 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
The adhesion of cell membranes is mediated by the binding of membrane-anchored receptor and ligand proteins. In this article, we review recent results from simulations and theory that lead to novel insights on how the binding equilibrium and kinetics of these proteins is affected by the membranes and by the membrane anchoring and molecular properties of the proteins. Simulations and theory both indicate that the binding equilibrium constant and the on- and off-rate constants of anchored receptors and ligands in their 2-dimensional (2D) membrane environment strongly depend on the membrane roughness from thermally excited shape fluctuations on nanoscales. Recent theory corroborated by simulations provides a general relation between
and the binding constant
of soluble variants of the receptors and ligands that lack the membrane anchors and are free to diffuse in 3 dimensions (3D).
Introduction
Cell adhesion processes and the adhesion of vesicles to the membranes of cells or organelles depend sensitively on the binding constant and binding kinetics of the membrane-anchored receptor and ligand molecules that mediate adhesion. Since the binding equilibrium constant and the on- and off-rate constants of these receptor and ligand molecules are difficult to measure in their natural 2-dimensional (2D) membrane environment, a central question is how they are related to the binding equilibrium constant
and the on- and off-rate constants of soluble variants of the receptors and ligands that lack the membrane anchors and are free to diffuse in 3 dimensions (3D).Citation1-10 The binding constant
and on- and off-rate constants of these soluble receptors and ligands can be quantified with standard experimental methods.Citation11-13
The binding equilibrium constant of membrane-anchored receptor and ligand molecules has units of area, while the binding constant
of soluble variants of these molecules has units of volume. Bell and co-workersCitation14 therefore suggested the relation
between the binding constants with a characteristic confinement length
that balances the different units of these constants. However, experimental data for
and
of several receptor and ligand pairs lead to values of the confinement length
that can differ by orders of magnitude, depending on whether
is determined with fluorescence methods or with mechanical methods.Citation1 Fluorescence methodsCitation15-21 probe the binding equilibrium of receptors and ligands in equilibrated adhesion zones of cells and lead to values of
of the order of nanometers. In contrast, mechanical methodsCitation22-31 probe the binding kinetics of anchored receptors and ligands during initial contacts and typically lead to values of
between tens of micrometers and millimeters in cell adhesion experiments.Citation1
In this article, we review recent results from computational model systems and theory that provide general and novel insights into the relation between the binding equilibrium and kinetics of membrane-anchored receptor and ligand molecules in 2D and the binding of soluble variants of these molecules in 3D. A central aspect of these computational and theoretical results is that the relation between the binding equilibrium constants and
involves 4 characteristic lengths, rather than a single confinement length.Citation9 Two of these 4 lengths are characteristic lengths of the receptor-ligand complex that reflect variations in the binding site, and how strongly the local membrane separation at the location of the complex is constrained by the complex. The remaining 2 lengths are the average separation and relative roughness of the apposing membranes and, thus, characteristic lengths of the membranes. The relative membrane roughness is the local standard deviation of the membranes from their average separation due to thermally excited shape fluctuations on nanoscales.
The binding equilibrium constant strongly depends both on the average membrane separation and the relative membrane roughness, which helps to understand why mechanical methods that probe the binding kinetics of membrane-anchored proteins during initial membrane contacts can lead to values for
that are orders of magnitude smaller than the values obtained from fluorescence measurements in equilibrated adhesion zones.Citation9 In equilibrated adhesion zones that are dominated by a single species of receptors and ligands, the average membrane separation is close to the preferred average separation for receptor-ligand binding at which
is maximal, and the relative membrane roughness is reduced by receptor-ligand bonds.Citation3,7 During initial membrane contacts, in contrast, both the average membrane separation and relative membrane roughness are larger, which can lead to significantly smaller values of
.
Characteristic lengths of membranes and membrane-anchored receptors and ligands
A membrane-anchored receptor can only bind to an apposing membrane-anchored ligand if the local membrane separation at the site of the receptor and ligand is within an appropriate range. This local separation
of the membranes varies – along the membranes, and in time – because of thermally excited membrane shape fluctuations. Experiments that probe the binding equilibrium constant
or the on- and off-rate constants
and
imply averages in space and time over membrane adhesion regions and measurement durations. Our recent simulations and theories indicate that these averages can be expressed asCitation9,10
(1)
(1)
(2)
(2) where
and
are the binding equilibrium constant and on-rate constant as functions of the local membrane separation
, and
is the distribution of local membrane separations that reflects the spatial and temporal variations of
. The single-peaked functions
and
are maximal at the preferred local separation of the receptors and ligands for binding, and have characteristic widths that depend on the anchoring, length, and flexibility of the receptors and ligands.Citation9,10 The off-rate constant follows from Eqs. (Equation1
(1)
(1) ) and (Equation2
(2)
(2) ) as
. Our simulations also show that the distribution
of the local separation is well approximated by the Gaussian distribution
(3)
(3) in situations in which the adhesion of 2 apposing membranes, or membrane segments, is mediated by a single type of receptors and ligands.Citation9,10 Here,
is the average separation of the membranes or membrane segments, and
is the relative roughness of the membranes. The relative roughness is the standard deviation of the local membrane separation
, i.e. the width of the distribution
. The distribution
describes both the spatial and temporal variations of the local membrane separation
of 2 apposing membranes, or membrane segments. Related temporal averages for the on-rate constant
and off-rate constant
at fixed membrane locations have been employed by Bihr et al.Citation32
The Eqs. (Equation1(1)
(1) ) and (Equation3
(3)
(3) ) illustrate 3 characteristic lengths of the binding constant
. These lengths are the width
of the single-peaked function
, which reflects how strongly the local separation
is constrained by a receptor-ligand (RL) complex, and the average separation
and relative roughness
of the membranes. A fourth characteristic length that affects the relation of the binding constants
and
in our theory is the ratio
of the translational space phase volume
of a bound soluble receptor in 3D and the translational phase space area
of a bound membrane-anchored receptor in 2D, relative to their ligands (see below). Similarly, 3 characteristic lengths of the on-rate constant
are the width
of the single-peaked function
, which reflects variations of the local separation
in the transition-state (TS) complex for binding, the average membrane separation
, and the relative membrane roughness
, according to Eqs. (Equation2
(2)
(2) ) and (Equation3
(3)
(3) ).
In equilibrated membrane adhesion zones that are dominated by a single type of receptors and ligands, the average membrane separation is close to the preferred average separation of these receptors and ligands for binding. Our simulations indicate that the relative membrane roughness then is determined by the concentration [RL] of the receptor-ligand bonds, which constrain the membrane shape fluctuationsCitation9,33:
(4)
(4) Here,
is the effective bending rigidity of the 2 apposing membranes with bending rigidities
and
, and
is the thermal energy, the driving force of membrane shape fluctuations. For a concentration
of receptor-ligand bonds and for typical values of the bending rigidities
and
of lipid membranesCitation34,35 and cell membranesCitation36,37 between 20
and 80
, we obtain estimates for the relative membrane roughness
between 3 nm and 6 nm from Eq. (Equation4
(4)
(4) ). For a 4 times larger bond concentration
, these roughness estimates are decreased by a factor of 2, according to Eq. (Equation4
(4)
(4) ). For a 4 times smaller bond concentration
, the roughness estimates are increased by a factor of 2, compared to the bond concentration
. The scaling relation (4) results from the fact that the membrane shape fluctuations on the relevant lateral length scales up to
, i.e., on length scales of the order of 10 or 100 nanometers, are dominated by the bending energy of the membranes. In contrast, the overall shape of cells on length scales of micrometers is dominated by the membrane tension and the cell cytoskeleton. The bending energy dominates over the membrane tension
on length scales smaller than the crossover length
, which adopts values of 100 or a few 100 nanometers for typical values of the bending rigidity
and tension
of cell membranes.Citation36
If the relative membrane roughness is much smaller than the widths
and
of the functions
and
, the binding of membrane-anchored receptors and ligands is only weakly affected by
. Such situations may occur in focal contacts or adherens junctions, which consist of clusters of integrin and cadherin complexes, respectively.Citation4,Citation38-41 In cell adhesion zones of immune cells and in the equilibrated adhesion zones probed with fluorescence methods,Citation15-21 in contrast, the relative membrane roughness is likely of the same order or larger than
and
. The computational model systems and theory described in the next sections indicate that the binding equilibrium and kinetics of the membrane-anchored receptors and ligands is then strongly affected both by the relative membrane roughness
and the average membrane separation
. If the relative membrane roughness
is significantly larger than
and
, the binding equilibrium constant
and on-rate constant
are both inversely proportional to
at the preferred average separation for binding.Citation9,10 Together with Eq. (Equation4
(4)
(4) ), these inverse proportionalities lead to a quadratic dependence of the bond concentration [RL] and the overall reaction rate on the concentrations [R] and [L] of unbound membrane-anchored receptors R and ligands L, which reflects the binding cooperativity caused by the membrane roughness on nanoscales.Citation3,7,10
Results from computational model systems of biomembrane adhesion
We have recently developed 2 computational model systems to investigate the binding of anchored receptors and ligands in their 2D membrane environment and the binding of soluble variants of the receptors and ligands that are fully mobile in 3D.Citation7,9,Citation10 First, we have developed a coarse-grained molecular model of biomembrane adhesionCitation7,10 (see ). In this model, lipid molecules consist of 3 hydrophobic head beads and 2 hydrophobic tails of 4 beads each, and the receptors and ligands are represented as cylindrical rods of beads, which are either anchored rather rigidly to a cylindrical transmembrane domain, or more flexibly to lipid molecules. We have investigated the binding equilibrium and kinetics of both these transmembrane and lipid-anchored receptors and ligands with molecular dynamics (MD) simulations, as well as the binding equilibrium and kinetics of soluble variants of the receptors and ligands that lack the membrane anchors. Related coarse-grained molecular models of biomembranes have been previously used to investigate the self-assembly,Citation42-45 fusion,Citation46-51 and lipid domainsCitation52-56 of membranes as well as the diffusion,Citation57,58 aggregation,Citation59 and curvature generationCitation60,61 of membrane proteins with MD simulations.
