ABSTRACT
Lim Domain and Actin Binding protein1 (lima1) influence cancer cell function. Thus far, functional role of lima1 in cholangiocarcinoma remains unknown. We used public databases, in vitro experiments, and multi-omics analysis to investigate the Lima1 in cholangiocarcinoma. Our results showed that lima1 expression is significantly upregulated and high levels of lima1 are significantly associated with vascular invasion in cholangiocarcinoma. Furthermore, lima1 knocking out inhibits the RBE cell invasion. Multi-omics data suggest that lima1 affect a broad spectrum of cancer related pathways, promoting tumor progression and metastatic ability in cholangiocarcinoma. This study provides insights into molecular associations of lima1 with tumorigenesist and establishes a preliminary picture of the correlation network in cholangiocarcinoma.
Introduction
Cholangiocarcinoma is an aggressive epithelial cell malignancy with late diagnosis and poor outcomes for afflicted patients [Citation1]. Cholangiocarcinoma arises from varying anatomical locations within the biliary tree which includes intrahepatic (ICC), perihilar(pCCA), and distal cholangiocarcinoma [Citation1]. Surgical treatment is considered the preferred option for all types of cholangiocarcinoma, however, when it comes to contemplated, the desmoplastic nature of cholangiocarcinoma such as lymph nodes and vascular structures needs to be evaluated, which may account for the increasing mortality rates and therapeutic resistance in this cancer [Citation2]. In addition, the 5-year survival rate of the radical cholecystectomy is extremely low and is not sensitive to radiotherapy or chemotherapy [Citation3]. Therefore, much attention has been dedicated to explore new methods as it is found to be necessary and most interesting topic in recent decades.
Lim Domain And Actin Binding protein1 (lima1, also known as EPLIN or SREBP3) has drawn more attention over the past decades and the regulation of lima1 extends from cytoskeletal dynamics and cell motility to cell division, gene regulation, apical extrusion, angiogenesis, cellular metabolism [Citation4,Citation5]. Previous studies suggested that lima1 is dysregulated in cancers and influence cancer cell proliferation and invasion, increasing the metastatic probability of cancers [Citation6]. Zhang et al first reported lima1 mutation promotes low plasma LDL cholesterol and decreases intestinal cholesterol absorption [Citation7]. In mice, intestine-specific lima1 deficiency causes cholesterol absorption disorder and were resistant to diet-induced hypercholesterolemia, explaining the phenotype of the lima1 variant in humans [Citation7]. Previous studies also have observed that lima1 have impact on cellular aggressiveness, clinical outcome, and that lima1 may participate in the angiogenic process in prostate and ovarian cancer [Citation8,Citation9]. Jiang et al. find that lima1 expression is aberrant and closely related to the clinical outcome of patients with esophageal cancer [Citation10]. Gong et al. find that lima1 appears to be associated with gastric patient sensitivity to neoadjuvant chemotherapy [Citation11]. Some studies also suggested that the loss of lima1 may involve in genomic instability in cancer cells [Citation12]. All these findings indicate that lima1 may have important role in tumorigenesis.
Given that lima1 as a possible therapeutic target as well as a possible regulator of tumor microenvironment affecting the efficacy of immunotherapy in cancers. Moreover, functional role of lima1 in cholangiocarcinoma remains largely unknown. In the present study, we sought to understand expression pattern of lima1 and possible biological role in cholangiocarcinoma. First, lima1 expression is evaluated by microarray datasets and several cholangiocarcinoma specimens with paired para-cancerous tissues. Then, the correlation between lima1 expression and clinical pathological features, cancer associated phenotypes (vascular invasion, neural invasion, lymph node metastasis), tumor size, TNM stages, overall survival (OS) of patients with cholangiocarcinoma were discussed. Correlation between lima1 expression and angiogenesis and ECM related genes, immune infiltration, immunomodulators was analyzed. Further, we simply examined functional role of lima1 in vitro. Finally, we exploited RNA sequencing, TMT-labeled quantitative proteomics and non-targeted metabolomics. These findings may aid for the development of possible therapeutic target as well as progenesis biomarker in cholangiocarcinoma.
Materials and methods
Study population and tissue samples
In this study, 93 samples from patients with pCCA or ICC who underwent surgical resection between January 2015 and December 2021 were analyzed. Paired paraffin‑embedded tumor and para‑tumor tissues were obtained from the Hospital of Traditional Chinese Medicine Affiliated to the Fourth Clinical Medical College of Xinjiang Medical University (Urumqi, China). Para‑tumor tissue was confined to tissue that was ~2 cm from the tumor margin. All patient’s baseline clinical characteristics and parameters were collected. All patients provided written informed consent admitted to the hospital between January 2015 and December 2021. The study was in compliance with the principles outlined in the Declaration of Helsinki Declaration and approved by the institutional review board of Hospital of Traditional Chinese Medicine Affiliated to the Fourth Clinical Medical College of Xinjiang Medical University. Human tumor and para‑tumor tissue samples were immediately snap-frozen in liquid nitrogen and stored at − 80°C.
Protein extraction and western blot analysis
Human tumor and para‑tumor tissues were lysed in RIPA lysis buffer (Beyotime) supplemented with protease and phosphatase inhibitors (complete Mini and Phosstop, Roche) on ice for 2 hours. After centrifugation, protein concentration of each sample was determined using the bicinchoninic acid method (BCA, ThermoFischer).
