ABSTRACT
Circadian clocks enable organisms to anticipate daily changes in the environment and coordinate temporal rhythms in physiology and behavior with the 24-h day-night cycle. The robust cycling of circadian gene expression is critical for proper timekeeping, and is regulated by transcription factor binding, RNA polymerase II (RNAPII) recruitment and elongation, and post-transcriptional mechanisms. Recently, it has become clear that dynamic alterations in chromatin landscape at the level of histone posttranslational modification and nucleosome density facilitate rhythms in transcription factor recruitment and RNAPII activity, and are essential for progression through activating and repressive phases of circadian transcription. Here, we discuss the characterization of the BRAHMA (BRM) chromatin-remodeling protein in Drosophila in the context of circadian clock regulation. By dissecting its catalytic vs. non-catalytic activities, we propose a model in which the non-catalytic activity of BRM functions to recruit repressive factors to limit the transcriptional output of CLOCK (CLK) during the active phase of circadian transcription, while the primary function of the ATP-dependent catalytic activity is to tune and prevent over-recruitment of negative regulators by increasing nucleosome density. Finally, we divulge ongoing efforts and investigative directions toward a deeper mechanistic understanding of transcriptional regulation of circadian gene expression at the chromatin level.
Introduction
The circadian clock is an endogenous timer that enables organisms from all kingdoms of life to anticipate environmental changes associated with the 24-h rotation of the Earth and maintains daily rhythms in physiology and behavior. The molecular design of the circadian oscillator and the mechanisms that relay the temporal information to output molecular pathways underlying biological rhythms have continued to fascinate researchers and have been studied extensively over the past two decades.Citation1-5 As in other organisms studied to date, the Drosophila circadian oscillator relies on a transcriptional-translational feedback mechanism to maintain endogenous cycling.Citation1,3,6 Two basic helix-loop-helix PER-ARNT-SIM (bHLH-PAS) transcription factors CLOCK (CLK) and CYCLE (CYC) are the key activators of circadian transcription. Within the core oscillator, they form heterodimers and bind to the E-box sequences of period (per) and timeless (tim) in a time-of-day specific manner, initiating transcription of these negative elements in the late day. CLK-activated gene expression subsequently peaks in the early evening. After a time-delay established by post-transcriptional and post-translational mechanisms, which are critical to extend the oscillatory cycle to a full 24 hour, PER and TIM enter the nucleus and inhibit the transcriptional activity of CLK-CYC later in the evening.Citation1,3,6 In a second interlocked feedback loop,Citation7 CLK-CYC heterodimers activate PAR Domain Protein 1ϵ (PDP1ϵ) and VRILLE (VRI), two transcription factors responsible for activating and repressing Clk expression, respectively.Citation8,9 As PDP1ϵ and VRI both bind to D-box elements (also called V/P box) on the clk promoter, the interplay between these two factors, and the delayed expression of PDP1ϵ relative to that of VRI, restrict the timing of prominent clk expression between early to mid-day. Two other transcription factors, clockwork orange (cwo)Citation10-13 and nejire (nej)/ CREB-binding protein (CBP)Citation14,15 have also been observed to modulate CLK-activated transcription and affect the core oscillator function, but their roles still need to be further clarified. Outside of the core oscillator, CLK has been observed to bind to at least 800 loci within the Drosophila genome based on chromatin immunoprecipitation (ChIP)-seq analysis, illustrating the mechanism by which the oscillator relays temporal cues and controls circadian cycling of output genes.Citation16
While the interactions between the key transcription factors of the Drosophila circadian oscillator, namely PER, TIM, CLK, CYC, and their rhythmic association with target circadian gene promoters are well characterized,Citation16-19 much less is known about the regulation at the chromatin level to facilitate daily rhythms in transcription until recently. Using ChIP followed by quantitative PCR (qPCR) and ChIP-seq analysis in Drosophila, mammalian systems, as well as in other clock models, a number of targeted and genome-wide studies have shown that clock-regulated genes undergo extensive daily chromatin remodeling with respect to histone modifications as well as nucleosome condensation.Citation18-22 In Drosophila, daily rhythms of H3K9 acetylation, H3K9 dimethylation, and H3K4 trimethylation are tightly linked to rhythmic per and tim expression.Citation19
It is now widely recognized that modulating genomic accessibility for transcription factor binding and RNAPII activity in a time-dependent manner to enable cycling gene expression involves the collaborative action of multiple regulatory protein complexes. Consequently, much effort has been dedicated to uncovering these histone modifiers and ATP-dependent chromatin remodelers.Citation20-36 For example, in mammals, multiple histone modifiers have been identified to regulate interactions between DNA and key clock transcription factors through posttranslational modifications. Positive regulators that were identified include the methyltransferase MLL1 (Mixed Lineage Leukemia protein 1), which methylates H3K4,Citation25 and the histone demethylase JARID1A (Jumonji/ARID domain-containing protein 1A), which was found to prevent histone deacetylase (HDAC) activity and promote circadian transcriptional activity.Citation26 The search for negative transcriptional regulators has identified SIRT1 (Sirtuin 1), an NAD+-dependent HDAC that counterbalances the putative histone acetyltransferase (HAT) activity of mammalian CLK.Citation27,28 More examples of histone modifiers that act as negative regulators of circadian transcription are discussed below. In contrast, less is known about the role of ATP-dependent chromatin remodelers in circadian transcription, except in the Neurospora clock. Four ATP-dependent chromatin remodelers have now been characterized and have been shown to modulate nucleosome density at different phases of Neurospora circadian transcription, with some promoting the activation phase (CLOCK ATPase (CATP) and SWItch/Sucrose Non-Fermentable (SWI/SNF)) while others are critical to repression (CLOCKSWITCH-1 (CSW-1)).Citation29-32 The role of chromodomain helicase DNA-binding protein 1 (CHD1) is a bit more enigmatic. It is required for remodeling at the frequency (frq) locus, which influences DNA methylation status and consequently affect the phasing of the clock.Citation32 We anticipate the same scenario in animal systems, including in Drosophila, in which different ATP-dependent remodelers function at different phases of circadian transcription to enable smooth transitions between permissive and repressive chromatin landscapes and vice versa to facilitate cycling transcription. So far, identification of these histone modifiers and chromatin remodelers have already led to significant progress and improved our understanding of circadian transcriptional control at the chromatin level.