Figure 1. (A) Snapshot from a molecular dynamics (MD) simulation of our coarse-grained molecular model of biomembrane adhesion. In this snapshot, the 2 apposing membranes both have an area of 120 × 120 nm2 and contain 25 transmembrane receptors and ligands. (B) Snapshot from a Monte Carlo (MC) simulation of our elastic-membrane model of biomembrane adhesion. The snapshot shows membrane segments of area 200 × 200 nm2 from simulations with overall membrane area 800 × 800 nm2 and 200 receptors and ligands of anchoring strength and length 20 nm.
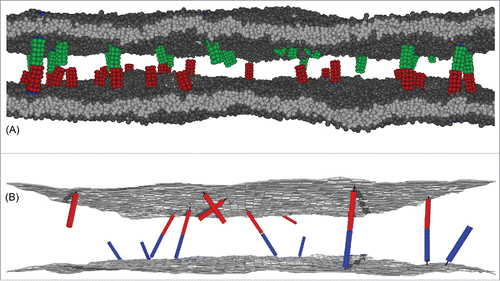
Second, we have developed an elastic-membrane model of biomembrane adhesion in which the membranes are represented as discretized elastic surfaces, and the receptors and ligands as anchored rigid or semi-flexible rods that diffuse continuously along the membranes and rotate around their anchoring points.Citation9 Using Monte Carlo (MC) simulations, we have determined both the binding constant of these anchored receptors and ligands as well as the binding constant
of soluble variants of the receptors and ligands. In previous elastic-membrane models of biomembrane adhesion, determining both
and
and the molecular characteristics affecting these binding constants has not been possible because the receptors and ligands are not explicitly represented as anchored molecules. Instead, the binding of receptors and ligands has been described implicitly by interactions that depend on the membrane separation.Citation32,62-69 In other previous elastic-membrane models, receptors and ligands are described by concentration fields rather than individual molecules,Citation70-79 or receptor-ligand bonds are treated as constraints on the local membrane separation.Citation33,Citation80-83
An important aspect for the binding of membrane-anchored receptors and ligands is the flexibility of the membrane anchoring. In our computational model systems, the anchoring flexibility of unbound membrane-anchored receptors and ligands can be described by the harmonic anchoring energy(5)
(5) with anchoring strength
and anchoring angle
, which is the angle between the direction of the receptors and ligands and the local membrane normal. An anchoring angle of zero thus corresponds to a perpendicular orientation of the receptors and ligands relative to the membrane. For our coarse-grained molecular model of biomembrane adhesion, the effective anchoring strength
can be determined by fitting the anchoring-angle distributions of unbound receptors and ligands observed in the MD simulations, which leads to the values
for our lipid-anchored receptors and ligands and
for our transmembrane receptors and ligands.Citation10 In our elastic-membrane model of biomembrane adhesion, the anchoring energy (5) of receptors and ligands is part of the overall configurational energy of the model, and the anchoring strength
thus can be ‘set’ as a parameter. We have performed MC simulations with the 3 values
,
and
.
The illustrate MC results for the binding constant of membrane-anchored receptors and ligands from 2 different simulation scenarios.Citation9 In the first scenario, the 2 apposing membranes are parallel and planar (see ). The local separation of the membranes is then identical at all membrane sites and, thus, identical to the average separation
of the membranes. By varying the membrane separation
in this scenario, we obtain the binding constant
as a function of the local membrane separation
from MC simulations in which the receptors and ligands diffuse along the planar membranes and rotate at their anchor points. In the second scenario, the 2 apposing membranes are flexible, and the local membrane separation
varies because of thermally excited shape fluctuations of the membranes (see ). These variations can be quantified by the relative roughness
of the membranes, which is the standard deviation of the local separation. In this scenario, the membranes are ‘free to choose’ an optimal average separation
at which the overall free energy is minimal, and we obtain
as a function of the membrane roughness
at the average membrane separation
from MC simulations that differ in the numbers of receptors and ligands, and in the membrane tension. In both MC simulations scenarios, the binding constant of the membrane-anchored receptors and ligands is obtained as
from the average area concentrations
,
, and
of the bound receptor-ligand complexes, unbound receptors, and unbound ligands observed in the simulations. The binding constant of soluble variants of the receptors and ligands can be obtained as
from the volume concentrations of the receptors and ligands observed in MC simulations. The binding constant
is determined by the binding potential of our model, and does not depend on the length of the complexes.Citation9
Figure 2. (A) Snapshot from a MC simulation with parallel and planar membranes. (B) and (C) Ratio of the binding constants of membrane-anchored and soluble receptors and ligands versus local membrane separation l for different anchoring strengths
and complex lengths
of the receptors and ligands of our elastic-membrane model of biomembrane adhesion. The data points represent MC data, and the lines theoretical results based on Eqs. (Equation7
(7)
(7) ) and (Equation8
(8)
(8) ). The binding constant
of soluble variants of the receptors and ligand is determined by the binding potential of the receptors and ligands and does not depend on the complex length
.
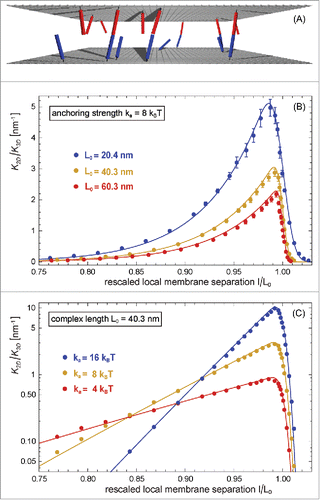
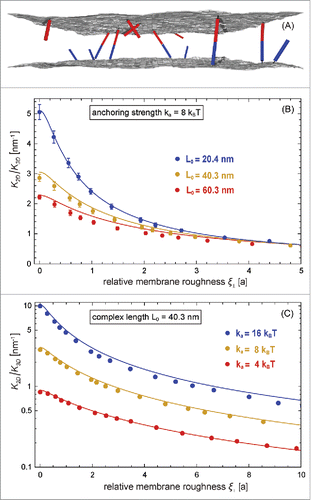
Figure 3. (A) Snapshot from a MC simulation with fluctuating membranes. (B) and (C) Ratio of the binding constants of membrane-anchored and soluble receptors and ligands vs. relative membrane roughness
of 2 equilibrated fluctuating membranes with preferred average separation for different anchoring strengths
and complex lengths
of the receptors and ligands. The data points represent MC data, and the lines represent theoretical results based on Eqs. (Equation1
(1)
(1) ), (Equation7
(7)
(7) ), and (Equation8
(8)
(8) ).