Briefly, equal amounts of protein homogenates were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), transferred onto nitrocellulose membranes. Membranes were blocked in Blocking Buffer and probed with primary antibody. Following primary antibodies were used lima1(50311, Cell signaling), GAPDH (5174, Cell signaling) β-actin(4967, Cell signaling). Membranes were subsequently incubated with anti-mouse or anti-rabbit secondary antibodies (Cell signaling). Signals were detected by the ECL detection method (ThermoFischer) and quantified by densitometry (image j).
Quantitative reverse transcription PCR (qRT-PCR)
Total RNA was isolated from fresh frozen tissue using Trizol RNA isolation kit (ThermoFischer).1ug of total RNA was transcribed reversely into cDNA according to the manufacturer’s instructions (Vazyme Biotech). First strand cDNA was generated biological triplicates were analyzed using SYBR green (Vazyme Biotech) and a Roche LightCycler 480 (Roche, LC480). Calculations were performed using the 2-ΔΔCT method. Experiment was conducted in triplicate for each sample, and detected gene expression was normalized to that of GAPDH. Lima1 mRNA levels were assessed using primers: lima1 (Forward: GACTCCCAGGTTAAGAGTGAGG Reverse: TTGCAGGTGCCTGAAACTTCT), GAPDH (Forward:GGAGCGAGA TCCCTCCAAAAT, Reverse: GGCTGTTGTCATACTTCTCATGG);
Immunohistochemistry
Tumor and para‑tumor tissues were fixed with formaldehyde and embedded in paraffin. After continuous sectioning, immunohistochemical staining was performed to evaluate the lima1 expression levels. In this study, we subjected a total of 93 pairs of formalin-fixed paraffin-embedded ICC or pCCA tissues to immunohistochemical staining tissue chip. The following antibodies were used in this study (lima1,50311, Cell signaling). The staining results were determined by the frequency of lima1-positive cells in tissues and were classified into four scoring categories as follows: The frequency of positive staining was rated from 0% to 100%:(0%, negative; 1+, 1% to 30%; 2+, 30% to50%; and 3+, >50%). The specimens were evaluated by two experienced researchers blinded to the clinicopathological variables. In statistical analysis, samples were then split into high expression group and low expression group according to the immunohistochemistry score. Scores with 0 and 1+ were categorized as ‘low expression’; Scores with 2+ and 3+were categorized as ‘high expression’.
Cell culture and, stable cell line and transfection
Human cholangiocarcinoma cell line RBE were obtained from the cell bank of Shanghai Academy of Chinese Sciences and cultured in containing RPMI1640 (ThermoFischer) medium containing 10% fetal bovine serum (Wisent), 100 U per ml penicillin (Wisent) and 100 µg per ml streptomycin (Wisent). The cells were cultured on cell culture plastics and placed in a 37°C, 5% CO2 incubator (ThermoFischer). Lentiviral infection: lenti-CRISPRV2 plasmid were acquired from Shanghai Academy of Chinese Sciences. Two targeting lima1 lentiCRISPR cas-9 plasmids were constructed, and the sequence of sgRNA was as follows: sgRNA-1: Forward, ACGGGAGTCATCTTCGGCAGGGG, sgRNA-2 Forward, AAGACGATCT TCTTCACTGAAGG).
We made stable lima1 KO RBE cell line with a lentivirus using 3rd generation packaging systems. Then protein was extracted from lima1 KO RBE cells for western blot verification and the highest efficiency was screened for subsequent experiments. For siRNA transfection, chemically synthetic siRNAs for Lima1 and their control siRNAs were purchased from Gene Pharma and transferred into RBE cells using Lipofectamine 3000 (ThermoFisher) following the manufacturer’s protocol.
Cell Counting Kit-8 assay
The cell proliferation assay was analyzed using a Cell Counting Kit-8 (CCK-8, Beyotime) following the manufacturer’s protocol. RBE cells were seeded in 96-well plates and the optical density at 450 nm wavelength (OD450) was measured every 24 h for 5 days.
Edu cell proliferation
Edu cell proliferation staining was analyzed using EdU Cell Proliferation Kit (EdU Cell Proliferation Kit with Alexa Fluor 555, epizyme) following the manufacturer’s protocol. Briefly, RBE cells were seeded in 12-well plates and cultured for 48 h. Subsequently, cells were incubated with EdU for 2 h, fixed with 4% paraformaldehyde for 10 min, and permeated with 0.3% Triton X-100 for another 10 min. The cells were incubated with the Click Reaction Mixture for 30 min and Hoechst for 10 min at room temperature, and then viewed using fluorescence microscope.
Cell invasion assay
Cell invasion was performed using Matrigel invasion chambers (CORNING 354,480) according to the manufacturer’s protocol. Briefly, cells were washed with PBS and then were resuspended in culture medium. Equal volumes of the cell suspensions were seeded in the Matrigel invasion chambers. After a 24 h incubation, cells were stained and analyzed by counting the cells from three microscopic fields.
Measurement of caspase-3 activity
Caspase-3 activity in RBE cell was measured using of caspase-3 activity kit (Solarbio, Beijing, China). Control and Lima1 KO cells were planted into the 96-well plates for indicated times and then culture with the caspase-3 substrate with reaction buffer for 4 h. The microplate reader was applied to detect the mixture at 405 nm.