The next challenge is to understand the mechanisms by which histone modifiers and ATP-dependent remodelers interact with each other as well as with circadian transcription factors to establish time-dependent changes in chromatin landscape, regulate RNAPII activity, and maintain robust cycling gene expression. For example, PER proteins have been found to be in complex with the SIN3A-HDAC1 histone deacetylase, Heterochromatin protein 1 (HP1)-Suv39h histone methyltransferase, and subunits of the Mi-2/nucleosome remodeling and deacetylase (NuRD) corepressor complexes in mouse liver extracts.Citation24,33,34 Knockdown of these histone modifiers and remodeling proteins in mammalian cells expressing a bioluminescence reporter using siRNAs suggests that PER may recruit these protein complexes to circadian promoters to orchestrate rhythmic repression of CLK-BMAL1 (BMAL1 is the mammalian homolog of CYC) activity. In addition, mammalian REV-ERBα is known to interact with the nuclear receptor corepressor 1 (NCOR1)-HDAC3 complex to negatively regulate bmal1 expression.Citation35 Here, we describe our recent identification of the BRAHMA (BRM) chromatin-remodeling complex as a CLK interactor in Drosophila, and discuss its regulation of circadian transcription by limiting CLK activity through both catalytic and non-catalytic mechanisms.Citation36
A chromatin remodeler regulates the Drosophila circadian clock
Using a mass spectrometry (MS) label-free quantitative proteomics approach, we observed that the core subunits of the BRM complex, including BRM, MOIRA, Bap55, BAP60, BAP111, and SNR1, interact with CLK in the nucleus of Drosophila Schneider (S2) cells (). Protein lysate was extracted from stable Drosophila S2 cell lines expressing N- or C-terminally 3XFLAG-tagged CLK. This was followed by subcellular fractionation and affinity purification using FLAG resins prior to MS analysis to identify native CLK-containing complexes in the nucleus. Raw MS data were first searched against the Uniprot database for D. melanogaster proteins followed by SAINT (Significance Analysis of Interactome) to statistically distinguish true interactors from background interactors. A stable cell line that expresses an empty vector was used for background deduction using a False Discovery Rate (FDR) of <0 .002.Citation37
Table 1. Subunits of the BRM chromatin-remodeling complex interact with CLK in the nucleus of Drosophila Schneider (S2) cells as detected by FLAG affinity purification followed by mass spectrometry.
The BRM complex in Drosophila is part of the SWI/SNF chromatin-remodeling family with members in both prokaryotes and eukaryotes.Citation38 These complexes have roles in the repositioning, reconfiguring, and ejecting of nucleosomes, and have been proposed to be key regulators of the expression of many genes.Citation38-42 Knocking down the BRM complex in mammalian cells resulted in both upregulation as well as downregulation of genes as assayed by ChIP-seq, supporting the notion that BRM functions as either transcriptional activator or repressor in a gene-specific manner.Citation40 The BRM complex is named for its catalytic subunit, the BRM protein, which contains an ATPase domain to enable the hydrolysis of ATP for mobilization of nucleosomes. This catalytic activity can facilitate or restrict access of protein regulators to the chromatin. The BRM complex is also known to modulate the function of RNA polymerase II (RNAPII)Citation39 as more condensed chromatin likely impedes the progression of the transcriptional machinery. Finally and not surprisingly, since splicing efficiency is also known to be associated with the chromatin landscape, the BRM complex has been implicated in pre-mRNA splicing regulation of developmental genes.Citation41,42
Following the identification of BRM and associated proteins as CLK interactors, we confirmed and characterized the role of BRM in clock regulation using two classes of genetic mutants: transgenic flies expressing brm double-stranded RNA (dsRNA) in tim-expressing clock neurons (), and transgenic flies expressing a catalytic-inactive brmK804R variant, also in tim-expressing clock neurons ().Citation36 This mutation produces BRMK804R with a lysine to arginine substitution in its ATPase site.Citation43 We expect that both catalytic activity (i.e. ability to condense or decondense chromatin) and non-catalytic activity (i.e., ability to bind protein interactors) of BRM would be reduced in the brm RNAi mutants, whereas only the catalytic activity would be abolished in the brmK804R mutants. We therefore hypothesized that characterizing both brm RNAi and brmK804R mutants in comparison to control flies () will allow us to differentiate between the catalytic vs. non-catalytic role of the BRM complex in regulating circadian gene expression. Our results are summarized in .
Figure 1. The role of BRM in Drosophila circadian transcription. (A) Flies in which brm RNAi is used to knock down brm expression are expected to exhibit both decrease in catalytic and non-catalytic activity. Decrease in the latter is postulated to result in decreased recruitment of BRM-interacting proteins to CLK-regulated promoter regions. Interacting proteins are designated as Px, Py, and Pz. Reduction in catalytic activity results in decrease in nucleosome density. (B) Flies expressing the brmK804R allele express a catalytically-inactive BRM protein that is unable to hydrolyze ATP to remodel chromatin (increase nucleosome density), yet retains interactions with other proteins.Citation43 (C) Control “wild type” flies expressing native BRM protein with intact catalytic and non-catalytic activity. (D) An overview of findings from Kwok et al,Citation36 including the phenotypes of the two brm mutants with respect to circadian locomotor activity rhythm, expression of core clock genes, nucleosome density, and CLK localization at the per promoter.
