitation128 showed that both loss and gain of function of fmi leads to overextension of dorsal md dendrites and an aberrant lack of repulsion between 2 md dendritic fields when they meet at the midline. In contrast to its role in PCP, fmi appears to act independently of frizzledCitation128 to regulate the proper timing and extent of dorsal dendrite arborization in md neurons,Citation129 though. Grueber et al.Citation130 showed that the da (dendritic arborization) subset of md neurons exhibits a phenomenon called “tiling," the complete coverage of a receptive area by dendritic branches without any overlap of branches from adjacent neurons of the same class. It was shown that fmi mutant da dendrites occasionally disrupted tiling by extending into neighboring territories. A screen for defects in mushroom body neurons also identified a dendritic over-reach phenotype in fmi mutants, extending the role of this gene to CNS neurons.Citation131
Through what mechanisms does fmi regulate dendrite extension and branching in Drosophila neurons? Kimura et al.Citation120 performed a structure-function analysis and found that the overgrowth of da neuron dendrites seen in fmi mutants could be partially rescued by an fmi transgene lacking the N-terminal cadherin repeats and EGF domains, but not by one lacking the C-terminal cytoplasmic tail. The aberrant dorsal overlap of adjacent dendrites, however, was not rescued by either construct, suggesting that the cadherin repeats, required for adhesion, are also required for subsequent heteroneuronal avoidance.Citation120 In some of its functions, fmi collaborates with Golden Goal (Gogo), a single-pass transmembrane protein that interacts with Flamingo in cis. In addition to conferring laminar synaptic specificity of R8 photoreceptor axons,Citation132 gogo regulates dendrite arborization in md-da neurons in collaboration with fmi. Hakeda and SuzukiCitation133 recently presented evidence that Gogo can negatively regulate dendrite outgrowth, possibly by responding to an inhibitory cue from the midline, and this can be enhanced by overexpression of fmi. Tiling involves repulsive self-avoidance of isoneuronal dendritic branches, which allows for the proper spread of a neuron's dendritic field. Matsubara et al.Citation134 show that class IV da neurons lacking fmi exhibit defective self-avoidance, manifested as increased self-crossing of dendritic branches. The LIM domain protein Espinas binds to an intracellular juxtamembrane domain of Flamingo, and this interaction is required for proper isoneuronal dendritic branch repulsion.Citation134
While Flamingo is generally a negative regulator of dendritic growth in Drosophila neurons, the different mammalian Celsr proteins have diverged in function. Shima et al.Citation119,121 have revealed opposing roles for Celsr2 and Celsr3 in the regulation of dendrite arborization by using RNAi knockdown in organotypic brain slice cultures. Knockdown of Celsr2 leads to greatly reduced dendrite arborization in both cortical pyramidal neurons and cerebellar Purkinje cells. This knockdown phenotype could be rescued by re-expression of a Celsr2 construct lacking the EGF and hormone receptor domains, but not by constructs lacking the cadherin repeats or the Celsr2B splice variant lacking a portion of the C-terminal cytoplasmic domain.Citation119 In contrast, knockdown of Celsr3 had the opposite effect: dendrite arborization was increased, with more basal dendrites and greater branching complexity.Citation121 These experiments, as well as those co-culturing neurons with non-neuronal cells expressing individual Celsr proteins, indicate that Celsr2 promotes, while Celsr3 inhibits, dendrite growth and arborization. Through elegant use of chimeric Celsr2/3 molecules, Shima et al.Citation121 surprisingly show that this functional reversal is controlled by a single amino acid in the cytoplasmic loop between TM1 and TM2. Flamingo, Celsr1, and Celsr3 share a conserved histidine in this domain, while Celsr2 uniquely has an arginine residue at this position. Neurons expressing Celsr2 exhibit longer neurites than controls when cultured with non-neuronal cells expressing Celsr2, but neurons expressing Celsr2 with the arginine mutated to a histidine exhibit shorter neurites than controls. Conversely, neurons expressing Celsr3 exhibit shorter neurites than controls when cultured with non-neuronal cells expressing Celsr3, but neurons expressing Celsr3 with the histidine mutated to an arginine exhibit longer neurites than controls.Citation121 In all cases these effects required homophilic matching between cadherin repeats present either in co-cultured cells or as soluble ectodomain proteins. Shima et al.Citation121 go on to present evidence that Celsr2 exerts its effect on dendrite growth through CaMKII while Celsr3 acts through calcineurin. Together these data support an evolutionarily conserved role for Flamingo and the Celsrs in the control of dendrite arborization, and uncover a fascinating divergence of function among the Celsrs due to a single amino acid difference that affects downstream signaling.
Concluding Remarks
Though Pcdhs were first identified in 1993, their first in vivo functions were not demonstrated until nearly a decade later, and only in the past few years have the molecular mechanisms underlying their cis and trans interactions and their intracellular signaling begun to be elucidated. As this review of the literature demonstrates, we are in a period of rapid growth in the understanding of this intriguing family of molecules, and, in particular, in the discovery of their important roles in the formation of neuronal circuitry during development, particularly in regulating the arborization of dendrites. Though the sheer diversity of the molecules discussed under the “Pcdh” (and atypical cadherin) banner ensures that many mechanisms will be unique to each family, subfamily, or even individual protein, we note here a few unifying points for future research.
First is the importance of Pcdh/atypical cadherin intracellular signaling, outside of any adhesive role they may have. For instance, the 7TM cadherins can clearly act as signaling receptors, even if not as bona fide G-protein coupled receptors (which remains unclear); the clustered Pcdhs can regulate adhesion and cytoskeleton-associated kinase pathways involving FAK, PYK2, and Rac1; some δ-Pcdhs such as Pcdh8 can activate kinases (such as TAO2β) to initiate cascades of intracellular signaling; and other δ-Pcdhs such as Pcdhs 10, 17, and 18b interact with Nap1 and the WAVE complex to regulate actin dynamics.Citation142-144 It is not clear in many cases whether such signaling is regulated constitutively by Pcdhs, or whether it is initiated upon homophilic trans-interactions, a key question that should be addressed in the future. Similarly, though progress has been made as described above, we still know relatively little about the intracellular binding partners and signaling pathways affected by many of the Pcdhs. Screens for Pcdh protein interactors should open up new vistas into the roles of these CAMs in neural development.