Flow cytometry for apoptosis
Annexin V-FITC cell apoptosis detection kit (Beyotime, C1062S) used for detection, the experimental steps were carried out according to the manufacturer’s instructions, and the results were analyzed by flow cytometry and GraphPad Prism 9.
Cell titer assay
The CellTiter-Glo® Luminescent Cell Viability Assay (G7570, Promega) is used to evaluate cell survival in Control and Lima1 KO cells. The experimental steps were carried out according to the manufacturer’s instructions, and the results were analyzed by flow cytometry and GraphPad Prism 9.
Multi-omics data analysis
We exploited RNA sequencing, TMT labeling quantitative proteomics and non-targeted metabolomics to compare the changes in RBE cells after Lima1 RNA interference with siRNA. RBE cells transfected with scramble or Lima1-targeting siRNA and extracted total RNA with TRIzol respectively. Then RNA quality was tested and quantified. Paired-end RNA-seq sequencing library was sequenced by Shanghai Yueda Technology Co., Ltd.W. RNA-seq data analysis for cell samples was performed and using DAVID database and GraphPad Prism 9 were used to analyze functional annotation clustering (https://david.ncifcrf.gov). Supplementary data contains the full RNA-seq dataset. Genes with absolute log2[fold change]≥2; adjusted p < 0.05 defined as statistically significant.
RBE cells transfected with scramble or Lima1-targeting siRNA and performed TMT labeling quantitative proteomics. Total of 6232 proteins were identified. Differentially expressed proteins screened based on the expression fold change of 1.2 times or more (upregulation greater than 1.2 times or downregulation less than 0.83 times) and p-value <.05. Proteomics and analysis were performed by Shanghai Yueda Technology Co., Ltd.W. DAVID database, Venn diagrams (https://bioinformatics.psb.ugent.be/webtools/Venn/) and GraphPad Prism 9 were also used to analyze functional annotation clustering, co-expression analysis.
HILIC UHPLC-Q-EXACTIVE MS technology was used to analyze the metabolic profile changes with scramble or Lima1-targeting siRNA samples. non-targeted metabolomics and analysis were performed by Shanghai Yueda Technology Co., Ltd.W.
Bioinformatic analysis
Data for levels of Cholangiocarcinoma expression were obtained from (http://GEPIA2.cancer-pku.cn/index. html), Gene Expression Omnibus (GEO) with accession number GSE26566, Tumor IMmune Estimation Resource (TIMER) and an integrated repository portal for tumor-immune system interactions (TISIDB) database. We used GraphPad Prism 9 software to analyze the results. Calculated absolute logFC (fold-change) ≥1.0 was used standard and p value of < 0.05 was defined as statistically significant.
Statistical analysis
All statistical analysis was conducted using SPSS version26.0 (IBM). Values were presented as proportions for categorical variables. Student’s t-test (normally distributed) or Mann – Whitney test (non-normally distributed) was used for continuous variables, and χ2 test was utilized for categorical variables. The associations between the continuous variables were assessed using a Spearman rank correlation test. p < 0.05 was considered statistically significant, and all statistical tests were two-sided.
Results
Lima1 expression is upregulated in cholangiocarcinoma
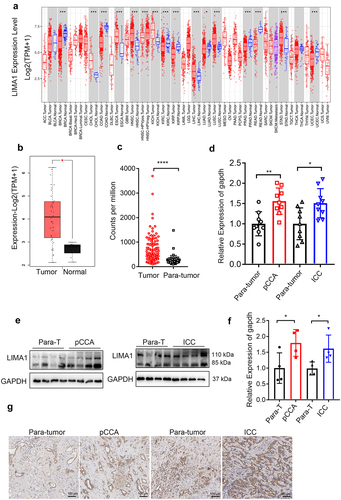
We examined gene expression profiles of lima1 in different tumor samples and paired normal tissues by analyzing the GEPIA2 database (). Further, we evaluate mRNA expression of lima1 in patients with cholangiocarcinoma using GEO database (). Based on the results from database, we compared nine samples of pCCA, nine samples of ICC tissues with nine samples of para-tumor tissues, and our results showed that expression of lima1 is significantly upregulated in cholangiocarcinoma (). Western blot and immunohistochemical analysis also indicated that a 1.8-fold increase in lima1 protein levels in cholangiocarcinoma as compared with the para-tumors (). Taken together, these results indicated that lima1 is upregulated in cholangiocarcinoma.
Figure 1. Lima1 expression is upregulated in cholangiocarcinoma.
Correlation analysis of lima1 expression with survival outcomes in cholangiocarcinoma
To assess the relationship between lima1 expression and cancer tumorigenesis, we used tissue microarray to analyze lima1 expression in cholangiocarcinoma specimens by immunohistochemical staining. A paraffin-embedded tissue array containing 93 paired tumor and para-tumor specimens were analyzed. Patients’ clinical features are presented in . Results from immunohistochemistry staining analysis revealed that lima1 level displayed low and high expression pattern, and the high level of lima1 is significantly associated with vascular invasion in pCCA and ICC (, p = 0.029 and p = 0.016).In addition, there were no significant associations between lima1 expression with age, sex, histological type, tumor size,cancer stage and neural invasion or lymphatic invasion (, p > 0.05).Taken together, this evidence indicated that lima1 expression and activation was closely related to vascular invasion in cholangiocarcinoma.