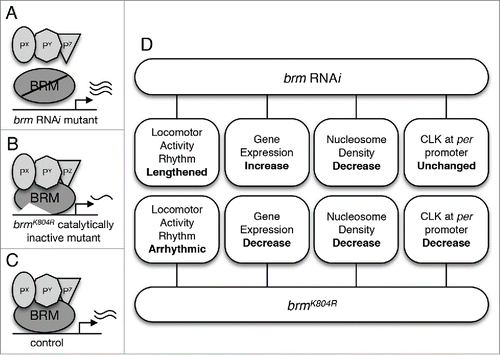
First, we verified the functionality of the BRM complex in regulating the circadian oscillator by examining the locomotor activity rhythms of BRM complex mutants using Drosophila Activity Monitoring System (DAMS).Citation44 Impaired clock function can be detected as changes in clock-controlled free-running rhythms, i.e. activity rhythms that are only driven by the endogenous clock when flies are held in constant darkness, as compared to wild type control. Knockdown of brm expression via RNAi resulted in the lengthening of circadian period (). Interestingly, independent knockdown of other subunits of the BRM complex also result in similar period-lengthening phenotype, suggesting that the whole protein complex is required for normal circadian clock function.Citation36 On the other hand, flies expressing brmK804R in clock neurons exhibited lengthened rhythms upon transition to free-running condition, but their phenotype quickly deteriorated to arrhythmicity (). The more severe phenotype in the catalytic-inactive brmK804R mutant cannot be explained easily, but it is possible that increasing the level of brm knockdown in the RNAi mutant (i.e., decrease in functional BRM protein) could lead to locomotor activity rhythm defect that is more in line with the brmK804R mutant. The observed brm RNAi knockdown in fly heads in our experiments was 40% on average.Citation36 Nevertheless, activity rhythm phenotypes of the two brm mutants support an important role of BRM in clock regulation. This is further supported by our observation that BRM binds to per and tim promoters as assayed by ChIP-qPCR.Citation36 Although we only investigated binding at per and tim promoters, it is likely BRM also binds to other CLK-activated target genes. The extent of the role of BRM in regulating circadian transcription will need to be confirmed by comprehensive ChIP-seq analysis.
The most surprising finding of our study was perhaps the fact that the two classes of brm mutants we examined exhibited opposing effects on circadian gene expression, with brm RNAi displaying increased clock gene expression compared to control and brmK804R showing downregulation of clock gene expression (). Since both classes of brm mutants were predicted to show a decrease in catalytic activity and exhibited a reduction in nucleosome density at per and tim loci as assayed by histone H3 ChIP-qPCR, our results suggested that the BRM complex normally functions to condense the chromatin at clock gene loci. Moreover, our results strongly suggested that it is likely the intact non-catalytic activity of BRMK804R that led to the differential effects exhibited by the two classes of brm mutants with respect to clock gene expression. We postulate that when brm is knocked down via RNAi, the chromatin is more open due to a loss of nucleosome remodeling catalytic activity, allowing for an aberrant elevation in gene expression that results partly from an increased recruitment of general transcription machinery (). While this explains the increase in clock gene expression and apparent decrease in RNAPII stalling in flies expressing brm RNAi, the opposite effect in clock gene expression observed in flies expressing the catalytically-inactive brmK804R remains puzzling, especially since the chromatin is also more relaxed in brmK804R flies as compared to wild type (). Fortunately, investigation of changes in CLK binding to the per and tim promoters promptly provided us with valuable clues to indicate that the non-catalytic activity retained by BRMK804R, which is knocked down in brm RNAi mutant, is the driving force for decreased clock gene expression in flies expressing brmK804R. ChIP-qPCR revealed that CLK binding to per and tim promoters in brmK804R mutants is lower compared to that of control flies, despite the more open chromatin in these mutants (). This suggests that the catalytically-inactive BRMK804R protein can influence CLK occupancy independent of ATP-dependent modifications in chromatin structure, perhaps through recruitment of repressive factors or factors that negatively impact CLK levels.
Figure 2. The effects of two classes of brm mutants on CLK-activated transcription. (A) Knockdown of brm via RNAi results in the reduction of both catalytic and non-catalytic functions. The decrease in catalytic activity leads to lower nucleosome density as compared to control, and decrease in non-catalytic function impairs recruitment of repressive complexes. As a result, CLK-activated gene expression is elevated. (B) Expression of catalytically-inactive BRMK804R protein results in decreased nucleosome density. The more open chromatin allows for an augmented effect of the non-catalytic function retained by BRMK804R, which may include recruitment of repressors. Experimental data suggests that interactors of BRM have a negative effect on CLK binding to the per promoter, and per expression is downregulated as a result of decreased CLK binding.Citation36 (C) Wild type BRM possesses both non-catalytic and catalytic functions that balance each other and serve to fine-tune CLK-activated transcription. Non-catalytic function of BRM may promote recruitment of proteins that have repressive effects on transcription. The over-recruitment of repressive factors is prevented by the catalytic function of BRM to maintain nucleosome density.