As a function of the local separation , the binding constant
is maximal at a local membrane separation
that is slightly smaller than the length
of the receptor-ligand complexes, and is asymmetric with respect to
(see ). This asymmetry reflects that the receptor-ligand complexes can tilt at local separations
smaller than
, but need to stretch at local separations larger than
. The maximum of the function
decreases with increasing length
of the rigid receptor-ligand complexes (see ), and strongly increases with increasing anchoring strength
of the receptors and ligands (see ). The width of the function
increases with decreasing anchoring strength
. These features of the function
can be understood from our general theory presented in the next section, which agrees with the MC data without any fit parameters (see full lines in ).
The MC data in and the corresponding MD data of illustrate that the binding constant of receptors and ligands anchored to fluctuating membranes decreases with increasing relative membrane roughness
at the optimal average membrane separation
for binding. In , the ratio
of the binding constant, the inverse ‘confinement length’, varies between 0.2 and 10 nm−1, depending on the relative roughness
of the membranes and on the anchoring strength and length of the receptors and ligands.
Figure 4. Ratio of the binding constants of membrane-anchored and soluble receptors and ligands versus relative membrane roughness
at the preferred average separation for (A) transmembrane and (B) lipid-anchored receptors and ligands of our coarse-grained molecular model of biomembrane adhesion. The MD data points result from a variety of membrane systems. In these systems, the area of the 2 apposing membranes ranges from 14 × 14 nm2 to 120 × 120 nm2, and the number of receptors (R) and ligands (L) varies between 1 and 25 (see figure legends). For membrane systems with several receptors and ligands, we obtain multiple data points for states that differ in the number of bound receptor-ligand complexes.Citation7,10 The red data points in (b) result from simulations with confining membrane potentials that restrict membrane shape fluctuations. In experiments, such a situation occurs for membranes bound to apposing surfaces as, e.g., in the surface force apparatus.Citation84,85 The full line in (A) represents a fit to Eq. (Equation11
(11)
(11) ) for the average membrane separation
with fit parameter
. We fit to Eq. (Equation11
(11)
(11) ) because the characteristic length
of our transmembrane receptors and ligands is about 0.38 nm and thus smaller than the values of the relative membrane roughness of all membrane systems in (a). The full line in (b) results from a fit based on Eqs. (Equation1
(1)
(1) ), (Equation7
(7)
(7) ), and (Equation8
(8)
(8) ) with fit parameters
nm2,
nm, and
for the anchoring strength
of lipid-anchored receptors and ligands obtained from the anchoring-angle distributions of the unbound receptors and ligands.
 ) for the average membrane separation l¯=l¯0 with fit parameter c˜2D=2.6±0.2. We fit to Eq. (Equation11(11) K2D≃c∼2DK3Dξ⊥exp[−(l¯−l¯0)22ξ⊥2](11) ) because the characteristic length ξRL of our transmembrane receptors and ligands is about 0.38 nm and thus smaller than the values of the relative membrane roughness of all membrane systems in (a). The full line in (b) results from a fit based on Eqs. (Equation1(1) K2D=∫K2D(l)P(l)dl(1) ), (Equation7(7) HRL(l, θa)≃kaθa2+12kRL(l/cosθa−L0)2(7) ), and (Equation8(8) K2D(l)=2πc2D∫0π/2e−HRL(l, θa)/kBTsinθadθa(8) ) with fit parameters c2D=420±40 nm2, L0=10.35±0.05 nm, and kRL=6.0±1.0 kBT/nm2 for the anchoring strength ka≃2.5 kBT of lipid-anchored receptors and ligands obtained from the anchoring-angle distributions of the unbound receptors and ligands.](/cms/asset/9cb65896-7a47-4e2a-b3cd-3d733090318c/kcam_a_1180487_f0004_oc.gif)
In , the values of range from 0.5 to 5 nm−1, depending on the relative membrane roughness
and on whether the receptors and ligands have a transmembrane anchor or a lipid anchor. The MD data points in result from a variety of membrane systems that differ in membrane area, in the number of receptors and ligands, or in the membrane potential.Citation10 The roughness depends on the area
of the membranes in the MD simulations because the periodic boundaries of the simulation box suppress membrane shape fluctuations with wavelength larger than
where
is the linear membrane size. In membrane systems with several anchored receptors and ligands, the roughness is affected by the number of receptor-ligand bonds because the bonds constrain the membrane shape fluctuations. For the small numbers of receptors and ligands in our MD simulations, the binding constants can be determined from the times spent in bound and unbound states.Citation7,10
The binding kinetics of the transmembrane and lipid-anchored receptors and ligands of our coarse-grained molecular model of biomembrane adhesion can be determined from the frequencies of binding and unbinding events observed in MD simulations.Citation7 The binding potential is identical for both types of receptors and ligands and has no barrier to ensure an efficient sampling of binding and unbinding events of receptors and ligands in our simulations. The kinetics of these events is then strongly enhanced compared with protein binding events in experiments.Citation19-21,Citation30,86 However, this rate enhancement does not affect our main results, which concern the dependence of the rate constants and equilibrium constant on the membrane separation and roughness. At the preferred average separation for binding, the 2D on-rates of the anchored receptors and ligands decrease with the relative membrane roughness, while the 2D off-rates increase with the relative roughness.Citation7,10 For our transmembrane receptors and ligands, the 2D off-rate
increases from about 90/ms to about 140/ms with an increase of the relative membrane roughness from 0.5 nm to 1.8 nm for the membrane systems of . For our lipid-anchored receptors and ligands, the 2D off-rate
increases from about 245/ms to about 290/ms with an increase of the relative membrane roughness from 0.2 nm to 1.7 nm for the membrane systems of . The 3D off-rate of soluble variants of these receptors and ligands with the same binding potential is
. This 3D off-rate is slightly larger than the off-rates of the lipid-anchored receptors and ligands, and about 3 to 5 times larger than the off-rates of the transmembrane receptors and ligands at the preferred average separation for binding. These results appear to indicate that the 2D off-rates of the receptors and ligands in our coarse-grained molecular model are smaller than the 3D off-rate due to constraints on the rotational motion from membrane anchoring, which are more pronounced for our transmembrane receptors and ligands. 2D off-rates that are slightly smaller than 3D off-rates have also been observed for the binding of T-cell receptors to MHC-peptides in experiments in which the T-cell cytoskeleton is disrupted.Citation19 In experiments with intact T-cell cytoskeleton, the 2D off-rates are affected by ATP-driven cytoskeletal forces exerted on TCR-MHC-peptide complexes.Citation19,21,Citation30,31,Citation87
General theory for the binding equilibrium and kinetics of membrane-anchored receptors and ligands
We have derived a general theory for the binding equilibrium and kinetics of membrane-anchored receptors and ligands that agrees with the results from our computational model systems. In this theory, the binding constants and
of membrane-anchored and soluble receptors and ligands can be calculated from the translational and rotational free-energy change upon binding. As a function of the local membrane separation
, the binding constant
has the general formCitation9
(6)
(6) in this theory. Here,
,
, and
are the rotational phase space volumes of the unbound receptors R, unbound ligands L, and bound receptor-ligand complex RL relative to the membranes, and
and
are the translational phase space area and translational phase space volume of the bound ligand relative to the receptor in 2D and 3D. The ratio
in Eq. (Equation6
(6)
(6) ) represents a characteristic length for the binding interface of the receptor-ligand complex and can be estimated as the standard deviation of the binding-site distance in the direction of the complex.Citation9 The rotational phase space volumes of the unbound receptors and ligands can be calculated as
. The remaining, theoretically ‘challenging’ term in Eq. (Equation6
(6)
(6) ) is the rotational phase space volume
of the bound complex, which determines the shape of the function
.