Second is the apparent dual adhesive function of the Pcdhs, in which depending on the cell type and situation, they can mediate attractive adhesion between cells, repulsive signaling among processes of the same neuron, or primarily affect the adhesion of other CAMs. This is most striking for the γ-Pcdhs, which can mediate adhesion as well as self-avoidance, but is also true for some of the δ-Pcdhs, which predominantly modulate adhesion of classical cadherins, as well as for Flamingo. It is possible that a switch from a primarily attractive adhesive event, to a repulsive event following initial adhesion, could be mediated by cis interactions with different partners at the membrane. In this way, certain Pcdhs could have distinct cellular effects in different neuronal subtypes depending on which interaction partners are present. Again, further identification of Pcdh-interacting proteins, particularly by focusing on distinct brain regions or neuronal cell types, will be helpful in this regard.
Third is the intriguing fact that all of the Pcdh families discussed here have some reported link to signaling by the Wnt family of growth factors. Though Flamingo appears to influence dendrite branching and outgrowth without requiring Wnt pathway interaction in flies, it does collaborate with this pathway in PCP and a recent report in C. elegans suggest it may do so in neurite outgrowth as well.Citation135 Two recent reports indicate that several γ-Pcdhs can inhibit the Wnt pathway in multiple cancer cell types,Citation136,137 though the mechanisms involved are unknown, and any relevance to neuronal Wnt signaling is not established. Several δ-Pcdhs were isolated in a large screen for proteins interacting with the Wnt receptor RYK, though again, it remains to be confirmed whether any such interactions are meaningful in a neuronal context.Citation138 Nevertheless, as it is known that Wnt signaling can affect dendrite arborizationCitation139,140 and synaptogenesis,Citation141 these hints of Pcdh-Wnt pathway interactions are worthy of further analysis in neuronal contexts.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Vaughn JE, Barber RP, Sims TJ. Dendritic development and preferential growth into synaptogenic fields: A quantitative study of Golgi-impregnated spinal motor neurons. Synapse 1988; 2:69-78; PMID:2458630; http://dx.doi.org/10.1002/syn.890020110
- Vaughn J, Anonymous. Fine structure of synaptogenesis in the vertebrate central nervous system. Synapse 1989; 3:255; PMID:2655146; http://dx.doi.org/10.1002/syn.890030312
- Cline H, Haas K. The regulation of dendritic arbor development and plasticity by glutamatergic synaptic input: a review of the synaptotrophic hypothesis. J Physiol 2008; 586:1509; PMID:18202093; http://dx.doi.org/10.1113/jphysiol.2007.150029
- Jan YN, Jan LY. Branching out: mechanisms of dendritic arborization. Nat Rev Neurosci 2010; 11:316; PMID:20404840; http://dx.doi.org/10.1038/nrn2836
- Dierssen M, Ramakers GJ. Dendritic pathology in mental retardation: from molecular genetics to neurobiology. Genes, Brain, and Behav 2006; 5 Suppl 2):48; PMID:16681800; http://dx.doi.org/10.1111/j.1601-183X.2006.00224.x
- Kishi N, Macklis JD. MeCP2 functions largely cell-autonomously, but also non-cell-autonomously, in neuronal maturation and dendritic arborization of cortical pyramidal neurons. Exp Neurol 2010; 222:51; PMID:20025874; http://dx.doi.org/10.1016/j.expneurol.2009.12.007
- Kwon CH, Luikart BW, Powell CM, Zhou J, Matheny SA, Zhang W, Li Y, Baker SJ, Parada LF. Pten regulates neuronal arborization and social interaction in mice. Neuron 2006; 50:377; PMID:16675393; http://dx.doi.org/10.1016/j.neuron.2006.03.023
- Hutsler JJ, Zhang H. Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain Res 2010; 1309:83; PMID:19896929; http://dx.doi.org/10.1016/j.brainres.2009.09.120
- Redies C, Hertel N, Hubner CA. Cadherins and neuropsychiatric disorders. Brain Res 2012; 1470:130-44; PMID:22765916; http://dx.doi.org/10.1016/j.brainres.2012.06.020
- Singh SK, Eroglu C. Neuroligins provide molecular links between syndromic and nonsyndromic autism. Sci Signal 2013; 6:re4.
- Penagarikano O, Geschwind DH. What does CNTNAP2 reveal about autism spectrum disorder? Trends Mol Med 2012; 18:156-63; PMID:22365836; http://dx.doi.org/10.1016/j.molmed.2012.01.003
- Kim S-Y, Yasuda S, Tanaka H, Yamagata K, Kim H. Non-clustered protocadherin. Cell Adhesion & Migration 2011; 5:97-105; PMID:21173574; http://dx.doi.org/10.4161/cam.5.2.14374
- Yagi T. Molecular codes for neuronal individuality and cell assembly in the brain. Front Mole Neurosci 2012; 5:45; PMID:22518100; http://dx.doi.org/10.3389/fnmol.2012.00045
- Weiner JA, Jontes JD. Protocadherins, not prototypical: a complex tale of their interactions, expression, and functions. Front Mole Neurosci 2013; 6:4; PMID:23515683
- Kahr I, Vandepoele K, van Roy F. Delta-protocadherins in health and disease. Elsevier, 2013:169-92.