Table 1. Baseline characteristics of patients with cholangiocarcinoma.
Table 2. Association between lima1 expression and the clinicopathologic variables of perihilar cholangiocarcinoma patients.
Table 3. association between lima1 expression and the clinicopathologic variables of intrahepatic cholangiocarcinoma patients.
Correlation analysis of lima1 expression with survival outcomes in cholangiocarcinoma
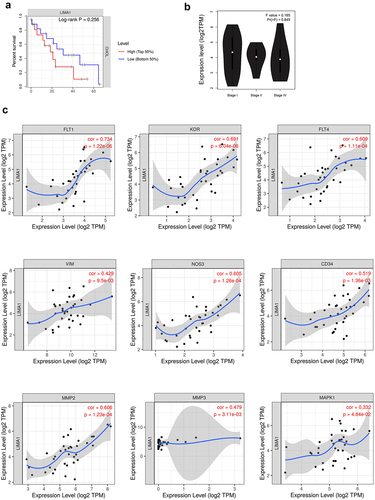
To assess the prognostic value of lima1 expression, the data of 36 patients with cholangiocarcinoma were analyzed from GEPIA2 and TIMER database. Based on the patient’s gene expression profile data and optimal threshold of overall survival, they were separated into low and high lima1 expression groups. Kaplan-Meier plotter was used to investigate the overall survival outcome. As shown in , elevated expression of lima1 was partly linked to poor overall survival in cholangiocarcinoma, but there were no significant differences between two groups in overall survival outcome when patients were stratified by the expression of lima1 (). Moreover, there were no significant correlation between lima1 and TNM stages (). With regard to vascular invasion, we explore the correlation of lima1 expression with angiogenesis and metastasis related genes by using TIMER database. Our analysis revealed that there was strong positive correlation of lima1 with FLT1 (VEGFR, r = 0.734, p = 1.22e-06), KDR (VEGFR2, r = 0.691, p = 6.04e-06), FLT4 (VEGFR3, r = 609, p = 1.11e-04), NOS3 (r = 0.605, p = 1.26e-04), and CD34 (r = 0.519, p = 1.36e-03). Lima1 expression was also positive correlation with expression of MMP-2 (r = 0.606, p = 1.23e-04), MMP-3 (r = 0.479, p = 3.11e-03), Vimentin (r = 0.429, p = 9.5e-03) and MAPK1 (r = 0.332, p = 4.84e-02) (). All this evidence indicated that lima1 expression was closely associated with ECM organization, cell migration and may be participated tumor progression.
Figure 2. Correlation analysis of lima1 expression with survival outcomes in cholangiocarcinoma.
Correlation between lima1 expression and immune infiltration, immunomodulators and chemokines in cholangiocarcinoma
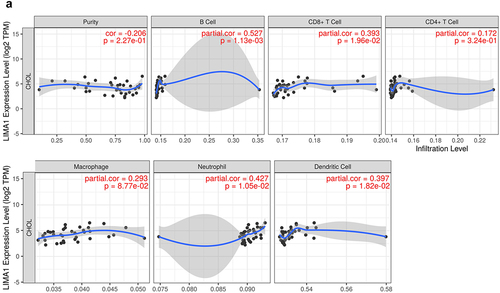
Next, we systematically depicted the relationship between lima1 and a various type of infiltrating immune cells such as B cells, CD8+T cells, CD4+T cells, macrophages, neutrophils and dendritic cells by processing the TIMER database. The results suggested that the expression level of lima1 was positively correlated with the infiltration degree of B cells (r = 0.527, p = 1.13e-03), CD8+ T cells (r = 0.398, p = 1.96e-02), CD4+ T cells (r = 0.172, p = 3.24e-01), macrophages (r = 0.293, p = 8.77e-02), neutrophils (r = 0.427, p = 1.05e-02) and dendritic cells (r = 0.397, p = 1.82e-02) (). In addition, we further assessed the relations between immunomodulators and chemokines (or receptors) and expression of lima1 using the TISIDB database. Lima1 expression with different immunomodulators and chemokines (or receptors) correlation was displayed in Supplementary Table S1 and . There was strong positive correlation with lima1 and a number of immunomodulators and chemokines (or receptors) such as PDCD1LG2 (PD-L2, CD273, from gene PDCD1LG2) (r = 0.512, p = .001), IL2RA (r = 0.502, p = .001), TMEM173 (r = 0.592, p = .0001), CCL8 (r = 0.580, p = .0002), CCL13 (r = 0.632, p = .00005), CCL18 (r = 0.520, p = .001) (Supplementary Table S1 and ). All this explain that lima1 has also important role in immunomodulation which manifested different patterns between normal tissues and cholangiocarcinoma.
Figure 3. Correlation between lima1 expression and immune infiltration in cholangiocarcinoma.