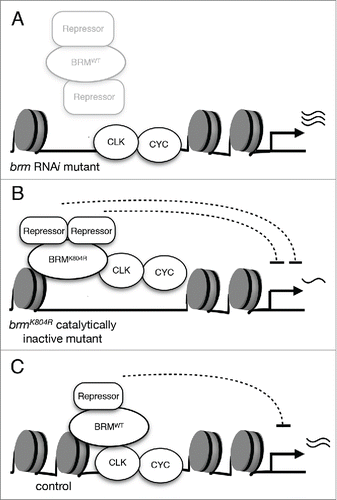
In light of this observation, along with the contrasting effects on circadian gene expression observed in brm RNAi versus brmK804Rmutants, we formulated a model that could explain the molecular phenotypes observed in the two classes of brm mutants and describe the catalytic and non-catalytic function of BRM (). We propose that the BRM complex recruits repressive complexes to negatively regulate CLK binding and/or restrain CLK activity during the active phase of clock gene transcription, and that over-accumulation of these repressive complexes is prevented by its ATP-dependent remodeling activity that serves to increase nucleosome density, thus counterbalancing and fine-tunes its non-catalytic function. As a result, in transgenic flies expressing BRMK804R, the loss of catalytic activity to maintain or increase nucleosome density leads to the over-recruitment of repressive factors (). The end result is a decrease in clock gene expression in brmK804R mutants. The initial period-lengthening phenotype of brmK804R mutants that quickly deteriorates into arrhythmicity in free-running conditions could potentially be explained by the fact that rhythmic circadian gene expression is highly dependent on rhythmic CLK binding to per loci, which is significantly diminished in this mutant.Citation36 Moreover, it is plausible to predict that the brmK804R mutation may impact output molecular pathways affecting activity rhythms, which could also contribute to the observed behavioral phenotype. In flies expressing brm RNAi, knockdown of brm expression results in the decrease of both catalytic and non-catalytic functions, leading to a twofold relief of repression when compared to the brmK804R mutant (). The reduction in non-catalytic activity leads to the decrease in repressive complexes that normally localize to circadian loci through interactions with BRM, and the loss of catalytic activity results in a decrease in nucleosome density, which may allow for potential increase of factors that have an activating effect on gene transcription. This leads to an outcome of increased circadian gene expression. Specifically, per mRNA expression was shown to be elevated in flies expressing brm RNAi (). The consequent increase in PER protein levels could explain the period-lengthening effect of brm RNAi on circadian activity rhythms,Citation36 as previous observations of increased or more stable PER proteins in other circadian mutants also led to period-lengthening phenotypes.Citation45-47
Emergent non-catalytic role of chromatin remodelers
One of the most illuminating findings in Kwok et al. was that the non-catalytic activity of BRM plays a critical role in regulating circadian transcription independent of ATP-dependent catalytic activity, possibly by recruiting repressive factors.Citation36 While the role of SWI/SNF complexes and other chromatin remodelers as transcriptional regulators has come to light in recent years, many studies focused on characterizing the role of their catalytic activity in regulating gene expression by modulating nucleosome density.Citation39,48,49 These studies largely utilized gene knockdown as the sole method for impairing the function of the BRM complex. Only a few studies have investigated its non-catalytic role in gene regulation. For instance, by using a dominant negative mutation in an evolutionarily conserved core BRM complex subunit SNR1, Marenda et al. showed that the interaction of SNR1 with wing-specific repressor NET and HDACs represses rhomboid expression and wing vein development.Citation50 Similarly to our findings, these repressive functions are independent of ATP-dependent catalytic activity. In addition, lysine-specific demethylase 1 (LSD1) has been found to bind and function as a corepressor of SNR1 during Drosophila wing development.Citation51 Such findings provide strong support that BRM has non-catalytic functions to repress gene expression through association with repressive factors.
Another relevant example that illustrates the importance of non-catalytic activity of chromatin remodelers is the recruitment of a repressive complex by the NuRD complex to regulate the mammalian clock.Citation24 The NuRD complex is an ATP-dependent chromatin remodeler that has been shown to repress gene expression in numerous cancer cell types.Citation52,53 Multiple subunits of the NuRD complex, including chromodomain helicase DNA binding protein 4 (CHD4), metastasis-associated protein 2 (MTA2), methyl-CpG-binding domain protein 2 (MBD2), GATAD2a, HDAC1, and RbAp48 have been observed to coimmunoprecipitate with PER2 from mouse liver extracts.Citation24 The catalytic subunit CHD4 is found to bind to CLK-BMAL1 during the active phase of circadian transcription and increase transcriptional activity by recruiting RNAPII. Whereas CHD4 and MTA2 were observed to interact with CLK-BMAL1 during both the activation and repression phases of circadian transcription independent of PER2, other subunits of the NuRD complex that have roles in conferring repression (HDAC1, MBD2, GATAD2a, RbAp48) are exclusively localized with PER2 until the repression phase. The split NuRD complex is therefore only fully assembled when PER2 binds to CLK-BMAL1, thereby initiating full repression. The NuRD complex is also a candidate repressor that may interact with BRM in the regulation of circadian clock genes, since subunits of SWI/SNF complexes have been shown to interact with the NuRD complex.Citation24,54
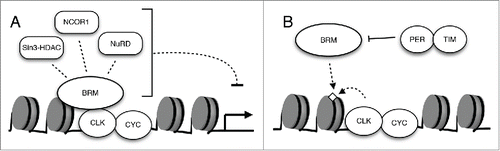
Additional proteomic and genetic analysis will be necessary to fully uncover repressive factors that interact with BRM to modulate CLK binding and activity. One class of histone modifying enzymes that are likely candidates are HDACs, as many HDACs have been found to be under circadian control as well as having direct interactions with clock proteins.Citation33,35,55,56 The repressive Sin3-HDAC complex, which is an evolutionarily conserved protein complex that includes HDAC1 and HDAC2,Citation55 has been found to coprecipitate with mammalian PER complexes and aid in the repression of circadian transcription.Citation33 BRG1 (mammalian homolog of Drosophila BRM protein) and other components of the SWI/SNF complex have also been found to interact with components of Sin3-HDAC,Citation57-59 pointing to the likelihood that BRM can directly associate with proteins involved in histone deacetylation and repression of clock genes. In addition to interacting with HDAC1- and HDAC2-containing complexes, BRG1 and other BRM complex related proteins have been observed to associate with corepressor complex NCOR1, which contains HDAC3.Citation60 This particular corepressor complex has been shown to repress gene expression in the mammalian circadian clock.Citation35,56 Taken together, there is ample evidence that the BRM complex can interact with repressive factors. This supports our model in which BRM limits CLK target gene output in part through non-catalytic interactions with repressive factors (). It is noteworthy that while our results suggest that BRM has a repressive role in Drosophila clock, recent investigation into the role of a Neurospora SWI/SNF chromatin remodeler revealed that it promotes transcriptional activation of the clock gene frequency.Citation30 While the non-catalytic effects were not investigated in the Neurospora study, varying effects of SWI/SNF complexes may be partly attributed to interaction with protein complexes that have activating and/or repressive functions through the non-catalytic activity of these remodelers. To extend this idea a little further, is it possible that the primary function of the catalytic activity of these chromatin remodelers is to tune the effects exerted by their non-catalytic activity to recruit positive or negative regulators, similar to a rheostat?