- Sharma P, McNeill H. Fat and dachsous cadherins. Prog Mol Biol Transl Sci 2013; 116:215-35; PMID:23481197; http://dx.doi.org/10.1016/B978-0-12-394311-8.00010-8
- Feng J, Han Q, Zhou L. Planar cell polarity genes, Celsr1-3, in neural development. Neurosci Bull 2012; 28:309-15; PMID:22622831; http://dx.doi.org/10.1007/s12264-012-1232-8
- Berger-Muller S, Suzuki T. Seven-pass transmembrane cadherins: roles and emerging mechanisms in axonal and dendritic patterning. Mol Neurobiol 2011; 44:313-20; PMID:21909747; http://dx.doi.org/10.1007/s12035-011-8201-5
- Nollet F, Kools P, van Roy F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J Mol Biol 2000; 299:551-72; PMID:10835267; http://dx.doi.org/10.1006/jmbi.2000.3777
- Morishita H, Yagi T. Protocadherin family: diversity, structure, and function. Curr Opin Cell Biol 2007; 19:584; PMID:17936607; http://dx.doi.org/10.1016/j.ceb.2007.09.006
- Hulpiau P, van Roy F. Molecular evolution of the cadherin superfamily. Int J Biochem Cell Biol 2009; 41:349-69; PMID:18848899; http://dx.doi.org/10.1016/j.biocel.2008.09.027
- Takeichi M. The cadherins: cell-cell adhesion molecules controlling animal morphogenesis. Development (Cambridge, England) 1988; 102:639; PMID:3048970
- Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mole Cell Biol 2005; 6:622; PMID:16025097; http://dx.doi.org/10.1038/nrm1699
- Halbleib JM, Nelson WJ. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev 2006; 20:3199; PMID:17158740; http://dx.doi.org/10.1101/gad.1486806
- Leckband D, Prakasam A. Mechanism and dynamics of cadherin adhesion. Ann Rev Biomed Eng 2006; 8:259; PMID:16834557; http://dx.doi.org/10.1146/annurev.bioeng.8.061505.095753
- Sano K, Tanihara H, Heimark RL, Obata S, Davidson M, St John T, Taketani S, Suzuki S. Protocadherins: a large family of cadherin-related molecules in central nervous system. Embo J 1993; 12:2249-56; PMID:8508762
- Obata S, Sago H, Mori N, Rochelle JM, Seldin MF, Davidson M, St John T, Taketani S, Suzuki ST. Protocadherin Pcdh2 shows properties similar to, but distinct from, those of classical cadherins. J Cell Sci 1995; 108 (Pt 12):3765-73; PMID:8719883
- Sago H, Kitagawa M, Obata S, Mori N, Taketani S, Rochelle JM, Seldin MF, Davidson M, St John T, Suzuki ST. Cloning, expression, and chromosomal localization of a novel cadherin-related protein, protocadherin-3. Genomics 1995; 29:631-40; PMID:8575755; http://dx.doi.org/10.1006/geno.1995.9956
- Suzuki ST. Protocadherins and diversity of the cadherin superfamily. J Cell Sci 1996; 109 (Pt 11):2609-11; PMID:8937978
- Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell 1999; 97:779-90; PMID:10380929; http://dx.doi.org/10.1016/S0092-8674(00)80789-8
- Wu Q, Zhang T, Cheng JF, Kim Y, Grimwood J, Schmutz J, Dickson M, Noonan JP, Zhang MQ, Myers RM, et al. Comparative DNA sequence analysis of mouse and human protocadherin gene clusters. Genome Res 2001; 11:389-404; PMID:11230163; http://dx.doi.org/10.1101/gr.167301
- Wu Q. Comparative Genomics and Diversifying Selection of the Clustered Vertebrate Protocadherin Genes. Genetics 2005; 169(4):2179-88; PMID: 15744052
- Tada MN, Senzaki K, Tai Y, Morishita H, Tanaka YZ, Murata Y, Ishii Y, Asakawa S, Shimizu N, Sugino H, et al. Genomic organization and transcripts of the zebrafish Protocadherin genes. Gene 2004; 340:197-211; PMID:15475161; http://dx.doi.org/10.1016/j.gene.2004.07.014
- Sugino H, Hamada S, Yasuda R, Tuji A, Matsuda Y, Fujita M, Yagi T. Genomic organization of the family of CNR cadherin genes in mice and humans. Genomics 2000; 63:75-87; PMID:10662547; http://dx.doi.org/10.1006/geno.1999.6066
- Ribich S, Tasic B, Maniatis T. Identification of long-range regulatory elements in the protocadherin-alpha gene cluster. Proc Natl Acad Sci U S A 2006; 103:19719; PMID:17172445; http://dx.doi.org/10.1073/pnas.0609445104
- Kehayova P, Monahan K, Chen W, Maniatis T. Regulatory elements required for the activation and repression of the protocadherin-alpha gene cluster. Proc Natl Acad Sci 2011; 108(41):17195-200; PMID: 21949399
- Yokota S, Hirayama T, Hirano K, Kaneko R, Toyoda S, Kawamura Y, Hirabayashi M, Hirabayashi T, Yagi T. Identification of the Cluster Control Region for the Protocadherin-b Genes Located beyond the Protocadherin-g Cluster. J Biol Chem 2011; 286:31885-95; PMID:21771796; http://dx.doi.org/10.1074/jbc.M111.245605
- Tasic B, Nabholz CE, Baldwin KK, Kim Y, Rueckert EH, Ribich SA, Cramer P, Wu Q, Axel R, Maniatis T. Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol Cell 2002; 10:21-33; PMID:12150904; http://dx.doi.org/10.1016/S1097-2765(02)00578-6
- Wang X, Su H, Bradley A. Molecular mechanisms governing Pcdh-gamma gene expression: evidence for a multiple promoter and cis-alternative splicing model. Genes Dev 2002; 16:1890-905; PMID:12154121; http://dx.doi.org/10.1101/gad.1004802