Lima1 KO inhibits RBE cell invasion in vitro
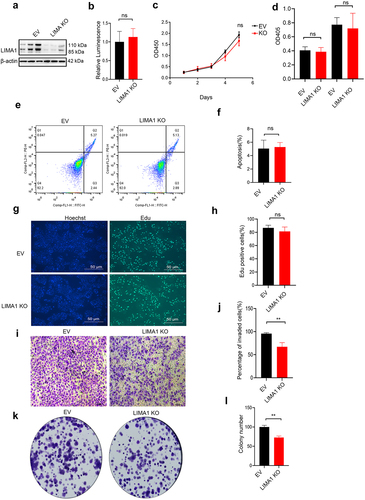
Given the important role of lima1, we further explored its function in vitro. We sought to determine whether Lima1 loss impacted proliferation capacity or survival. Cell Titer assay, CCK-8 and Edu cell proliferation staining were performed to evaluate cell survival and growth. After knocking out (KO) the expression of Lima1 in RBE cells, no changes in cell viability and cell proliferation were observed (). Caspase 3 Assay, Flow Detection analysis was performed to detect cancer cell apoptosis and there are no significant differences between lima1 KO cells relative to control cells (). Further, we determined whether lima1 KO affect the cancer cell invasion ability. As we expected, lima1 KO inhibits the RBE cell invasion and colony formation (). Thus, lima1 may promote cholangiocarcinoma tumorigenesis by affecting cancer cell invasion.
Figure 4. Lima1 KO inhibits RBE cell invasion in vitro.
Analysis of multi-omics data in vitro
Next, we assessed how downregulation of Lima1 in RBE cells impacted the cancer cell invasion. We exploited RNA sequencing to compare the changes at the mRNA level in RBE cells after Lima1 RNA interference with siRNA (). DAVID database was used to analyze functional annotation clustering. RNA Seq analysis identified total of 717 genes were significantly changed after Lima1 RNAi(log2[fold change]≥2; adjusted p < .05;).Further KEGG analysis identified that metabolism, ECM-receptor interaction, Focal adhesion,TGF-β pathway,PI3K-Akt signaling pathway, cell cycle were significantly enriched after Lima1 downregulation (). The mRNA level associated with ECM-receptor interaction, Focal adhesion, TGF-β pathway,(VTN,LAMA4,FN1,ITGA10,GRB2,EMP2,BAMBI,ID3,ID4,NOG,
Figure 5. Analysis of multi-omics data in vitro.
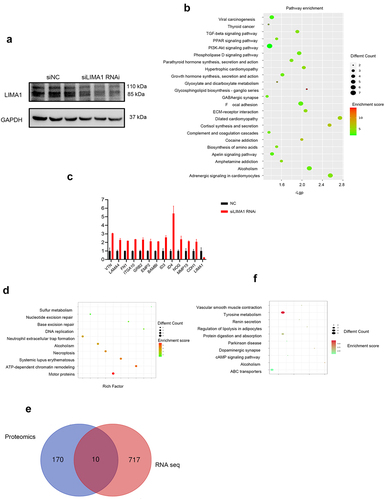
MMP15, CDH1) was assessed (). No change in the expression of apoptosis, stemness and differentiation related genes was detected. Most cell proliferation genes were unaffected, except for PCNA. This gene, a key factor in cell proliferation, DNA replication and cell cycle regulation, showed a significant downregulation in Lima1 RNAi as compared with the Control cells (log2FoldChange: −1.68; p < .0001). TMT-labeled quantitative proteomics of either Lima1 RNAi or Control RBE cells was performed followed by differential expression analysis using DAVID database. 170 protein were significantly changed after Lima1 RNAi(log2[fold change]≥1.2; adjusted p < .05;). GO enrichment and KEGG enrichment analysis showed that these differentially expressed proteins were mainly involved in the regulation of important metabolic pathways such as Motor proteins, ATP-dependent chromatin remodeling, Systemic lupus erythematosus, Necroptosis and Alcoholism (). Overlap analysis was performed to evaluate co-changes at the mRNA and protein levels. We identified that PHF6, HSPD1, CDK1, CHEK1, SUB1, HBS1L PCNA, STAMBP, LAMA4, PPP4R4 are changed synchronously at the mRNA and protein levels and most of them are closely related to the cell cycle as well as cancer cell metastasis (). Furthermore, we performed non-targeted metabolomics and results showed that tyrosine metabolic pathway was significantly affected after Lima1 RNAi (). Multi-omics data suggest that Lima1 may participate in the metabolism, ECM-receptor interaction, Focal adhesion,TGF-β pathway,PI3K-Akt signaling pathway, cell cycle and affect a broad spectrum of cancer related pathways, promoting tumor progression and metastatic ability in cholangiocarcinoma.
Discussion
cholangiocarcinoma is prominently characterized by uncontrolled tumor cell proliferation, lymph node metastasis and robust neovascularization which is not only key mechanism of tumorigenesis and cancer development, but also important factor of poor prognosis [Citation3]. Although great efforts of early diagnosis and optimum anti-cancer therapy have been made in the past decades, there is still lack of effective methods for early diagnosis as well as preferred treatment, resulting in low survival rates for cholangiocarcinoma patients [Citation2]. Rich tumor microenvironment, profound genetic heterogeneity and intricate biological process made the management of cholangiocarcinoma more difficult and challenging. To improve poor outcomes, much attention should take into consideration for early and adequate diagnosis as well as optimum therapies with improvement in patient survival based on the underlying biological mechanisms of tumorigenesis and progression.