We have found that the rotational phase space volume of the bound receptor-ligand complex can be calculated from an effective configurational energy
of the bound receptor-ligand complex. In our computational model systems, the binding angles and binding angle variations of the rigid, rod-like receptor and ligand molecules are small compared to their anchoring-angle variations. A receptor and ligand then have an approximately collinear orientation in the complex, and approximately equal anchoring angles
. The effective configurational energy is thenCitation9
(7)
(7) The first term of this effective energy is the sum of the anchoring energies (5) for the receptor and ligand in the complex, and the second term is a harmonic approximation for variations in the length
of the receptor-ligand complex, i.e. in the distance between the 2 anchoring points of the complex. For parallel membranes with separation
and approximately identical anchoring angles
of the RL complex in these membranes, the length of the complex, i.e., the distance between the 2 anchoring points in the membranes, is
. With the effective configurational energy (7), the rotational phase space volume of the bound complex can be calculated as
, which leads to
(8)
(8) with
.
The theoretical result for of Eq. (Equation8
(8)
(8) ) agrees with MC data for our elastic-membrane model of biomembrane adhesion without any fit parameters (see lines in ). For our elastic-membrane model, the effective spring constant
and preferred length
of the receptor-ligand complex in the effective configurational energy (7) can be calculated from the standard deviations of the binding angle and binding-site distance and from the lengths of the receptors and ligands.Citation9 By combining the Eqs. (Equation1
(1)
(1) ), (Equation3
(3)
(3) ), and (Equation8
(8)
(8) ), we obtain general results for the binding constant
of receptors and ligands anchored to fluctuating membranes that agree with MC data without fit parameters (see lines in ). Our general theory for the binding constant
thus captures the essential features of the ‘dimensionality reduction’ from 3D to 2D due to membrane anchoring.
In analogy to Eq. (Equation7(7)
(7) ) for the bound receptor-ligand complex, we have postulated the effective configurational energyCitation10
(9)
(9) for the transition-state complex of the binding reaction of membrane-anchored receptors and ligands, with the same anchoring strength
as in Eq. (Equation7
(7)
(7) ). This effective configurational energy reflects that a receptor and ligand molecule can only bind at appropriate relative orientations and separations. The effective spring constant
for the length variations of the transition-state complex is smaller than the corresponding spring constant
of the RL complex, because the variations in the binding-site distance and binding angle, which affect the effective spring constants, are larger in the transition state.Citation10 The preferred effective length
of the transition-state complex, in contrast, is in general close to the preferred length
of the bound RL complex. In analogy to Eq. (Equation8
(8)
(8) ), the on-rate constant is
(10)
(10) for a given separation
of the planar and parallel membranes. The integration over the angle
in Eq. (Equation10
(10)
(10) ) can be interpreted as an integration over the transition-state ensemble of the binding reaction. The on-rate constant
of receptors and ligands anchored to fluctuating membranes can then be obtained from an average over the local membrane separation
(see Eq. (Equation2
(2)
(2) )). This average over local separations for the on-rate constant
relies on characteristic timescales for membrane fluctuations that are significantly smaller than the timescales for the diffusion of the anchored molecules on the relevant length scales.Citation10,32 In contrast, the average in Eq. (Equation1
(1)
(1) ) for the binding constant
is independent of these timescales because
is an equilibrium quantity that does not depend on dynamic aspects.
The effective configurational energies (7) and (9) describe the bound complex and the transition-state complex of membrane-anchored receptors and ligands as effective harmonic springs that can tilt. In contrast, classical theories describe these complexes as simple harmonic springs.Citation32,88,89 As functions of the local membrane separation , the binding equilibrium constant
and on-rate constant
then have a symmetric, Gaussian shape in this classical theory (see Appendix). However, the MC data of illustrate that the function
is clearly asymmetric, in agreement with Eq. (Equation8
(8)
(8) ) of our theory. In , both our theory (full lines) and the classical theory (dashed lines) are compared to data from MD simulations.Citation10 In these simulations of our smallest model system with membrane area
nm2 and a single lipid-anchored receptor and ligand, the average separation
of the membranes is varied by varying the number of water beads between the membranes. The relative membrane roughness in this system is determined by the membrane area and attains the value
nm. Our theoretical results (full lines) are in good agreement with the MD data. The results for the classical theory (dashed lines) deviate from the data because they do not reflect the asymmetry of
and
as functions of the average membrane separation
, which results from the asymmetry of
and
.
Figure 5. (A) Ratio of the binding constants of lipid-anchored and soluble receptors and ligands and (B) on-rate constant
of lipid-anchored receptors and ligands vs. average membrane separation
of 2 membranes with area 14 × 14 nm2 and a single lipid-anchored receptor and ligand in our coarse-grained molecular model. The relative membrane roughness is determined by the membrane area in this system and attains the value
nm. The data points result from MD simulations. The full lines in (A) result from a fit of our general theoretical results for
from Eqs. (Equation1
(1)
(1) ), (Equation7
(7)
(7) ), and (Equation8
(8)
(8) ) with fit parameters
nm2,
nm, and
for the anchoring strength
of our lipid-anchored receptors and ligands. The full lines in (b) result from a fit of our general theoretical results for
from Eqs. (Equation9
(9)
(9) ), (Equation10
(10)
(10) ), and (Equation2
(2)
(2) ) with fit parameters
,
nm, and
. The dashed lines represent fits to Eqs. (Equation17
(17)
(17) ) and (Equation18
(18)
(18) ) obtained for the classical Gaussian theory with fit parameters (a)
nm2,
nm, and
nm and (b)
,
nm, and
nm.
 ) and (Equation18(18) kon=konmaxξkξ⊥2+ξk2exp[−(l¯−lk)22(ξ⊥2+ξk2)](18) ) obtained for the classical Gaussian theory with fit parameters (a) K2Dmax=146±9 nm2, lK=9.36±0.06 nm, and ξK=1.08±0.04 nm and (b) konmax=42.1±1.4 μm2/s, lk=9.35±0.05 nm, and ξk=1.41±0.05 nm.](/cms/asset/304103e3-a35c-420e-bdfe-6bca31d9517e/kcam_a_1180487_f0005_b.gif)
For a relative membrane roughness that is much larger than the widths
and
of the functions
and
, the distribution
of local membrane separations
is nearly constant over the range of local separations
for which
and
are not negligibly small. The Eqs. (Equation1
(1)
(1) ) and (Equation2
(2)
(2) ) of our theory then simplify to Citation9,10
(11)
(11) with
and
(12)
(12) with
for a Gaussian distribution
of the local membrane separation
(see Eq. (Equation3
(3)
(3) )). Here,
and
are the preferred average separations for large roughnesses. For such large roughnesses, the dependence of
and
on the average separation
is dominated by the shape of the distribution
, and the asymmetry of
and
are ‘averaged out’ in Eqs. (Equation1
(1)
(1) ) and (Equation2
(2)
(2) ). At the preferred average separations for binding, i.e. at the average separations for which the Gaussian functions in Eqs. (Equation11
(11)
(11) ) and (Equation12
(12)
(12) ) are maximal, the binding constant
and on-rate constant
are inversely proportional to the relative membrane roughness
.
In our theory, the widths and
of the functions
and
depends on the anchoring strength
of the receptors and ligands, and the preferred lengths and effective spring constants of the bound complex and the transition-state complexCitation9,10:
(13)
(13)
(14)
(14) For the lipid-anchored receptors and ligands of our coarse-grained molecular model, these widths are
nm and
nm. For the transmembrane receptors and ligands, we have
nm and
nm. For the receptors and ligands of our elastic-membrane model, the width
of the function
ranges between
nm and
nm, depending on the anchoring strength
and complex length
of the receptors and ligands. For receptor-ligand complexes of length
nm, we have
nm,
nm, and
nm for the anchoring strengths
,
, and
. For receptors and ligands with anchoring strength
, we have
nm,
nm, and
nm for the complex lengths
nm,
nm, and
nm.
Conclusions and outlook
The computational model systems and theories reviewed in this article indicate that the relative roughness of 2 adhering membranes plays an important role for the binding of membrane-anchored receptors and ligands. For concentrations [RL] of receptor-ligand bonds around
, the relative membrane roughness
obtained from Eq. (Equation4
(4)
(4) ) is of the same magnitude or larger than the characteristic lengths
and
of the receptors and the ligands in our computational model systems, which reflect how strongly the local separation of the membranes is constrained by the receptor-ligand and transition-state complexes. The binding constant
and on-rate constant
of the receptors and ligands then decreases with increasing relative membrane roughness
in equilibrated membrane adhesion zones in which the average separation
of the membranes is close to the preferred average separation
of the receptors and ligands for binding.