- Esumi S, Kakazu N, Taguchi Y, Hirayama T, Sasaki A, Hirabayashi T, Koide T, Kitsukawa T, Hamada S, Yagi T. Monoallelic yet combinatorial expression of variable exons of the protocadherin-alpha gene cluster in single neurons. Nat Genet 2005; 37:171-6; PMID:15640798; http://dx.doi.org/10.1038/ng1500
- Kaneko R, Kato H, Kawamura Y, Esumi S, Hirayama T, Hirabayashi T, Yagi T. Allelic gene regulation of Pcdh-alpha and Pcdh-gamma clusters involving both monoallelic and biallelic expression in single Purkinje cells. J Biol Chem 2006; 281:30551-60; PMID:16893882; http://dx.doi.org/10.1074/jbc.M605677200
- Noguchi Y, Hirabayashi T, Katori S, Kawamura Y, Sanbo M, Hirabayashi M, Kiyonari H, Nakao K, Uchimura A, Yagi T. Total expression and dual gene-regulatory mechanisms maintained in deletions and duplications of the Pcdha cluster. J Biol Chem 2009; 284:32002-14; PMID:19797050; http://dx.doi.org/10.1074/jbc.M109.046938
- Hirano K, Kaneko R, Izawa T, Kawaguchi M, Kitsukawa T, Yagi T. Single-neuron diversity generated by Protocadherin-b cluster in mouse central and peripheral nervous systems. Front Mole Neurosci 2012; 5:90; PMID:22969705
- Schreiner D, Weiner JA. Combinatorial homophilic interaction between gamma-protocadherin multimers greatly expands the molecular diversity of cell adhesion. Proc Natl Acad Sci U S A 2010; 107:14893; PMID:20679223; http://dx.doi.org/10.1073/pnas.1004526107
- Zipursky SL, Sanes JR. Chemoaffinity revisited: dscams, protocadherins, and neural circuit assembly. Cell 2010; 143:343; PMID:21029858; http://dx.doi.org/10.1016/j.cell.2010.10.009
- Murata Y, Hamada S, Morishita H, Mutoh T, Yagi T. Interaction with protocadherin-gamma regulates the cell surface expression of protocadherin-alpha. J Biol Chem 2004; 279:49508-16; PMID:15347688; http://dx.doi.org/10.1074/jbc.M408771200
- Han MH, Lin C, Meng S, Wang X. Proteomics analysis reveals overlapping functions of clustered protocadherins. Mole Cell Proteomics 2010; 9:71; http://dx.doi.org/10.1074/mcp.M900343-MCP200
- Thu CA, Chen WV, Rubinstein R, Chevee M, Wolcott HN, Felsovalyi KO, Tapia JC, Shapiro L, Honig B, Maniatis T. Single-cell identity generated by combinatorial homophilic interactions between alpha, beta, and gamma protocadherins. Cell 2014; 158:1045-59; PMID:25171406; http://dx.doi.org/10.1016/j.cell.2014.07.012
- Kohmura N, Senzaki K, Hamada S, Kai N, Yasuda R, Watanabe M, Ishii H, Yasuda M, Mishina M, Yagi T. Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron 1998; 20:1137-51; PMID:9655502; http://dx.doi.org/10.1016/S0896-6273(00)80495-X
- Blank M, Triana-Baltzer GB, Richards CS, Berg DK. Alpha-protocadherins are presynaptic and axonal in nicotinic pathways. Mol Cell Neurosci 2004; 26:530-43; PMID:15276155; http://dx.doi.org/10.1016/j.mcn.2004.04.008
- Morishita H, Murata Y, Esumi S, Hamada S, Yagi T. CNR/Pcdhalpha family in subplate neurons, and developing cortical connectivity. Neuroreport 2004; 15:2595-9; PMID:15570159; http://dx.doi.org/10.1097/00001756-200412030-00007
- Morishita H, Kawaguchi M, Murata Y, Seiwa C, Hamada S, Asou H, Yagi T. Myelination triggers local loss of axonal CNR/protocadherin alpha family protein expression. Eur J Neurosci 2004; 20:2843-7; PMID:15579137; http://dx.doi.org/10.1111/j.1460-9568.2004.03803.x
- Wang X, Weiner JA, Levi S, Craig AM, Bradley A, Sanes JR. Gamma protocadherins are required for survival of spinal interneurons. Neuron 2002; 36:843-54; PMID:12467588; http://dx.doi.org/10.1016/S0896-6273(02)01090-5
- Phillips GR, Huang JK, Wang Y, Tanaka H, Shapiro L, Zhang W, Shan WS, Arndt K, Frank M, Gordon RE, et al. The presynaptic particle web: ultrastructure, composition, dissolution, and reconstitution. Neuron 2001; 32:63-77; PMID:11604139; http://dx.doi.org/10.1016/S0896-6273(01)00450-0
- Phillips GR, Tanaka H, Frank M, Elste A, Fidler L, Benson DL, Colman DR. Gamma-protocadherins are targeted to subsets of synapses and intracellular organelles in neurons. J Neurosci 2003; 23:5096-104; PMID:12832533
- Li Y, Serwanski DR, Miralles CP, Fiondella CG, Loturco JJ, Rubio ME, De Blas AL. Synaptic and nonsynaptic localization of protocadherin-gammaC5 in the rat brain. J Comp Neurol 2010; 518:3439-63; PMID:20589908; http://dx.doi.org/10.1002/cne.22390
- Garrett AM, Weiner JA. Control of CNS synapse development by gamma-protocadherin-mediated astrocyte-neuron contact. J Neurosci 2009; 29:11723; PMID:19776259; http://dx.doi.org/10.1523/JNEUROSCI.2818-09.2009
- Lefebvre JL, Zhang Y, Meister M, Wang X, Sanes JR. gamma-Protocadherins regulate neuronal survival but are dispensable for circuit formation in retina. Development 2008; 135:4141-51; PMID:19029044; http://dx.doi.org/10.1242/dev.027912
- Kallenbach S, Khantane S, Carroll P, Gayet O, Alonso S, Henderson CE, Dudley K. Changes in subcellular distribution of protocadherin gamma proteins accompany maturation of spinal neurons. J Neurosci Res 2003; 72:549-56; PMID:12749019; http://dx.doi.org/10.1002/jnr.10618