Lima1 was initially identified as an actin cross-linking protein that is preferentially expressed in human epithelia and has important role in epithelial cell junctions. A series of previous studies showed that lima1 expression is lost in some cancers, such as lung, gastric, breast, prostate and esophageal cancers, involving in cancer cell migration and invasion by regulating the actin cytoskeleton and membrane ruffling [Citation6,Citation8–10].
There is also growing evidence that lima1 is linked to extracellular signal-regulated kinase (ERK) signaling and epidermal growth factor (EGF) promotes lima1 protein turnover via phosphorylation, ubiquitination and degradation [Citation13]. Tomoko et al. find that lima1is a target of the p53 family and a potential therapeutic target in n osteosarcoma and colon cancers [Citation14]. Ross J et al. showed that lima1level was reduced in clinical prostate cancer and overexpression of lima1 reduced prostate cancer cell growth, migration and invasion indicating lima1 has important role in regulating the aggressive characteristics of prostate cancer cells [Citation8]. Another study has showed that lima1 expression was significantly associated with responsiveness to chemotherapy contributing to overall survival in gastric cancer [Citation11]. Kong et al. showed that lima1 is upregulated in cholangiocarcinoma and may associated with cholangiocarcinoma development, progression or poor prognosis [Citation15]. All of this pave the way for lima1 to be used in diagnosis and treatment biomarker with cancer patients.
In conjunction with the literature and our work, different expression pattern of lima1 was observed in a variety of human tumors including lung, breast, testicular, and bladder cancers, indicating that lima1 may play diverse role in the tumorigenesis various kinds of cancer.
In cholangiocarcinoma, lima1 expression is different from other tumors and role of lima1 remains largely unknown. In the present study, we sought to explore expression pattern of lima1 and possible biological role in cholangiocarcinoma. Lima1 expression level examined using clinical cholangiocarcinoma cancer tissues and our results showed that lima1 is significantly upregulated as compared with para-tumor tissues. It is interesting to observe that the expression pattern and inhibitory effect of lima1 on cholangiocarcinoma is contrast to the findings in prostate, ovarian cancer cell lines [Citation6–8].
Next, we find that lima1 expression is closely related to vascular invasion. Further, TIMER database analysis revealed that there was strong positive correlation of lima1 with angiogenesis and metastasis related genes such as VEGFR1, VEGFR3, NOS3, CD34, MMP-2, MMP-3, Vimentin and MAPK1 [Citation16–19]. Taken together these observations indicated that lima1 may regulate the expression of VEGF, and ECM related genes, promoting tumor progression and metastatic ability in cholangiocarcinoma. Although, longer survival time tendency for patients with low expression of lima1was observed, there is no statistically significant between two groups as is shown in . The small sample size and short follow-up time are probably attributed to the results.
In cholangiocarcinoma, stimulatory and inhibitory factors released by tumor microenvironment result in the formation of tumor-associated blood and lymph vessel [Citation20]. The invasiveness of cholangiocarcinoma is closely related to the formation of a dense network of lymphatic vessels and a reduced number of blood vessels [Citation21,Citation22]. Thus, gaining insights on intervention that slowdown of vascular network generation may shed light on new targets in cholangiocarcinoma. Several studies have also established a potential link between lima1 and the process of angiogenesis [Citation5]. Herein, we demonstrated that lima1 expression was closely related to with angiogenesis and metastasis related genes and may be participated tumor progression. Although, the precise molecular mechanisms by which lima1 involves angiogenesis and invasion remains unknown, our studies provide conclusive evidence that lima1 could be regarded as a potential therapeutic target and biomarker for cholangiocarcinoma.
Next, the correlation of lima1 with tumor immune environment, immune infiltration landscape and different immunomodulators, chemokines (or receptors) were assessed. Lima1 expression is positively correlated with the infiltration of B cells, CD8+ T cells, neutrophils, dendritic cells and also associated with a number of immunomodulators, chemokines (or receptors) such as PDCD1LG2 (PD-L2), IL2RA, TMEM173, CCL8, CCL13, CCL18, CXCL5. These immunomodulators and chemokines (or receptors) increase in many cancers and involved in the recruitment of immune cells, cancer cell migration, epithelial-to-mesenchymal transition (EMT) and chemotherapy resistance, regulating tumor micro-environment (TME) [Citation20,Citation23–29]. Our results indicated that lima1 may regulate or interact with immune cell infiltration and has also important role in immunomodulation which may have pave the way for development of immunotherapeutic strategies for cholangiocarcinoma.
Finally, we have demonstrated that Lima1 KO inhibits cell invasion compared to control cells and has no significant effect on cholangiocarcinoma cancer cell growth or apoptosis. Our in vitro studies suggest that decreased expression of Lima1 may be associated with a better clinical prognosis by reducing cancer cell invasion, consistent with data from GEPIA2 showing improved overall survival outcomes. Additionally, our RNA-seq analysis identified significant enrichment in pathways related to metabolism, ECM-receptor interaction, focal adhesion, TGF-β signaling, PI3K-Akt signaling, and the cell cycle following Lima1 downregulation. No changes were detected in the expression of genes related to apoptosis, stemness, or differentiation. TMT-labeled quantitative proteomics analysis showed that the differentially expressed proteins were mainly involved in regulating important metabolic pathways. Non-targeted metabolomics revealed a significant impact on the tyrosine metabolic pathway after Lima1 RNAi. These multi-omics data suggest that Lima1 may be involved in metabolism, ECM-receptor interaction, focal adhesion, TGF-β signaling, and PI3K-Akt signaling pathways, affecting a broad spectrum of cancer-related pathways and promoting tumor progression and metastatic ability in cholangiocarcinoma.