In the next years, experimental model systems of biomembrane adhesion may confirm the effect of the relative membrane roughness on the binding constant
of membrane-anchored receptors and ligands. In such model systems, the adhesion of reconstituted membranes is mediated by anchored adhesion proteins,Citation68,90-100 by anchored saccharides,Citation101,102 or by anchored DNA.Citation103-106 The roughness-dependence of
can be confirmed by demonstrating that
increases with the concentration [RL] of bound receptor-ligand complexes, because the relative membrane roughness
decreases with increasing bond concentration [RL]. Measuring the relative membrane roughness requires a spatial resolution in the nanometer range both in the directions parallel and perpendicular to the membranes, which is beyond the scope of current optical methods used to probe membrane shape fluctuations.Citation107,108 However, the relative membrane roughness can be measured in neutron scattering experiments on stacks of oriented membranes that interact via anchored molecules.Citation102
Our general theories for the binding constant and binding kinetics of membrane-anchored molecules reviewed in this article are in good agreement with simulation data for our computational model systems. These theories identify characteristic properties of the receptor and ligand molecules and of the apposing membranes that determine the binding equilibrium and kinetics. In the general Eqs. (Equation1
(1)
(1) ) and (Equation2
(2)
(2) ), the molecular properties of the receptors and ligands, including their membrane anchoring, are reflected in the functions
and
, and the properties of the membranes are reflected in the distribution
of the local membrane separation
. The distribution
has the Gaussian shape (3) with the average membrane separation
and relative membrane roughness
as characteristic lengths if the adhesion is dominated by a single type of receptors and ligands.Citation9,10 In our detailed theories for
and
reviewed in Section IV, the receptor-ligand complex and the transition-complex are described as elastic springs that can tilt, which results in asymmetric, non-Gaussian functions
for
. Our theoretical results for the ratio of the binding constants
and
of membrane-anchored and soluble receptors and ligands agree with MC data without any fit parameters (see ), which indicates that our theory captures the essential features of the ‘dimensionality reduction’ from 3D to 2D due to membrane anchoring, for both planar and fluctuating membranes. Other theories concern the binding of receptors and ligands anchored to essentially planar membranes,Citation4,8 the binding of DNA immobilized on apposing nanoparticle surfaces,Citation109,110 or the binding of flexible receptor and ligand polymers.Citation111-114
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Appendix: Gaussian theory for membrane-anchored receptors and ligands
In classical theories,Citation32,88,89 the effective configurational energies and
of membrane-anchored receptor-ligand and transition-state complexes depend only on the membrane separation
. In harmonic approximation, such effective configurational energies lead to Gaussian functions
(15)
(15)
(16)
(16) Here,
and
are the widths of the functions
and
. For a Gaussian distribution
of the local membrane separation as in Eq. (Equation3
(3)
(3) ), the averages over all local separation
in Eqs. (Equation1
(1)
(1) ) and (Equation2
(2)
(2) ) can be calculated explicitly, which leads to
(17)
(17)
(18)
(18) From these 2 equations, we obtain the off-rate constant
(19)
(19) with
. Related expressions for averages at fixed membrane locations in the special case
have been derived by Bihr et al.Citation32
The dependence of the off-rate constant on the average membrane separation
can be understood from the first and second derivative of
with respect to
. The first derivative
vanishes at the average membrane separation
(20)
(20) The value of the second derivative at
at this membrane separation is positive for
, and negative for
. As a function of
, the off-rate constant thus exhibits a minimum at
for
, and a maximum for
. Depending on the values of
,
,
,
, and
, the location
of this minimum or maximum can adopt values that differ strongly from the locations
and
of the maxima of the Gaussian functions (15) and (16). Negative values of
imply that the off-rate constant
is monotonously increasing at positive average separations
for
, and monotonously decreasing at such average separations for
. Because the membranes cannot intersect, the average separation
of the membranes does not attain negative values.
References
- Dustin ML, Bromley SK, Davis MM, Zhu C. Identification of self through two-dimensional chemistry and synapses. Annu Rev Cell Dev Biol 2001; 17:133-57; PMID:11687486; http://dx.doi.org/10.1146/annurev.cellbio.17.1.133
- Orsello CE, Lauffenburger DA, Hammer DA. Molecular properties in cell adhesion: a physical and engineering perspective. Trends Biotechnol 2001; 19:310-6; PMID:11451473; http://dx.doi.org/10.1016/S0167-7799(01)01692-4
- Krobath H, Rozycki B, Lipowsky R, Weikl TR. Binding cooperativity of membrane adhesion receptors. Soft Matter 2009; 5:3354-61; http://dx.doi.org/10.1039/b902036e
- Wu Y, Vendome J, Shapiro L, Ben-Shaul, A, Honig, B. Transforming binding affinities from three dimensions to two with application to cadherin clustering. Nature 2011; 475:510-3; PMID:21796210; http://dx.doi.org/10.1038/nature10183
- Leckband D, Sivasankar S. Cadherin recognition and adhesion. Curr Opin Cell Biol 2012; 24:620-7; PMID:22770731; http://dx.doi.org/10.1016/j.ceb.2012.05.014
- Zarnitsyna V, Zhu C. T cell triggering: insights from 2D kinetics analysis of molecular interactions. Phys Biol Aug 2012; 9:045005; PMID:22871794; http://dx.doi.org/10.1088/1478-3975/9/4/045005
- Hu J, Lipowsky R, Weikl TR. Binding constants of membrane-anchored receptors and ligands depend strongly on the nanoscale roughness of membranes. Proc Natl Acad Sci USA 2013; 110(38):15283-8; PMID:24006364; http://dx.doi.org/10.1073/pnas.1305766110
- Wu Y, Honig B, Ben-Shaul A. Theory and simulations of adhesion receptor dimerization on membrane surfaces. Biophys J Mar 2013; 104:1221-9; PMID:23528081; http://dx.doi.org/10.1016/j.bpj.2013.02.009
- Xu G-K, Hu J, Lipowsky R, Weikl TR. Binding constants of membrane-anchored receptors and ligands: A general theory corroborated Monte Carlo simulations. J Chem Phys 2015; 143:243136; PMID:26723621
- Hu J, Xu G-K, Lipowsky R, Weikl TR. Binding kinetics of membrane-anchored receptors and ligands: Molecular dynamics simulations and theory. J Chem Phys 2015; 143:243137; PMID:26723622
- Schuck P. Use of surface plasmon resonance to probe the equilibrium and dynamic aspects of interactions between biological macromolecules. Annu Rev Biophys Biomol Struct 1997; 26:541-66; PMID:9241429; http://dx.doi.org/10.1146/annurev.biophys.26.1.541
- Rich RL, Myszka DG. Advances in surface plasmon resonance biosensor analysis. Curr Opin Biotechnol 2000; 11:54-61; PMID:10679342; http://dx.doi.org/10.1016/S0958-1669(99)00054-3