- Fernandez-Monreal M, Kang S, Phillips GR. Gamma-protocadherin homophilic interaction and intracellular trafficking is controlled by the cytoplasmic domain in neurons. Mole Cell Neurosci 2009; 40:344; PMID:19136062; http://dx.doi.org/10.1016/j.mcn.2008.12.002
- Fernández-Monreal M, Oung T, Hanson HH, O'Leary R, Janssen WG, Dolios G, Wang R, Phillips GR. g-protocadherins are enriched and transported in specialized vesicles associated with the secretory pathway in neurons. Eur J Neurosci 2010; 32:921-31; http://dx.doi.org/10.1111/j.1460-9568.2010.07386.x
- Junghans D, Heidenreich M, Hack I, Taylor V, Frotscher M, Kemler R. Postsynaptic and differential localization to neuronal subtypes of protocadherin b16 in the mammalian central nervous system. Euro J Neurosci 2008; 27:559; PMID:18279309; http://dx.doi.org/10.1111/j.1460-9568.2008.06052.x
- Puller C, Haverkamp S. Cell-type-specific localization of protocadherin b16 at AMPA and AMPA/kainate receptor-containing synapses in the primate retina. J Comp Neurol 2010; 519:467-79; http://dx.doi.org/10.1002/cne.22528
- Prasad T, Wang X, Gray PA, Weiner JA. A differential developmental pattern of spinal interneuron apoptosis during synaptogenesis: insights from genetic analyses of the protocadherin-gamma gene cluster. Development (Cambridge, England) 2008; 135:4153; PMID:19029045; http://dx.doi.org/10.1242/dev.026807
- Weiner JA, Wang X, Tapia JC, Sanes JR. Gamma protocadherins are required for synaptic development in the spinal cord. Proc Natl Acad Sci U S A 2005; 102:8-14; PMID:15574493; http://dx.doi.org/10.1073/pnas.0407931101
- Su H, Marcheva B, Meng S, Liang FA, Kohsaka A, Kobayashi Y, Xu AW, Bass J, Wang X. Gamma-protocadherins regulate the functional integrity of hypothalamic feeding circuitry in mice. Dev Biol 2010; 339:38; PMID:20025866; http://dx.doi.org/10.1016/j.ydbio.2009.12.010
- Lin YC, Koleske AJ. Mechanisms of synapse and dendrite maintenance and their disruption in psychiatric and neurodegenerative disorders. Annu Rev Neurosci 2010; 33:349-78; PMID:20367247; http://dx.doi.org/10.1146/annurev-neuro-060909-153204
- Li Y, Xiao H, Chiou TT, Jin H, Bonhomme B, Miralles CP, Pinal N, Ali R, Chen WV, Maniatis T, et al. Molecular and functional interaction between protocadherin-gammaC5 and GABAA receptors. J Neurosci 2012; 32:11780-97; PMID:22915120; http://dx.doi.org/10.1523/JNEUROSCI.0969-12.2012
- Garrett AM, Schreiner D, Lobas MA, Weiner JA. gamma-protocadherins control cortical dendrite arborization by regulating the activity of a FAK/PKC/MARCKS signaling pathway. Neuron 2012; 74:269; PMID:22542181; http://dx.doi.org/10.1016/j.neuron.2012.01.028
- Chen J, Lu Y, Meng S, Han MH, Lin C, Wang X. alpha- and gamma-Protocadherins negatively regulate PYK2. J Biol Chem 2009; 284:2880; PMID:19047047; http://dx.doi.org/10.1074/jbc.M807417200
- Beggs HE, Schahin-Reed D, Zang K, Goebbels S, Nave KA, Gorski J, Jones KR, Sretavan D, Reichardt LF. FAK deficiency in cells contributing to the basal lamina results in cortical abnormalities resembling congenital muscular dystrophies. Neuron 2003; 40:501; PMID:14642275; http://dx.doi.org/10.1016/S0896-6273(03)00666-4
- Rico B, Beggs HE, Schahin-Reed D, Kimes N, Schmidt A, Reichardt LF. Control of axonal branching and synapse formation by focal adhesion kinase. Nat Neurosci 2004; 7:1059; PMID:15378065; http://dx.doi.org/10.1038/nn1317
- Gundlfinger A, Kapfhammer JP, Kruse F, Leitges M, Metzger F. Different regulation of Purkinje cell dendritic development in cerebellar slice cultures by protein kinase Calpha and -beta. JNeurobiol 2003; 57:95; http://dx.doi.org/10.1002/neu.10259
- Metzger F, Kapfhammer JP. Protein kinase C activity modulates dendritic differentiation of rat Purkinje cells in cerebellar slice cultures. Euro J Neurosci 2000; 12:1993; PMID:10886339; http://dx.doi.org/10.1046/j.1460-9568.2000.00086.x
- Schrenk K, Kapfhammer JP, Metzger F. Altered dendritic development of cerebellar Purkinje cells in slice cultures from protein kinase Cgamma-deficient mice. Neuroscience 2002; 110:675; PMID:11934475; http://dx.doi.org/10.1016/S0306-4522(01)00559-0
- Li H, Chen G, Zhou B, Duan S. Actin Filament Assembly by Myristoylated, Alanine-rich C Kinase Substrate-Phosphatidylinositol-4,5-diphosphate Signaling Is Critical for Dendrite Branching. Mole Biol Cell 2008; 19:4804; PMID:18799624; http://dx.doi.org/10.1091/mbc.E08-03-0294
- Suo L, Lu H, Ying G, Capecchi MR, Wu Q. Protocadherin clusters and cell adhesion kinase regulate dendrite complexity through Rho GTPase. J Mol Cell Biol 2012; 4:362; PMID:22730554; http://dx.doi.org/10.1093/jmcb/mjs034
- Ledderose J, Dieter S, Schwarz MK. Maturation of postnatally generated olfactory bulb granule cells depends on functional gamma-protocadherin expression. SciRep 2013; 3:1514.