She et al. found that silencing Lima1 suppresses the EMT process and tumor-associated pathways in vitro. Increased Lima1 expression indicates poor survival, identifying it as a prognostic biomarker in HNSC [Citation30,Citation31]. However, Li et al. found that low Lima1 levels correlate with worse survival for patients with cholangiocarcinoma, which is inconsistent with our results [Citation32]. The discrepancy between previous reports is likely due to the multiple and complex molecular mechanisms of Lima1 in different tumors and the unique features of cholangiocarcinoma. Additionally, our in vitro functional experiments used KO cell lines, while multiple omics studies used RNA interference cells. Differences in knockdown efficiency may lead to some functional and phenotypic differences, indicating Lima1’s widespread impact on cancer.
However, our research is based on small samples, databases, and in vitro experiments combined with multi-omics data. These are insufficient to determine precise mechanisms and conclusions, and further studies are required to explore the exact molecular mechanisms of Lima1.
In summary, we demonstrated that Lima1 expression is significantly upregulated, and high levels of Lima1 correlate with poorer overall survival in cholangiocarcinoma patients. Lima1 is closely related to vascular invasion and shows a strong positive correlation with angiogenesis-related genes and ECM-related pathways. Furthermore, knocking out Lima1 inhibits cholangiocarcinoma cell invasion. Multi-omics data suggest that Lima1 may participate in metabolism, ECM-receptor interaction, focal adhesion, TGF-β signaling, and PI3K-Akt signaling pathways, affecting a broad spectrum of cancer-related pathways and promoting tumor progression and metastatic ability in cholangiocarcinoma. This study provides insights into the molecular associations of Lima1 with cell invasion, ECM-related pathways, and tumor metabolism, establishing a preliminary picture of the correlation network in cholangiocarcinoma.
Authors’ contributions
HO, AA conducted the experiments and statistical analysis, YL collected and organized clinical data, SbD wrote the manuscript.
supplementary file 1.pdf
Download PDF (147.5 KB)sup table 2.docx
Download MS Word (13.7 KB)Sup table 1.docx
Download MS Word (13.7 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Data supporting the findings of this study are available within its supplementary materials.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19336918.2024.2383068
Additional information
Funding
References
- Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet. 2014;383(9935):2168–2179. doi: 10.1016/S0140-6736(13)61903-0
- Khan AS, Dageforde LA. Cholangiocarcinoma. Surg Clin North Am. 2019;99(2):315–335. doi: 10.1016/j.suc.2018.12.004
- Rizvi S, Khan SA, Hallemeier CL, et al. Cholangiocarcinoma — evolving concepts and therapeutic strategies. Nat Rev Clin Oncol. 2018;15(2):95–111. doi: 10.1038/nrclinonc.2017.157
- Zeng J, Jiang WG, Sanders AJ. Epithelial protein lost in neoplasm, EPLIN, the cellular and molecular prospects in cancers. Biomolecules. 2021;11(7):1038. doi: 10.3390/biom11071038
- Liang L, Zhao L, Zan Y, et al. MiR-93-5p enhances growth and angiogenesis capacity of HUVECs by down-regulating EPLIN. Oncotarget. 2017;8(63):107033–107043. doi: 10.18632/oncotarget.22300
- Collins RJ, Jiang WG, Hargest R, et al. EPLIN: a fundamental actin regulator in cancer metastasis? Cancer Metastasis Rev. 2015;34(4):753–764. doi: 10.1007/s10555-015-9595-8
- Zhang YY, Fu ZY, Wei J, et al. A LIMA1 variant promotes low plasma LDL cholesterol and decreases intestinal cholesterol absorption. Sci (New York, NY). 2018;360(6393):1087–1092. doi: 10.1126/science.aao6575
- Collins RJ, Morgan LD, Owen S, et al. Mechanistic insights of epithelial protein lost in neoplasm in prostate cancer metastasis. Int J Cancer. 2018;143(10):2537–2550. doi: 10.1002/ijc.31786
- Liu R, Martin TA, Jordan NJ, et al. Epithelial protein lost in neoplasm-α (EPLIN-α) is a potential prognostic marker for the progression of epithelial ovarian cancer. Int J Oncol. 2016;48(6):2488–2496. doi: 10.3892/ijo.2016.3462
- Liu Y, Sanders AJ, Zhang L, et al. EPLIN-α expression in human oesophageal cancer and its impact on cellular aggressiveness and clinical outcome. Anticancer Res. 2012;32(4):1283–1289.