Figure 3. The interactions of BRM with additional factors regulate its recruitment to promoter regions and modulate circadian transcription. (A) The regulation of circadian gene expression in Drosophila is likely orchestrated through the interaction of a chromatin remodeler such as BRM with multiple epigenetic modifiers. Examples of candidate BRM-interacting repressors are depicted. (B) The possible role of CLK as well as PER and TIM in the recruitment and localization of BRM to the per promoter.
The recruitment of BRM to CLK-activated promoters
In addition to identifying potential repressive factors that are recruited by BRM to regulate the circadian transcriptome, studies to understand the manner in which BRM itself is recruited to clock genes will also provide insights into the mechanisms by which BRM regulates Drosophila clock function. Although our published results did not reveal significant rhythmic BRM recruitment over the circadian cycle, the lack of rhythmic BRM binding to CLK-activated promoters could be attributed to the use of flies expressing epitope-tagged BRM under the control of the tim-GAL4 driver for ChIP analysis. We anticipate to revisit this question as ChIP analysis with a BRM antibody will likely provide a better representation of native temporal pattern of BRM recruitment. A ChIP quality BRM antibody is currently being generated.
In the mean time, the lack of a ChIP quality BRM antibody does not prevent the investigation of whether the key clock transcription factors may be involved in recruiting BRM to circadian promoters (). We have already shown that both CLK and TIM can interact with BRM in Drosophila S2 cells as well as in flies.Citation36 In addition, SWI/SNF complexes have previously been shown to be recruited to target sites through direct interactions with gene-specific transcription factors.Citation61-63 Since the role of BRM in regulating circadian gene expression appears to be repressive in nature, we investigated if BRM is recruited to clock promoters by the negative elements of the Drosophila circadian oscillator, PER and TIM. In the Drosophila clock, a critical event in clock progression is the heterodimerization of these two proteins.Citation1,3 Dimerization with TIM stabilizes PER in the cytoplasm and eventually leads to nuclear translocation, as TIM is responsible for shuttling PER from the cytoplasm into the nucleus upon nighttime to initiate the repression phase of CLK-activated transcription.Citation64 To investigate whether PER and/or TIM are responsible for recruiting BRM to target gene promoters, flies were subjected to constant light (LL) conditions following standard 12hr light: 12hr dark (LD) entrainment to reduce PER and TIM protein levels. Since TIM is targeted for degradation in the presence of light and PER is unstable without TIM, LL conditions should be effective in reducing the levels of both proteins. As predicted, examining PER and TIM protein levels from whole head protein extracts showed only basal levels throughout the day, with the lack of protein accumulation at ZT16 and ZT22, which we otherwise observed in LD conditions (). Mobility shift of PER and TIM due to progressive phosphorylation normally observed in flies housed in LD conditions is also lost in flies held in LL as assayed by western blots. As a separate verification that the LL condition was successful in disrupting the role of PER and TIM in maintaining periodicity of the clock, the behavioral phenotypes of the flies were examined (). Once conditions transitioned from LD to LL, flies exhibited loss of rhythmicity, closely reflecting the loss of PER and TIM protein cycling ().
Figure 4. Disruption in PER and TIM expression using constant light treatment (LL) affects BRM localization at circadian promoters. (A) Verification that circadian rhythms in PER (top panel) and TIM (bottom panel) accumulation and post-translational modifications are abolished in LL conditions. Protein expression patterns in control flies that were entrained for 3 d in 12hr light: 12 hr dark conditions at 25°C are compared to flies subjected to constant light conditions (LL) for one day following 3 d of LD entrainment. Protein was extracted from fly heads as described in Kwok et al.Citation36 and resolved by SDS-PAGE. Antibodies against PER and TIM have been previously described.Citation36 Loading was normalized using anti-HSP70 (Sigma, St. Louis, MO). (B) Locomotor activity rhythms verify loss of rhythmicity in LL conditions. Male flies were subjected to locomotor activity assays using the Drosophila Activity Monitoring System (DAMS) (Trikinetics, Waltham, MA) as described in Chiu et al.Citation44 Flies were entrained for 4 d in 12hr light: 12 hr dark conditions. On the fifth day, LD conditions were shifted +6 h to confirm that flies did not exhibit entrainment defects. Starting the 8th day, flies held in LL conditions and periodicity of the clock was assessed. Shaded areas = lights-off. TUG = tim-(UAS)-Gal4 driver.Citation70 (C) Chromatin immunoprecipitation (ChIP) assay showing BRM localization to the per promoter in LD vs. LL conditions. Transgenic flies expressing BRM-FLAG driven by TUG were harvested at the 4 indicated time points (ZT) in either LD or LL conditions. α-FLAG (Sigma, St. Louis, MO) was used in ChIP-qPCR experiments as described in Kwok et al.Citation36 Data shown are from 3 biological replicates with technical triplicates performed during qPCR. Error bars = SEM for biological replicates (n = 3). Two-tailed t-tests were used to determine statistical differences (P < 0.05) between LD and LL treatments at each ZT. Asterisks denote significant difference observed (P < 0.05). Experimental procedures for ChIP-qPCR are detailed in Kwok et al.Citation36
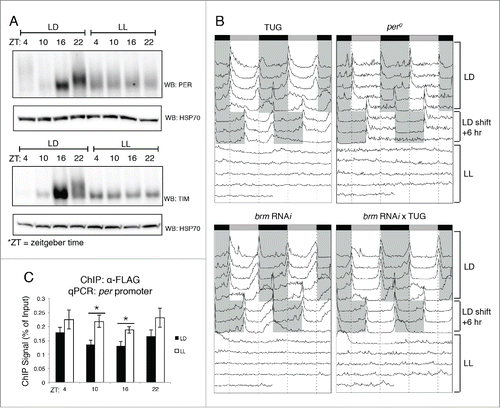
To determine if reduction in PER and TIM affects BRM recruitment to the per promoter, ChIP-qPCR experiments were performed to compare flies entrained in LD and subsequently held in LL conditions and control flies that were kept in LD throughout the experiment. Interestingly, it appears that PER and TIM may be somewhat antagonistic to BRM interaction with the chromatin, as flies exposed to LL conditions exhibited higher levels of BRM recruitment to the per promoter, particularly at ZT10 and ZT16 (). This suggests that PER and TIM may have roles in limiting BRM accumulation at the chromatin, or serve to actively remove BRM. This presents another possible mechanism of fine-tuning the precision of CLK-activated transcription. Future experiments are necessary to further dissect the interactions between BRM and PER:TIM.