- Millard SS, Zipursky SL. Dscam-mediated repulsion controls tiling and self-avoidance. Curr Opin Neurobiol 2008; 18:84; PMID:18538559; http://dx.doi.org/10.1016/j.conb.2008.05.005
- Hattori D, Millard SS, Wojtowicz WM, Zipursky SL. Dscam-mediated cell recognition regulates neural circuit formation. Ann Rev Cell Dev Biol 2008; 24:597; PMID:18837673; http://dx.doi.org/10.1146/annurev.cellbio.24.110707.175250
- Grueber WB, Sagasti A. Self-avoidance and Tiling: Mechanisms of Dendrite and Axon Spacing. Cold Spring Harbor Perspect Biol 2010; 2:a001750-a; PMID:20573716; http://dx.doi.org/10.1101/cshperspect.a001750
- Garrett AM, Tadenev AL, Burgess RW. DSCAMs: restoring balance to developmental forces. Front Mol Neurosci 2012; 5:86; PMID:22912601; http://dx.doi.org/10.3389/fnmol.2012.00086
- Fuerst PG, Bruce F, Tian M, Wei W, Elstrott J, Feller MB, Erskine L, Singer JH, Burgess RW. DSCAM and DSCAML1 function in self-avoidance in multiple cell types in the developing mouse retina. Neuron 2009; 64:484; PMID:19945391; http://dx.doi.org/10.1016/j.neuron.2009.09.027
- Fuerst PG, Koizumi A, Masland RH, Burgess RW. Neurite arborization and mosaic spacing in the mouse retina require DSCAM. Nature 2008; 451:470-4; PMID:18216855; http://dx.doi.org/10.1038/nature06514
- Hughes ME, Bortnick R, Tsubouchi A, Baumer P, Kondo M, Uemura T, Schmucker D. Homophilic Dscam interactions control complex dendrite morphogenesis. Neuron 2007; 54:417; PMID:17481395; http://dx.doi.org/10.1016/j.neuron.2007.04.013
- Matthews BJ, Kim ME, Flanagan JJ, Hattori D, Clemens JC, Zipursky SL, Grueber WB. Dendrite self-avoidance is controlled by Dscam. Cell 2007; 129:593; PMID:17482551; http://dx.doi.org/10.1016/j.cell.2007.04.013
- Soba P, Zhu S, Emoto K, Younger S, Yang SJ, Yu HH, Lee T, Jan LY, Jan YN. Drosophila sensory neurons require Dscam for dendritic self-avoidance and proper dendritic field organization. Neuron 2007; 54:403; PMID:17481394; http://dx.doi.org/10.1016/j.neuron.2007.03.029
- Lefebvre JL, Kostadinov D, Chen WV, Maniatis T, Sanes JR. Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature 2012; 488:517; PMID:22842903; http://dx.doi.org/10.1038/nature11305
- Prasad T, Weiner JA. Direct and indirect regulation of spinal cord Ia afferent terminal formation by the gamma-protocadherins. Front Mol Neurosci 2011; 4:54; PMID:22275881; http://dx.doi.org/10.3389/fnmol.2011.00054
- Gibson DA, Tymanskyj S, Yuan RC, Leung HC, Lefebvre JL, Sanes JR, Chedotal A, Ma L. Dendrite self-avoidance requires cell-autonomous slit/robo signaling in cerebellar purkinje cells. Neuron 2014; 81:1040-56; PMID:24607227; http://dx.doi.org/10.1016/j.neuron.2014.01.009
- Golan-Mashiach M, Grunspan M, Emmanuel R, Gibbs-Bar L, Dikstein R, Shapiro E. Identification of CTCF as a master regulator of the clustered protocadherin genes. Nucleic Acids Res 2012; 40(8):3378-91; PMID:22210889
- Guo Y, Monahan K, Wu H, Gertz J, Varley KE, Li W, Myers RM, Maniatis T, Wu Q. CTCF/cohesin-mediated DNA looping is required for protocadherin alpha promoter choice. Proc Natl Acad Sci U S A 2012; 109:21081-6; PMID:23204437; http://dx.doi.org/10.1073/pnas.1219280110
- Monahan K, Rudnick ND, Kehayova PD, Pauli F, Newberry KM, Myers RM, Maniatis T. Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of protocadherin-alpha gene expression. Proc Natl Acad Sci U S A 2012; 109:9125-30; PMID:22550178; http://dx.doi.org/10.1073/pnas.1205074109
- Hirayama T, Tarusawa E, Yoshimura Y, Galjart N, Yagi T. CTCF is required for neural development and stochastic expression of clustered Pcdh genes in neurons. Cell Reports 2012; 2:345; PMID:22854024; http://dx.doi.org/10.1016/j.celrep.2012.06.014
- Toyoda S, Kawaguchi M, Kobayashi T, Tarusawa E, Toyama T, Okano M, Oda M, Nakauchi H, Yoshimura Y, Sanbo M, et al. Developmental epigenetic modification regulates stochastic expression of clustered protocadherin genes, generating single neuron diversity. Neuron 2014; 82:94-108; PMID:24698270; http://dx.doi.org/10.1016/j.neuron.2014.02.005
- Wolverton T, Lalande M. Identification and characterization of three members of a novel subclass of protocadherins. Genomics 2001; 76:66-72; PMID:11549318; http://dx.doi.org/10.1006/geno.2001.6592
- Redies C, Vanhalst K, Roy F. delta-Protocadherins: unique structures and functions. Cell Mol Life Sci 2005; 62:2840; PMID:16261259; http://dx.doi.org/10.1007/s00018-005-5320-z
- Hirano S, Yan Q, Suzuki ST. Expression of a novel protocadherin, OL-protocadherin, in a subset of functional systems of the developing mouse brain. J Neurosci 1999; 19:995-1005; PMID:9920663
- Yamagata K, Andreasson KI, Sugiura H, Maru E, Dominique M, Irie Y, Miki N, Hayashi Y, Yoshioka M, Kaneko K, et al. Arcadlin is a neural activity-regulated cadherin involved in long term potentiation. J Biol Chem 1999; 274:19473-1979; PMID:10383464; http://dx.doi.org/10.1074/jbc.274.27.19473
- Hoshina N, Tanimura A, Yamasaki M, Inoue T, Fukabori R, Kuroda T, Yokoyama K, Tezuka T, Sagara H, Hirano S, et al. Protocadherin 17 regulates presynaptic assembly in topographic corticobasal Ganglia circuits. Neuron 2013; 78:839-54; PMID:23684785; http://dx.doi.org/10.1016/j.neuron.2013.03.031
- Tai K, Kubota M, Shiono K, Tokutsu H, Suzuki ST. Adhesion Properties and Retinofugal Expression of Chicken Protocadherin-19. Brain Res 2010; 1344:13-24; PMID: 20438721