- McDonnell JM. Surface plasmon resonance: towards an understanding of the mechanisms of biological molecular recognition. Curr Opin Chem Biol 2001; 5:572-7; PMID:11578932; http://dx.doi.org/10.1016/S1367-5931(00)00251-9
- Bell GI, Dembo M, Bongrand P. Cell adhesion. Competition between nonspecific repulsion and specific bonding. Biophys J 1984; 45:1051-64; PMID:6743742; http://dx.doi.org/10.1016/S0006-3495(84)84252-6
- Dustin ML, Ferguson LM, Chan PY, Springer TA, Golan DE. Visualization of CD2 interaction with LFA-3 and determination of the two-dimensional dissociation constant for adhesion receptors in a contact area. J Cell Biol 1996; 132:465-74; PMID:8636222; http://dx.doi.org/10.1083/jcb.132.3.465
- Dustin ML, Golan DE, Zhu DM, Miller JM, Meier W, Davies EA, van der Merwe PA. Low affinity interaction of human or rat T cell adhesion molecule CD2 with its ligand aligns adhering membranes to achieve high physiological affinity. J Biol Chem 1997; 272:30889-98; PMID:9388235; http://dx.doi.org/10.1074/jbc.272.49.30889
- Zhu D-M, Dustin ML, Cairo CW, Golan DE. Analysis of two-dimensional dissociation constant of laterally mobile cell adhesion molecules. Biophys J 2007; 92:1022-34; PMID:17085486; http://dx.doi.org/10.1529/biophysj.106.089649
- Tolentino TP, Wu J, Zarnitsyna VI, Fang Y, Dustin ML, Zhu C. Measuring diffusion and binding kinetics by contact area FRAP. Biophys J 2008; 95:920-30; PMID:18390627; http://dx.doi.org/10.1529/biophysj.107.114447
- Huppa JB, Axmann M, Mörtelmaier MA, Lillemeier BF, Newell EW, Brameshuber M, Klein LO, Schütz GJ, Davis MM. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature 2010; 463:963-7; PMID:20164930; http://dx.doi.org/10.1038/nature08746
- Axmann M, Huppa JB, Davis MM, Schütz GJ. Determination of interaction kinetics between the T cell receptor and peptide-loaded MHC class II via single-molecule diffusion measurements. Biophys J 2012; 103:L17-9; PMID:22853916; http://dx.doi.org/10.1016/j.bpj.2012.06.019
- O'Donoghue GP, Pielak RM, Smoligovets AA, Lin JJ, Groves JT. Direct single molecule measurement of TCR triggering by agonist pMHC in living primary T cells. Elife 2013; 2:e00778; PMID:23840928; http://dx.doi.org/10.7554/eLife.00778
- Kaplanski G, Farnarier C, Tissot O, Pierres A, Benoliel AM, Alessi MC, Kaplanski S, Bongrand P. Granulocyte-endothelium initial adhesion. Analysis of transient binding events mediated by E-selectin in a laminar shear flow. Biophys J 1993; 64:1922-33; PMID:7690258; http://dx.doi.org/10.1016/S0006-3495(93)81563-7
- Alon R, Hammer DA, Springer TA. Lifetime of the P-selectin-carbohydrate bond and its response to tensile force in hydrodynamic flow. Nature 1995; 374:539-42; PMID:7535385; http://dx.doi.org/10.1038/374539a0
- Piper JW, Swerlick RA, Zhu C. Determining force dependence of two-dimensional receptor-ligand binding affinity by centrifugation. Biophys J 1998; 74:492-513, ; PMID:9449350; http://dx.doi.org/10.1016/S0006-3495(98)77807-5
- Chesla SE, Selvaraj P, Zhu C. Measuring two-dimensional receptor-ligand binding kinetics by micropipette. Biophys J 1998; 75:1553-72; PMID:9726957; http://dx.doi.org/10.1016/S0006-3495(98)74074-3
- Merkel R, Nassoy P, Leung A, Ritchie K, Evans E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature 1999; 397:50-3; PMID:9892352; http://dx.doi.org/10.1038/16219
- Williams TE, Nagarajan S, Selvaraj P, Zhu C. Quantifying the impact of membrane microtopology on effective two-dimensional affinity. J Biol Chem 2001; 276:13283-8; PMID:11278674; http://dx.doi.org/10.1074/jbc.M010427200
- Chen W, Evans EA, McEver RP, Zhu C. Monitoring receptor-ligand interactions between surfaces by thermal fluctuations. Biophys J 2008; 94:694-701; PMID:17890399; http://dx.doi.org/10.1529/biophysj.107.117895
- Chien Y-H, Jiang N, Li F, Zhang F, Zhu C, Leckband D. Two stage cadherin kinetics require multiple extracellular domains but not the cytoplasmic region. J Biol Chem 2008; 283:1848-56; PMID:17999960; http://dx.doi.org/10.1074/jbc.M708044200
- Huang J, Zarnitsyna VI, Liu B, Edwards LJ, Jiang N, Evavold BD, Zhu C. The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness. Nature 2010; 464:932-6; PMID:20357766; http://dx.doi.org/10.1038/nature08944
- Liu B, Chen W, Evavold BD, Zhu C. Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell 2014; 157:357-68, ; PMID:24725404; http://dx.doi.org/10.1016/j.cell.2014.02.053
- Bihr T, Seifert U, Smith A-S. Nucleation of ligand-receptor domains in membrane adhesion. Phys Rev Lett 2012; 109:258101; PMID:23368503; http://dx.doi.org/10.1103/PhysRevLett.109.258101
- Krobath H, Schütz GJ, Lipowsky R, Weikl TR. Lateral diffusion of receptor-ligand bonds in membrane adhesion zones: Effect of thermal membrane roughness. Europhys Lett 2007; 78:p. 38003; http://dx.doi.org/10.1209/0295-5075/78/38003
- Nagle JF. Introductory lecture: Basic quantities in model biomembranes. Faraday Discuss 2013; 161:11-29; PMID:23805735; http://dx.doi.org/10.1039/C2FD20121F
- Dimova R. Recent developments in the field of bending rigidity measurements on membranes. Adv Colloid Interface Sci 2014; 208:225-34; PMID:24666592; http://dx.doi.org/10.1016/j.cis.2014.03.003
- Pontes B, Ayala Y, Fonseca ACC, Romao LF, Amaral RF, Salgado LT, Lima FR, Farina M, Viana NB, Moura-Neto V, et al. Membrane elastic properties and cell function. PLoS One 2013; 8:e67708; PMID:23844071
- Betz T, Lenz M, Joanny J-F, Sykes C. Atp-dependent mechanics of red blood cells. Proc Natl Acad Sci USA 2009; 106:15320-5; http://dx.doi.org/10.1073/pnas.0904614106
- Zamir E, Geiger B. Molecular complexity and dynamics of cell-matrix adhesions. J Cell Sci 2001; 114:3583-90; PMID:11707510
- Leckband DE, de Rooij J. Cadherin adhesion and mechanotransduction. Annu Rev Cell Dev Biol 2014; 30:291-315; PMID:25062360; http://dx.doi.org/10.1146/annurev-cellbio-100913-013212
- Biswas KH, Hartman KL, C-h Yu, Harrison OJ, Song H, Smith AW, WYC Huang, Lin W-C, Guo Z, Padmanabhan A, et al. E-cadherin junction formation involves an active kinetic nucleation process. Proc Natl Acad Sci USA Sep 2015; 112:10932-7; PMID:26290581; http://dx.doi.org/10.1073/pnas.1513775112
- Yap AS, Gomez GA, Parton RG. Adherens junctions revisualized: Organizing cadherins as nanoassemblies. Dev Cell 2015; 35:12-20; PMID:26460944; http://dx.doi.org/10.1016/j.devcel.2015.09.012
- Goetz R, Lipowsky R. Computer simulations of bilayer membranes: Self-assembly and interfacial tension. J Chem Phys 1998; 108:7397-409; http://dx.doi.org/10.1063/1.476160
- Shelley JC, Shelley MY, Reeder RC, Bandyopadhyay S, Klein ML. A coarse grain model for phospholipid simulations. J Phys Chem B 2001; 105:4464-70; http://dx.doi.org/10.1021/jp010238p
- Marrink SJ, de Vries AH, Mark AE. Coarse grained model for semiquantitative lipid simulations. J Phys Chem B 2004; 108:750-60; http://dx.doi.org/10.1021/jp036508g
- Shih AY, Arkhipov A, Freddolino PL, Schulten K. Coarse grained protein-lipid model with application to lipoprotein particles. J Phys Chem B 2006; 110:3674-84; PMID:16494423; http://dx.doi.org/10.1021/jp0550816
- Marrink SJ, Mark AE. The mechanism of vesicle fusion as revealed by molecular dynamics simulations. J Am Chem Soc 2003; 125(37):11144-5; PMID:16220905; http://dx.doi.org/10.1021/ja036138+