Since we showed that BRM binds to CLK in Drosophila S2 cells and in flies,Citation36 it is also possible that CLK itself is responsible for the recruitment of BRM to circadian promoters (). There is evidence that mammalian CLK possesses activity that surpasses transcriptional activation. CLK in mammals has been shown to function as a pioneer transcription factor, with the ability to interact with nucleosome-bound DNA and promote nucleosome removal.Citation65 CLK has also been shown to function as a histone acetyltransferase (HAT) in the mammalian clock.Citation66 Although loss of CLK acetyltransferase activity appears to exert the most significant effect on the acetylation status of BMAL1,Citation67 analysis of CLK activity has also uncovered H3K14 and H3K9 as target sites for acetylation,Citation66 both of which may serve as recognition sites for bromodomain-containing proteins such as BRM ().Citation68 These marks could consequently recruit the BRM complex, which we have shown to limit circadian transcription during peak transcription times through possible interactions with repressive factors. This limiting activity by BRM is perhaps terminated by PER and TIM (suggested earlier), which then continue to promote transcriptional repression via the removal of CLOCK-CYC from E-box regions.Citation18 While it has yet to be investigated whether Drosophila CLK also possesses pioneer transcription factor and HAT activity, its ability to recruit BRM can be examined by assaying BRM binding to circadian promoters using ChIP in clkout flies, a null clk mutant.Citation69
Perspectives
Daily cycling of gene expression is a hallmark of all circadian timekeeping systems studied to date. The circadian clock is not only a fundamentally important life supporting system that commands attention, but the dynamic nature of this oscillating system also offers an excellent opportunity to study the processes that lead to transitions from repressive to permissive chromatin landscapes and vice versa. With its versatile genetic tools and rich community resources, Drosophila will continue to serve as an excellent model to study this complex and multi-layered transcription regulatory system, and help us understand the functions and mechanisms of interactions between cellular protein complexes in the context of cellular oscillation and organismal timekeeping.
In summary, we reviewed our current understanding of the role of the chromatin-remodeling protein BRM in circadian transcription in Drosophila. Identification and characterization of additional ATP-dependent chromatin remodelers in animal clock systems will be necessary to generate a more complete mechanistic description of the roles of nucleosome remodeling in animal clocks. Furthermore, we highlighted future investigative directions that will aid in further elucidating the mechanisms by which BRM interacts with additional factors, including histone modifying enzymes and circadian transcription factors, to dynamically modulate the chromatin landscape and maintain robust cycling gene expression to enable timekeeping.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Andrew Dingwall for providing us with the UAS-brmK804R flies used in Kwok et al.Citation36 Responder lines expressing UAS-RNAi for Brahma subunits were obtained from Vienna Drosophila RNAi Center and Bloomington Drosophila Stock Center at Indiana University. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIGMS or NIH.
Funding
This work is supported by NIH R01GM102225, NSF MCB 1342603, and NSF IOS 1456297 awarded to JCC. RSK is supported by Grant Number T32-GM008799 from NIGMS-NIH.