- Chen X, Gumbiner BM. Paraxial protocadherin mediates cell sorting and tissue morphogenesis by regulating C-cadherin adhesion activity. J Cell Biol 2006; 174:301; PMID:16847104; http://dx.doi.org/10.1083/jcb.200602062
- Yasuda S, Tanaka H, Sugiura H, Okamura K, Sakaguchi T, Tran U, Takemiya T, Mizoguchi A, Yagita Y, Sakurai T, et al. Activity-induced protocadherin arcadlin regulates dendritic spine number by triggering N-cadherin endocytosis via TAO2beta and p38 MAP kinases. Neuron 2007; 56:456; PMID:17988630; http://dx.doi.org/10.1016/j.neuron.2007.08.020
- Biswas S, Emond MR, Jontes JD. Protocadherin-19 and N-cadherin interact to control cell movements during anterior neurulation. J Cell Biol 2010; 191:1029-41; PMID:21115806; http://dx.doi.org/10.1083/jcb.201007008
- Emond MR, Biswas S, Blevins CJ, Jontes JD. A complex of Protocadherin-19 and N-cadherin mediates a novel mechanism of cell adhesion. J Cell Biol 2011; 195:1115-21; PMID:22184198; http://dx.doi.org/10.1083/jcb.201108115
- Piper M, Dwivedy A, Leung L, Bradley RS, Holt CE. NF-protocadherin and TAF1 regulate retinal axon initiation and elongation in vivo. J Neurosci 2008; 28:100-5; PMID:18171927; http://dx.doi.org/10.1523/JNEUROSCI.4490-07.2008
- International League Against Epilepsy Consortium on Complex Epilepsies. Electronic address e-auea. Genetic determinants of common epilepsies: a meta-analysis of genome-wide association studies. Lancet Neurol 2014; 13:893-903; PMID:25087078; http://dx.doi.org/10.1016/S1474-4422(14)70171-1
- Tsai N-P, Wilkerson JR, Guo W, Maksimova MA, DeMartino GN, Cowan CW, Huber KM. Multiple Autism-Linked Genes Mediate Synapse Elimination via Proteasomal Degradation of a Synaptic Scaffold PSD-95. Cell 2012; 151:1581-94; PMID:23260144; http://dx.doi.org/10.1016/j.cell.2012.11.040
- Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, Hill RS, Mukaddes NM, Balkhy S, Gascon G, Hashmi A, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science (New York, NY) 2008; 321:218; http://dx.doi.org/10.1126/science.1157657
- Schreiner D, Muller K, Hofer HW. The intracellular domain of the human protocadherin hFat1 interacts with Homer signalling scaffolding proteins. FEBS letters 2006; 580:5295; PMID:16979624; http://dx.doi.org/10.1016/j.febslet.2006.08.079
- Ciani L, Patel A, Allen ND, ffrench-Constant C. Mice lacking the giant protocadherin mFAT1 exhibit renal slit junction abnormalities and a partially penetrant cyclopia and anophthalmia phenotype. Mol Cell Biol 2003; 23:3575-82; PMID:12724416; http://dx.doi.org/10.1128/MCB.23.10.3575-3582.2003
- Saburi S, Hester I, Goodrich L, McNeill H. Functional interactions between Fat family cadherins in tissue morphogenesis and planar polarity. Development 2012; 139:1806-20; PMID:22510986; http://dx.doi.org/10.1242/dev.077461
- Nakayama M, Nakajima D, Yoshimura R, Endo Y, Ohara O. MEGF1/fat2 proteins containing extraordinarily large extracellular domains are localized to thin parallel fibers of cerebellar granule cells. Mol Cell Neurosci 2002; 20:563; PMID:12213440; http://dx.doi.org/10.1006/mcne.2002.1146
- Nagae S, Tanoue T, Takeichi M. Temporal and spatial expression profiles of the Fat3 protein, a giant cadherin molecule, during mouse development. Dev Dynamics 2007; 236:534; PMID:17131403; http://dx.doi.org/10.1002/dvdy.21030
- Deans MR, Krol A, Abraira VE, Copley CO, Tucker AF, Goodrich LV. Control of neuronal morphology by the atypical cadherin fat3. Neuron 2011; 71:820; PMID:21903076; http://dx.doi.org/10.1016/j.neuron.2011.06.026
- Probst B, Rock R, Gessler M, Vortkamp A, Puschel AW. The rodent Four-jointed ortholog Fjx1 regulates dendrite extension. Dev Biol 2007; 312:461-70; PMID:18028897; http://dx.doi.org/10.1016/j.ydbio.2007.09.054
- Usui T, Shima Y, Shimada Y, Hirano S, Burgess RW, Schwarz TL, Takeichi M, Uemura T. Flamingo, a seven-pass transmembrane cadherin, regulates planar cell polarity under the control of Frizzled. Cell 1999; 98:585; PMID:10490098; http://dx.doi.org/10.1016/S0092-8674(00)80046-X
- Chae J, Kim MJ, Goo JH, Collier S, Gubb D, Charlton J, Adler PN, Park WJ. The Drosophila tissue polarity gene starry night encodes a member of the protocadherin family. Development 1999; 126:5421-9; PMID:10556066
- Shima Y, Kengaku M, Hirano T, Takeichi M, Uemura T. Regulation of dendritic maintenance and growth by a mammalian 7-pass transmembrane cadherin. Dev Cell 2004; 7:205-16; PMID:15296717; http://dx.doi.org/10.1016/j.devcel.2004.07.007
- Kimura H, Usui T, Tsubouchi A, Uemura T. Potential dual molecular interaction of the Drosophila 7-pass transmembrane cadherin Flamingo in dendritic morphogenesis. J Cell Sci 2006; 119:1118; PMID:16507587; http://dx.doi.org/10.1242/jcs.02832
- Shima Y, Kawaguchi SY, Kosaka K, Nakayama M, Hoshino M, Nabeshima Y, Hirano T, Uemura T. Opposing roles in neurite growth control by two seven-pass transmembrane cadherins. Nat Neurosci 2007; 10:963; PMID:17618280; http://dx.doi.org/10.1038/nn1933
- Chen PL, Clandinin TR. The cadherin Flamingo mediates level-dependent interactions that guide photoreceptor target choice in Drosophila. Neuron 2008; 58:26; PMID:18400160; http://dx.doi.org/10.1016/j.neuron.2008.01.007