- Shillcock JC, Lipowsky R. Tension-induced fusion of bilayer membranes and vesicles. Nat Mater 2005; 4:225-8; PMID:15711550; http://dx.doi.org/10.1038/nmat1333
- Grafmüller A, Shillcock J, Lipowsky R. Pathway of membrane fusion with two tension-dependent energy barriers. Phys Rev Lett 2007; 98, p. 218101; PMID:17677811; http://dx.doi.org/10.1103/PhysRevLett.98.218101
- Grafmüller A, Shillcock J, Lipowsky R. The fusion of membranes and vesicles: pathway and energy barriers from dissipative particle dynamics. Biophys J 2009; 96:2658-75; PMID:19348749; http://dx.doi.org/10.1016/j.bpj.2008.11.073
- Smirnova YG, Marrink S-J, Lipowsky R, Knecht V. Solvent-exposed tails as prestalk transition states for membrane fusion at low hydration. J Am Chem Soc 2010; 132:6710-8; PMID:20411937; http://dx.doi.org/10.1021/ja910050x
- Risselada HJ, Kutzner C, Grubmueller H. Caught in the act: Visualization of SNARE-mediated fusion events in molecular detail. Chem Bio Chem 2011; 12:1049-55; PMID:21433241; http://dx.doi.org/10.1002/cbic.201100020
- Illya G, Lipowsky R, Shillcock JC. Two-component membrane material properties and domain formation from dissipative particle dynamics. J Chem Phys 2006; 125:114710; PMID:16999504
- Risselada HJ, Marrink SJ. The molecular face of lipid rafts in model membranes. Proc Natl Acad Sci USA 2008; 105:17367-72; PMID:18987307; http://dx.doi.org/10.1073/pnas.0807527105
- de Meyer FJ-M, Benjamini A, Rodgers JM, Misteli Y, Smit B. Molecular simulation of the DMPC-cholesterol phase diagram. J Phys Chem B 2010; 114:10451-61; PMID:20662483; http://dx.doi.org/10.1021/jp103903s
- Apajalahti T, Niemela P, Govindan PN, Miettinen MS, Salonen E, Marrink S-J, Vattulainen I. Concerted diffusion of lipids in raft-like membranes. Faraday Discuss 2010; 144:411-30; PMID:20158041; http://dx.doi.org/10.1039/B901487J
- Bennett WFD, Tieleman DP. Computer simulations of lipid membrane domains. Biochim Biophys Acta-Biomembr 2013; 1828:1765-76; PMID:23500617; http://dx.doi.org/10.1016/j.bbamem.2013.03.004
- Gambin Y, Lopez-Esparza R, Reffay M, Sierecki E, Gov NS, Genest M, Hodges RS, Urbach W. Lateral mobility of proteins in liquid membranes revisited. Proc Natl Acad Sci USA 2006; 103(7):2098-102; PMID:16461891; http://dx.doi.org/10.1073/pnas.0511026103
- Guigas G, Weiss M. Size-dependent diffusion of membrane inclusions. Biophys J 2006; 91(7):2393-8; PMID:16829562; http://dx.doi.org/10.1529/biophysj.106.087031
- Reynwar BJ, Illya G, Harmandaris VA, Müller MM, Kremer K, Deserno M. Aggregation and vesiculation of membrane proteins by curvature-mediated interactions. Nature 2007; 447:461-4; PMID:17522680; http://dx.doi.org/10.1038/nature05840
- Arkhipov A, Yin Y, Schulten K. Four-scale description of membrane sculpting by BAR domains. Biophys J 2008; 95:2806-21; PMID:18515394; http://dx.doi.org/10.1529/biophysj.108.132563
- Simunovic M, Srivastava A, Voth GA. Linear aggregation of proteins on the membrane as a prelude to membrane remodeling. Proc Natl Acad Sci USA 2013; 110:20396-401; PMID:24284177; http://dx.doi.org/10.1073/pnas.1309819110
- Lipowsky R. Adhesion of membranes via anchored stickers. Phys Rev Lett 1996; 77:1652-5; PMID:10063132; http://dx.doi.org/10.1103/PhysRevLett.77.1652
- Weikl TR, Lipowsky R. Adhesion-induced phase behavior of multicomponent membranes. Phys Rev E 2001; 64:011903; PMID:11461284; http://dx.doi.org/10.1103/PhysRevE.64.011903
- Weikl TR, Groves JT, Lipowsky R. Pattern formation during adhesion of multicomponent membranes. Europhys Lett 2002; 59:916-22; http://dx.doi.org/10.1209/epl/i2002-00130-3
- Weikl TR, Lipowsky R. Pattern formation during T-cell adhesion. Biophys J 2004; 87:3665-78; PMID:15377531; http://dx.doi.org/10.1529/biophysj.104.045609
- Asfaw M, Rozycki B, Lipowsky R, Weikl TR. Membrane adhesion via competing receptor/ligand bonds. Europhys Lett 2006; 76:703-9; http://dx.doi.org/10.1209/epl/i2006-10317-0
- Tsourkas PK, Baumgarth N, Simon SI, Raychaudhuri S. Mechanisms of B-cell synapse formation predicted by Monte Carlo simulation. Biophys J 2007; 92:4196-208; PMID:17384077; http://dx.doi.org/10.1529/biophysj.106.094995
- Reister-Gottfried E, Sengupta K, Lorz B, Sackmann E, Seifert U, Smith AS. Dynamics of specific vesicle-substrate adhesion: From local events to global dynamics. Phys Rev Lett 2008; 101:208103; PMID:19113383; http://dx.doi.org/10.1103/PhysRevLett.101.208103
- Paszek MJ, Boettiger D, Weaver VM, Hammer DA. Integrin clustering is driven by mechanical resistance from the glycocalyx and the substrate. PLoS Comput Biol 2009; 5:e1000604; PMID:20011123; http://dx.doi.org/10.1371/journal.pcbi.1000604
- Komura S, Andelman D. Adhesion-induced lateral phase separation in membranes. Eur Phys J E 2000; 3:259-71; http://dx.doi.org/10.1007/s101890070018
- Bruinsma R, Behrisch A, Sackmann E. Adhesive switching of membranes: experiment and theory. Phys Rev E 2000; 61:4253-67; PMID:11088221; http://dx.doi.org/10.1103/PhysRevE.61.4253
- Qi SY, Groves JT, Chakraborty AK. Synaptic pattern formation during cellular recognition. Proc Natl Acad Sci USA 2001; 98:6548-6553; PMID:11371622; http://dx.doi.org/10.1073/pnas.111536798
- Chen H-Y. Adhesion-induced phase separation of multiple species of membrane junctions. Phys Rev E 2003; 67:031919; PMID:12689113; http://dx.doi.org/10.1103/PhysRevE.67.031919
- Raychaudhuri S, Chakraborty AK, Kardar M. Effective membrane model of the immunological synapse. Phys Rev Lett 2003; 91:208101; PMID:14683399; http://dx.doi.org/10.1103/PhysRevLett.91.208101
- Coombs D, Dembo M, Wofsy C, Goldstein B. Equilibrium thermodynamics of cell-cell adhesion mediated by multiple ligand-receptor pairs. Biophys J 2004; 86:1408-23; PMID:14990470; http://dx.doi.org/10.1016/S0006-3495(04)74211-3
- Shenoy VB, Freund LB. Growth and shape stability of a biological membrane adhesion complex in the diffusion-mediated regime. Proc Natl Acad Sci USA 2005; 102:3213-8.
- Wu J-Y, Chen H-Y. Membrane-adhesion-induced phase separation of two species of junctions. Phys Rev E 2006; 73:011914; PMID:16486192; http://dx.doi.org/10.1103/PhysRevE.73.011914
- Zhang C-Z, Wang Z-G. Nucleation of membrane adhesions. Phys Rev E 2008; 77:021906.
- Atilgan E, Ovryn B. Nucleation and growth of integrin adhesions. Biophys J 2009; 96:3555-72; PMID:19413961; http://dx.doi.org/10.1016/j.bpj.2009.02.023
- Zuckerman D, Bruinsma R. Statistical mechanics of membrane adhesion by reversible molecular bonds. Phys Rev Lett 1995; 74:3900-3; PMID:10058325; http://dx.doi.org/10.1103/PhysRevLett.74.3900
- Speck T, Reister E, Seifert U. Specific adhesion of membranes: Mapping to an effective bond lattice gas. Phys Rev 2010; 82:021923; PMID:20866853
- Weil N, Farago O. Entropy-driven aggregation of adhesion sites of supported membranes. Eur Phys J E 2010; 33:81-7; PMID:20848152; http://dx.doi.org/10.1140/epje/i2010-10646-7
- Dharan N, Farago O. Formation of adhesion domains in stressed and confined membranes. Soft Matter 2015; 11(19):3780-5; PMID:25833123; http://dx.doi.org/10.1039/C5SM00295H
- Israelachvili JN, Intermolecular and surface forces, 2nd ed. 1992, Academic Press.