References
- Allada R, Chung BY. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol 2010; 72:605-24; PMID:20148690; http://dx.doi.org/10.1146/annurev-physiol-021909-135815
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol 2010; 72:517-49; PMID:20148687; http://dx.doi.org/10.1146/annurev-physiol-021909-135821
- Hardin PE, Panda S. Circadian timekeeping and output mechanisms in animals. Curr Opin Neurobiol 2013; 23:724-31; PMID:23731779; http://dx.doi.org/10.1016/j.conb.2013.02.018
- Hsu PY, Harmer SL. Wheels within wheels: the plant circadian system. Trends Plant Sci 2014; 19:240-9; PMID:24373845; http://dx.doi.org/10.1016/j.tplants.2013.11.007
- Baker CL, Loros JJ, Dunlap JC. The circadian clock of Neurospora crassa. FEMS Microbiol Rev 2012; 36:95-110; PMID:21707668; http://dx.doi.org/10.1111/j.1574-6976.2011.00288.x
- Peschel N, Helfrich-Förster C. Setting the clock-by nature: circadian rhythm in the fruitfly Drosophila melanogaster. FEBS Lett 2011; 585:1435-42; PMID:21354415; http://dx.doi.org/10.1016/j.febslet.2011.02.028
- Glossop NP, Lyons LC, Hardin PE. Interlocked feedback loops within the Drosophila circadian oscillator. Science 1999; 286:766-8; PMID:10531060; http://dx.doi.org/10.1126/science.286.5440.766
- Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NR, Hardin PE, Young MW, Storti RV, Blau J. Vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 2003; 112:329-41; PMID:12581523; http://dx.doi.org/10.1016/S0092-8674(03)00074-6
- Glossop NR, Houl JH, Zheng H, Ng FS, Dudek SM, Hardin PE. VRILLE feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron 2003; 37:249-61; PMID:12546820; http://dx.doi.org/10.1016/S0896-6273(03)00002-3
- Lim C, Chung BY, Pitman JL, McGill JJ, Pradhan S, Lee J, Keegan KP, Choe J, Allada R. clockwork orange encodes a transcriptional repressor important for circadian-clock amplitude in Drosophila. Curr Biol 2007; 17:1802-9; http://dx.doi.org/10.1016/j.cub.2007.05.039
- Kadener S, Stoleru D, McDonald M, Nawathean P, Rosbash M. Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev 2007; 21:1675-86; PMID:17578907; http://dx.doi.org/10.1101/gad.1552607
- Matsumoto A, Ukai-Tadenuma M, Yamada RG, Houl J, Uno KD, Kasukawa T, Dauwalder B, Itoh TQ, Takahashi K, Ueda R et al. A functional genomics strategy reveals clockwork orange as a transcriptional regulator in the Drosophila circadian clock. Genes Dev 2007; 21:1687-700; PMID:17578908; http://dx.doi.org/10.1101/gad.1552207
- Richier B, Michard-Vanhee C, Lamouroux A, Papin C, Rouyer F. The clockwork orange Drosophila protein functions as both an activator and a repressor of clock gene expression. J Biol Rhythms 2008; 23:103-16; PMID:18375860; http://dx.doi.org/10.1177/0748730407313817
- Hung HC, Maurer C, Kay SA, Weber F. Circadian transcription depends on limiting amounts of the transcription co-activator nejire/CBP. J Biol Chem 2007; 282:31349-57; PMID:17635913; http://dx.doi.org/10.1074/jbc.M702319200
- Lim C, Lee J, Choi C, Kim J, Doh E, Choe J. Functional role of CREB-binding protein in the circadian clock system of Drosophila melanogaster. Mol Cell Biol 2007; 27:4876-90; PMID:17452464; http://dx.doi.org/10.1128/MCB.02155-06
- Abruzzi KC, Rodriguez J, Menet JS, Desrochers J, Zadina A, Luo W, Tkachev S, Rosbash M. Drosophila CLOCK target gene characterization: implications for circadian tissue-specific gene expression. Genes Dev 2011; 25:2374-2386
- Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev 2006; 20:723-33; PMID:16543224; http://dx.doi.org/10.1101/gad.1404406
- Menet JS, Abruzzi KC, Desrochers J, Rodriguez J, Rosbash M. Dynamic PER repression mechanisms in the Drosophila circadian clock: from on-DNA to off-DNA. Genes Dev 2010; 24:358-67; PMID:20159956; http://dx.doi.org/10.1101/gad.1883910
- Taylor P, Hardin PE. Rhythmic E-Box binding by CLK-CYC controls daily cycles in per and tim transcription and chromatin modifications. Mol Cell Biol 2008; 28:4642-52; PMID:18474612; http://dx.doi.org/10.1128/MCB.01612-07
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 2012; 338:349-54; PMID:22936566; http://dx.doi.org/10.1126/science.1226339
- Barneche F, Malapeira J, Mas P. The impact of chromatin dynamics on plant light responses and circadian clock function. J Exp Bot 2014; 65:2895-913; PMID:24520020; http://dx.doi.org/10.1093/jxb/eru011
- Hatori M, Panda S. Nucleosome dynamics regulate Neurospora circadian clock. EMBO Rep 2013; 14:854-55; PMID:24030278; http://dx.doi.org/10.1038/embor.2013.143
- Aquilar-Arnal L, Sassone-Corsi P. Chromatin landscape and circadian dynamics: Spatial and temporal organization of clock transcription. Proc Natl Acad Sci USA 2012; 122:6863-70
- Kim JY, Kwak PB, Weitz CJ. Specificity in circadian clock feedback from targeted reconstitution of the NuRD corepressor. Mol Cell 2014; 56:738-48; PMID:25453762; http://dx.doi.org/10.1016/j.molcel.2014.10.017
- Katada S, Sassone-Corsi P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat Struct Mol Biol 2010; 17:1414-1421; PMID:21113167; http://dx.doi.org/10.1038/nsmb.1961
- DiTacchio L, Le HD, Vollmers C, Hatori M, Witcher M, Secombe J, Panda S. Histone lysine demethylase JARID1a Activates CLOCK-BMAL1 and influences the circadian clock. Science 2011; 333:1881-1885; PMID:21960634; http://dx.doi.org/10.1126/science.1206022