- Gong W, Zeng J, Ji J, et al. EPLIN expression in gastric cancer and impact on prognosis and Chemoresistance. Biomolecules. 2021;11(4):547. doi: 10.3390/biom11040547
- Wu D. Epithelial protein lost in neoplasm (EPLIN): beyond a tumor suppressor. Genes & Dis. 2017;4(2):100–107. doi: 10.1016/j.gendis.2017.03.002
- Zhang S, Wang X, Iqbal S, et al. Epidermal growth factor promotes protein degradation of epithelial protein lost in neoplasm (EPLIN), a putative metastasis suppressor, during epithelial-mesenchymal transition. J Biol Chem. 2013;288(3):1469–1479. doi: 10.1074/jbc.M112.438341
- Ohashi T, Idogawa M, Sasaki Y, et al. p53 mediates the suppression of cancer cell invasion by inducing LIMA1/EPLIN. Cancer Lett. 2017;390:58–66. doi: 10.1016/j.canlet.2016.12.034
- Kong J, Shen S, Zhang Z, et al. Identification of hub genes and pathways in cholangiocarcinoma by coexpression analysis. cancer biomarkers: section a of disease markers. Cancer Biomarker. 2020;27(4):505–517. doi: 10.3233/CBM-190038
- Nakagawa SA, Lopes A, Lopes de Carvalho A, et al. Nitric oxide synthases, cyclooxygenase-2, nitrotyrosine, and angiogenesis in chondrosarcoma and their relation to prognosis. J Bone Joint Surg Am Volume. 2010;92:1738–1746.
- McColl BK, Stacker SA, Achen MG. Molecular regulation of the VEGF family – inducers of angiogenesis and lymphangiogenesis. APMIS. 2004;112(7–8):463–480. doi: 10.1111/j.1600-0463.2004.apm11207-0807.x
- Li XW, Tuergan M, Abulizi G. Expression of MAPK1 in cervical cancer and effect of MAPK1 gene silencing on epithelial-mesenchymal transition, invasion and metastasis. Asian Pac J Trop Med. 2015;8(11):937–943. doi: 10.1016/j.apjtm.2015.10.004
- Scheau C, Badarau IA, Costache R, et al. The role of matrix metalloproteinases in the epithelial-mesenchymal transition of hepatocellular carcinoma. Anal Cell Pathol (Amst). 2019;2019:9423907. doi: 10.1155/2019/9423907
- Fabris L, Sato K, Alpini G, et al. The tumor microenvironment in cholangiocarcinoma progression. Hepatol (Baltimore, Md). 2021;73(Suppl 1):75–85. doi: 10.1002/hep.31410
- Tang D, Nagano H, Yamamoto H, et al. Angiogenesis in cholangiocellular carcinoma: expression of vascular endothelial growth factor, angiopoietin-1/2, thrombospondin-1 and clinicopathological significance. Oncol Rep. 2006;15:525–532. doi: 10.3892/or.15.3.525
- Adams RH, Alitalo K. Molecular regulation of angiogenesis and lymphangiogenesis. Nat Rev Mol Cell Biol. 2007;8(6):464–478. doi: 10.1038/nrm2183
- Zhao J, Ou B, Han D, et al. Tumor-derived CXCL5 promotes human colorectal cancer metastasis through activation of the ERK/Elk-1/Snail and AKT/GSK3β/β-catenin pathways. Mol Cancer. 2017;16(1):70. doi: 10.1186/s12943-017-0629-4
- Nguyen CH, Schlerka A, Grandits AM, et al. IL2RA promotes aggressiveness and stem cell–related properties of acute myeloid leukemia. Cancer Res. 2020;80(20):4527–4539. doi: 10.1158/0008-5472.CAN-20-0531
- Nagarsheth N, Wicha MS, Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat Rev Immunol. 2017;17(9):559–572. doi: 10.1038/nri.2017.49
- Grippin AJ, Dyson KA, Qdaisat S, et al. Nanoparticles as immunomodulators and translational agents in brain tumors. J Neurooncol. 2021;151(1):29–39. doi: 10.1007/s11060-020-03559-9
- Lavelle EC, McLachlan JB. Editorial overview: immunomodulation: striking the right balance: using immunomodulators to target infectious diseases, cancer, and autoimmunity. Curr Opin Pharmacol. 2018;41:vii–ix. doi: 10.1016/j.coph.2018.07.013
- Mollica Poeta V, Massara M, Capucetti A, et al. Chemokines and chemokine receptors: new targets for cancer immunotherapy. Front Immunol. 2019;10:379. doi: 10.3389/fimmu.2019.00379
- Saeed A, Park R, Al-Jumayli M, et al. Biologics, immunotherapy, and future directions in the treatment of advanced cholangiocarcinoma. Clin Colorectal Cancer. 2019;18:81–90. doi: 10.1016/j.clcc.2019.02.005
- Ma W, Liao Y, Gao Z, et al. Overexpression of LIMA1 indicates poor prognosis and promotes epithelial-mesenchymal transition in head and neck squamous cell carcinoma. Clin Med Insights Oncol. 2022 Jul 8;16:11795549221109493. doi: 10.1177/11795549221109493 PMID: 35837368; PMCID: PMC9274436.
- Huang H, Du Y, Zhao D, et al. The relationship between the prognostic marker LIMA1 in head and neck squamous cell carcinoma and immune infiltration. J Oncol. 2022 Aug 31;2022:1040116. doi: 10.1155/2022/1040116 PMID: 37181789; PMCID: PMC10175016.
- Jiang W, Yang X, Shi K, et al. MAD2 activates IGF1R/PI3K/AKT pathway and promotes cholangiocarcinoma progression by interfering USP44/LIMA1 complex. Oncogene. 2023 Nov;42(45):3344–3357. doi: 10.1038/s41388-023-02849-6 Epub 2023 Sep 26. PMID: 37752233.