- Bayas MV, Kearney A, Avramovic A, van der Merwe PA, Leckband DE. Impact of salt bridges on the equilibrium binding and adhesion of human CD2 and CD58. J Biol Chem 2007; 282:5589-96; PMID:17172599; http://dx.doi.org/10.1074/jbc.M607968200
- Robert P, Nicolas A, Aranda-Espinoza S, Bongrand P, Limozin L. Minimal encounter time and separation determine ligand-receptor binding in cell adhesion. Biophys J 2011; 100:(11):2642-51; PMID:21641309; http://dx.doi.org/10.1016/j.bpj.2011.04.011
- Wang J-h, Reinherz EL. The structural basis of T-lineage immune recognition: TCR docking topologies, mechanotransduction, and co-receptor function. Immunol Rev 2012; 250:102-19; PMID:23046125; http://dx.doi.org/10.1111/j.1600-065X.2012.01161.x
- Dembo M, Torney DC, Saxman K, Hammer D. The reaction-limited kinetics of membrane-to-surface adhesion and detachment. Proc R Soc Lond B 1988; 234:55-83; PMID:2901109; http://dx.doi.org/10.1098/rspb.1988.0038
- Gao H, Qian J, Chen B. Probing mechanical principles of focal contacts in cell-matrix adhesion with a coupled stochastic-elastic modelling framework. J R Soc Interface 2011; 8:1217-32; PMID:21632610; http://dx.doi.org/10.1098/rsif.2011.0157
- Albersdörfer A, Feder T, Sackmann E. Adhesion-induced domain formation by interplay of long-range repulsion and short-range attraction force: A model membrane study. Biophys J 1997; 73:245-57; PMID:9199789; http://dx.doi.org/10.1016/S0006-3495(97)78065-2
- Kloboucek A, Behrisch A, Faix J, Sackmann E. Adhesion-induced receptor segregation and adhesion plaque formation: A model membrane study. Biophys J 1999; 77:2311-28; PMID:10512849; http://dx.doi.org/10.1016/S0006-3495(99)77070-0
- Maier CW, Behrisch A, Kloboucek A, Simson DA, Merkel R. Specific biomembrane adhesion - indirect lateral interactions between bound receptor molecules. Eur Phys J E 2001; 6:273-6; http://dx.doi.org/10.1007/s10189-001-8041-9
- Lorz BG, Smith A-S, Gege C, Sackmann E. Adhesion of giant vesicles mediated by weak binding of Sialyl-Lewis(x) to E-selectin in the presence of repelling poly(ethylene glycol) molecules. Langmuir 2007; 23:12293-300; PMID:17918980; http://dx.doi.org/10.1021/la701824q
- Purrucker O, Goennenwein S, Foertig A, Jordan R, Rusp M, Baermann M, Moroder L, Sackmann E, Tanaka M. Polymer-tethered membranes as quantitative models for the study of integrin-mediated cell adhesion. Soft Matter 2007; 3:333-6; http://dx.doi.org/10.1039/B612069E
- Streicher P, Nassoy P, Bärmann M, Dif A, Marchi-Artzner V, Brochard-Wyart F, Spatz J, Bassereau P. Integrin reconstituted in GUVs: a biomimetic system to study initial steps of cell spreading. Biochim Biophys Acta 2009; 1788:2291-300; PMID:19665445; http://dx.doi.org/10.1016/j.bbamem.2009.07.025
- Monzel C, Fenz SF, Merkel R, Sengupta K. Probing biomembrane dynamics by dual-wavelength reflection interference contrast microscopy. Chemphyschem 2009; 10:2828-38; PMID:19821476; http://dx.doi.org/10.1002/cphc.200900645
- Fenz SF, Smith A-S, Merkel R, Sengupta K. Inter-membrane adhesion mediated by mobile linkers: Effect of receptor shortage. Soft Matter 2011; 7(3):952-62; http://dx.doi.org/10.1039/C0SM00550A
- Bihr T, Fenz S, Sackmann E, Merkel R, Seifert U, Sengupta K, Smith A-S. Association rates of membrane-coupled cell adhesion molecules. Biophys J 2014; 107:L33-6; PMID:25468354; http://dx.doi.org/10.1016/j.bpj.2014.10.033
- Sackmann E, Smith A-S. Physics of cell adhesion: some lessons from cell-mimetic systems. Soft Matter 2014; 10(11):1644-59; PMID:24651316; http://dx.doi.org/10.1039/c3sm51910d
- Schmidt D, Bihr T, Fenz S, Merkel R, Seifert U, Sengupta K, Smith A-S. Crowding of receptors induces ring-like adhesions in model membranes. Biochim Biophys Acta-Mol Cell Res 2015; 1853:2984-91; PMID:26028591; http://dx.doi.org/10.1016/j.bbamcr.2015.05.025
- Gourier C, Pincet F, Perez E, Zhang Y, Mallet J-M, Sinay P. Specific and non specific interactions involving LeX determinant quantified by lipid vesicle micromanipulation. Glycoconjugate J 2004; 21:165-74; PMID:15483381; http://dx.doi.org/10.1023/B:GLYC.0000044847.15797.2e
- Schneck E, Deme B, Gege C, Tanaka M. Membrane adhesion via homophilic saccharide-saccharide interactions investigated by neutron scattering. Biophys J 2011; 100:2151-9; PMID:21539782; http://dx.doi.org/10.1016/j.bpj.2011.03.011
- Chan Y-HM, Lenz P, Boxer SG. Kinetics of DNA-mediated docking reactions between vesicles tethered to supported lipid bilayers. Proc Natl Acad Sci USA 2007; 104:18913-8.
- Beales PA, Vanderlick TK. DNA as membrane-bound ligand-receptor pairs: Duplex stability is tuned by intermembrane forces. Biophys J 2009; 96:1554-65; PMID:19217872; http://dx.doi.org/10.1016/j.bpj.2008.11.027
- Chung M, Koo BJ, Boxer SG. Formation and analysis of topographical domains between lipid membranes tethered by DNA hybrids of different lengths. Faraday Discuss 2013; 161:333-45; discussion 419–59; PMID:23805748; http://dx.doi.org/10.1039/C2FD20108A
- Parolini L, Mognetti BM, Kotar J, Eiser E, Cicuta P, Di L Michele. Volume and porosity thermal regulation in lipid mesophases by coupling mobile ligands to soft membranes. Nat Commun 2015; 6:5948; PMID:25565580
- Pierres A, Benoliel A-M, Touchard D, Bongrand P. How cells tiptoe on adhesive surfaces before sticking. Biophys J 2008; 94:4114-22; PMID:18234815; http://dx.doi.org/10.1529/biophysj.107.125278
- Monzel C, Schmidt D, Kleusch C, Kirchenbuechler D, Seifert U, Smith A-S, Sengupta K, Merkel R. Measuring fast stochastic displacements of bio-membranes with dynamic optical displacement spectroscopy. Nat Commun 2015; 6:8162; PMID:26437911.
- Leunissen ME, Frenkel D. Numerical study of DNA-functionalized microparticles and nanoparticles: explicit pair potentials and their implications for phase behavior. J Chem Phys 2011; 134:084702; PMID:21361551; http://dx.doi.org/10.1063/1.3557794
- Varilly P, Angioletti-Uberti S, Mognetti BM, Frenkel D. A general theory of DNA-mediated and other valence-limited colloidal interactions. J Chem Phys 2012; 137:094108; PMID:22957556; http://dx.doi.org/10.1063/1.4748100
- Jeppesen C, Wong JY, Kuhl TL, Israelachvili JN, Mullah N, Zalipsky S, Marques CM. Impact of polymer tether length on multiple ligand-receptor bond formation. Science 2001; 293:465-8; PMID:11463908; http://dx.doi.org/10.1126/science.293.5529.465
- Moreira AG, Marques CM. The role of polymer spacers in specific adhesion. J Chem Phys April 2004; 120:6229-37; PMID:15267510; http://dx.doi.org/10.1063/1.1651088
- Moore NW, Kuhl TL. The role of flexible tethers in multiple ligand-receptor bond formation between curved surfaces. Biophys J 2006; 91:1675-87; PMID:16751237; http://dx.doi.org/10.1529/biophysj.105.079871
- Zhang C-Z, Wang Z-G. Polymer-tethered ligand-receptor interactions between surfaces ii. Langmuir 2007; 23:13024-39; PMID:18001063; http://dx.doi.org/10.1021/la7017133