- Bao H, Berlanga ML, Xue M, Hapip SM, Daniels RW, Mendenhall JM, Alcantara AA, Zhang B. The atypical cadherin flamingo regulates synaptogenesis and helps prevent axonal and synaptic degeneration in Drosophila. Mol Cell Neurosci 2007; 34:662; PMID:17321750; http://dx.doi.org/10.1016/j.mcn.2007.01.007
- Tissir F, Bar I, Jossin Y, Goffinet AM. Protocadherin Celsr3 is crucial in axonal tract development. Nat Neurosci 2005; 8:451-7; PMID:15778712
- Lee RC, Clandinin TR, Lee CH, Chen PL, Meinertzhagen IA, Zipursky SL. The protocadherin Flamingo is required for axon target selection in the Drosophila visual system. Nat Neurosci 2003; 6:557-63; PMID:12754514; http://dx.doi.org/10.1038/nn1063
- Senti KA, Usui T, Boucke K, Greber U, Uemura T, Dickson BJ. Flamingo regulates R8 axon-axon and axon-target interactions in the Drosophila visual system. Curr Biol 2003; 13:828-32; PMID:12747830; http://dx.doi.org/10.1016/S0960-9822(03)00291-4
- Gao FB, Brenman JE, Jan LY, Jan YN. Genes regulating dendritic outgrowth, branching, and routing in Drosophila. Genes Deve 1999; 13:2549; PMID:10521399; http://dx.doi.org/10.1101/gad.13.19.2549
- Gao FB, Kohwi M, Brenman JE, Jan LY, Jan YN. Control of dendritic field formation in Drosophila: the roles of flamingo and competition between homologous neurons. Neuron 2000; 28:91; PMID:11086986; http://dx.doi.org/10.1016/S0896-6273(00)00088-X
- Sweeney NT, Li W, Gao FB. Genetic manipulation of single neurons in vivo reveals specific roles of flamingo in neuronal morphogenesis. Dev Biol 2002; 247:76-88; PMID:12074553; http://dx.doi.org/10.1006/dbio.2002.0702
- Grueber WB, Jan LY, Jan YN. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development (Cambridge, England) 2002; 129:2867; PMID:12050135
- Reuter JE, Nardine TM, Penton A, Billuart P, Scott EK, Usui T, Uemura T, Luo L. A mosaic genetic screen for genes necessary for Drosophila mushroom body neuronal morphogenesis. Development 2003; 130:1203-13; PMID:12571111; http://dx.doi.org/10.1242/dev.00319
- Hakeda-Suzuki S, Berger-Muller S, Tomasi T, Usui T, Horiuchi SY, Uemura T, Suzuki T. Golden Goal collaborates with Flamingo in conferring synaptic-layer specificity in the visual system. Nat Neurosci 2011; 14:314-23; PMID:21317905; http://dx.doi.org/10.1038/nn.2756
- Hakeda S, Suzuki T. Golden goal controls dendrite elongation and branching of multidendritic arborization neurons in Drosophila. Genes Cells 2013; 18:960-73; PMID:23919529; http://dx.doi.org/10.1111/gtc.12089
- Matsubara D, Horiuchi S-Y, Shimono K, Usui T, Uemura T. The seven-pass transmembrane cadherin Flamingo controls dendritic self-avoidance via its binding to a LIM domain protein, Espinas, in Drosophila sensory neurons. Genes Dev 2011; 25:1982-96; PMID:21937715; http://dx.doi.org/10.1101/gad.16531611
- Huarcaya Najarro E, Ackley BD. C. elegans fmi-1/flamingo and Wnt pathway components interact genetically to control the anteroposterior neurite growth of the VD GABAergic neurons. Dev Biol 2013; 377:224-235; PMID:23376536; http://dx.doi.org/10.1016/j.ydbio.2013.01.014
- Dallosso, AR, Hancock, AL, Szemes, M, Moorwood, K, Chilukamarri, L, Tsai, HH, Sarkar, A, Barasch, J, Vuononvirta, R, Jones, C, et al. Frequent long-range epigenetic silencing of protocadherin gene clusters on chromosome 5q31 in Wilms' tumor. PLoS Genetics 2009; 5:e1000745; PMID:19956686; http://dx.doi.org/10.1371/journal.pgen.1000745
- Dallosso AR, Oster B, Greenhough A, Thorsen K, Curry TJ, Owen C, Hancock AL, Szemes M, Paraskeva C, Frank M, et al. Long-range epigenetic silencing of chromosome 5q31 protocadherins is involved in early and late stages of colorectal tumorigenesis through modulation of oncogenic pathways. Oncogene 2012; 31:4409-4419; PMID:22249255; http://dx.doi.org/10.1038/onc.2011.609
- Berndt JD, Aoyagi A, Yang P, Anastas JN, Tang L, Moon RT. Mindbomb 1, an E3 ubiquitin ligase, forms a complex with RYK to activate Wnt/b-catenin signaling. J Cell Biol 2011; 194:737-750; PMID:21875946; http://dx.doi.org/10.1083/jcb.201107021
- Rosso SB, Sussman D, Wynshaw-Boris A, Salinas PC. Wnt signaling through Dishevelled, Rac and JNK regulates dendritic development. Nat Neurosci 2005; 8: 34-42; PMID:15608632; http://dx.doi.org/10.1038/nn1374
- Hiester BG, Galati DF, Salinas PC, Jones KR. Neurotrophin and Wnt signaling cooperatively regulate dendritic spine formation. Mol Cell Neurosci 2013; 56: 115-127; PMID:23639831; http://dx.doi.org/10.1016/j.mcn.2013.04.006
- Ciani L, Salinas PC. WNTs in the vertebrate nervous system: from patterning to neuronal connectivity. Nat Rev Neurosci 2005; 6:351-362; PMID:15832199; http://dx.doi.org/10.1038/nrn1665
- Nakao S, Platek A, Hirano S, Takeichi M. Contact-dependent promotion of cell migration by the OL-protocadherin-Nap1 interaction. J Cell Biol 2008; 182:395-410; PMID:18644894; http://dx.doi.org/10.1083/jcb.200802069
- Biswas S, Emond MR, Duy PQ, Hao le T, Beattie CE, Jontes JD. Protocadherin-18b interacts with Nap1 to control motor axon growth and arborization in zebrafish. Mol Biol Cell 2014; 25: 633-642; PMID:24371087; http://dx.doi.org/10.1091/mbc.E13-08-0475
- Hayashi S, Inoue Y, Kiyonari H, Abe T, Misaki K, Moriguchi H, Tanaka Y, Takeichi M. Protocadherin-17 mediates collective axon extension by recruiting actin regulator complexes to interaxonal contacts. Dev Cell 2014; 30: 673-687; PMID:25199687; http://dx.doi.org/10.1016/j.devcel.2014.07.015