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 2008; 134:317-328; PMID:18662546; http://dx.doi.org/10.1016/j.cell.2008.06.050
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 2008; 134:329-340; PMID:18662547; http://dx.doi.org/10.1016/j.cell.2008.07.002
- Cha J, Zhou M, Liu Y. CATP is a critical component of the Neurospora circadian clock by regulating the nucleosome occupancy rhythm at the frequency locus. EMBO Rep 2013; 14:923-30; PMID:23958634; http://dx.doi.org/10.1038/embor.2013.131
- Wang B, Kettenbach AN, Gerber SA, Loros JJ, Dunlap JC. Neurospora WC-1 recruits SWI/SNF to remodel frequency and initiate a circadian cycle. PLoS Genet 2014; 10:e1004599; PMID:25254987; http://dx.doi.org/10.1371/journal.pgen.1004599
- Belden WJ, Loros JJ, Dunlap JC. Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Mol Cell 2007; 25:587-600; PMID:17317630; http://dx.doi.org/10.1016/j.molcel.2007.01.010
- Belden WJ, Lewis ZA, Selker EU, Loros JJ, Dunlap JC. CHD1 remodels chromatin and influences transient DNA methylation at the clock gene frequency. PLoS Genet 2011; 7:e1002166; PMID:21811413; http://dx.doi.org/10.1371/journal.pgen.1002166
- Duong HA, Robles MS, Knutti D, Weitz CJ. A molecular mechanism for circadian clock negative feedback. Science 2011; 332:1436-39; PMID:21680841; http://dx.doi.org/10.1126/science.1196766
- Duong HA, Weitz CJ. Temporal orchestration of repressive chromatin modifiers by circadian clock Period complexes. Nat Struct Mol Biol 2014; 21:126-32; PMID:24413057; http://dx.doi.org/10.1038/nsmb.2746
- Yin L, Lazar MA. The orphan nuclear receptor rev-erbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol 2005; 19:1452-9; PMID:15761026; http://dx.doi.org/10.1210/me.2005-0057
- Kwok RS, Li YH, Lei AJ, Chiu JC. The catalytic and non-catalytic functions of the Brahma chromatin-remodeling protein collaborate to fine-tune circadian transcription in Drosophila. PLoS Genet 2015; 11:e1005307; PMID:26132408; http://dx.doi.org/10.1371/journal.pgen.1005307
- Choi H, Larsen B, Lin ZY, Breitkreutz A, Mellacheruvu D, Fermin D, Qin ZS, Tyers M, Gingras AC, Nesvizhskii AI. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat Methods 2011; 8:70-3; PMID:21131968; http://dx.doi.org/10.1038/nmeth.1541
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem 2009; 78:273-304; PMID:19355820; http://dx.doi.org/10.1146/annurev.biochem.77.062706.153223
- Vorobyeva NE, Nikolenko JV, Nabirochkina EN, Krasnov AN, Shidlovskii YV, Georgieva SG. SAYP and Brahma are important for ‘repressive’ and ‘transient’ Pol II pausing. Nucleic Acids Res 2012; 40:7319-31; PMID:22638575; http://dx.doi.org/10.1093/nar/gks472
- Tolstorukov MY, Sansam CG, Lu P, Koellhoffer EC, Helming KC, Alver BH, Tillman EJ, Evans JA, Wilson BG, Park PJ et al. Swi/Snf chromatin remodeling/tumor suppressor complex establishes nucleosome occupancy at target promoters. Proc Natl Acad Sci USA 2013; 110:10165-70; PMID:23723349; http://dx.doi.org/10.1073/pnas.1302209110
- Zraly CB, Dingwall AK. The chromatin remodeling and mRNA splicing functions of the Brahma (SWI/SNF) complex are mediated by the SNR1/SNF5 regulatory subunit. Nucleic Acids Res 2012; 40:5975-87; PMID:22467207; http://dx.doi.org/10.1093/nar/gks288
- Tyagi A, Ryme J, Brodin D, Ostlund Farrants AK, Visa N. SWI/SNF associates with nascent pre-mRNPs and regulates alternative pre-mRNA processing. PLoS Genet 2009; 5:e1000470; PMID:19424417; http://dx.doi.org/10.1371/journal.pgen.1000470
- Elfring LK, Daniel C, Papoulas O, Deuring R, Sarte M, Moseley S, Beek SJ, Waldrip WR, Daubresse G, De Pace A et al. Genetic analysis of brahma: the Drosophila homolog of the yeast chromatin remodeling factor SWI2/SNF2. Genetics 1998; 148:251-265; PMID:9475737
- Chiu JC, Low KH, Pike DH, Yildirim E, Edery I. Assaying locomoter activity to study circadian rhythms and sleep parameters in Drosophila. J Vis Exp 2010; 43:2157; PMID:20972399
- Chiu JC, Vanselow JT, Kramer A, Edery I. The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes Dev 2008; 22:1758-72; PMID:18593878; http://dx.doi.org/10.1101/gad.1682708
- Lamaze A, Lamouroux A, Vias C, Hung HC, Weber F, Rouyer F. The E3 ubiquitin ligase CTRIP controls CLOCK levels and PERIOD oscillations in Drosophila. EMBO Rep 2011; 12:549-557; PMID:21525955; http://dx.doi.org/10.1038/embor.2011.64
- Kim EY, Jeong EH, Park S, Jeong HJ, Edery I, Cho JW. A role for O-GlcNAcylation in setting the circadian clock speed. Genes Dev 2012; 26:490-502; PMID:22327476; http://dx.doi.org/10.1101/gad.182378.111
- Shi J, Zheng M, Ye Y, Li M, Chen X, Hu X, Sun J, Zhang X, Jiang C. Drosophila Brahma complex remodels nucleosome organizations in multiple aspects. Nucleic Acids Res. 2014; 42:9730-9; PMID:25081211; http://dx.doi.org/10.1093/nar/gku717
- Vorobyeva NE, Soshnikova NV, Nikolenko JV, Kuzmina JL, Nabirochkina EN, Georgieva SG, Shidlovskii YV. Transcriptional coactivator SAYP combines chromatin remodeler Brahma and transcription initiation factor TRIID into a single supercomplex. Proc Natl Acad Sci USA 2009; 106:11049-54; PMID:19541607; http://dx.doi.org/10.1073/pnas.0901801106
- Marenda DR, Zraly CB, Dingwall AK. The Drosophila Brahma (SWI/SNF) chromatin remodeling complex exhibits cell-type specific activation and repression functions. Dev Biol 2004; 267:279-93; PMID:15013794; http://dx.doi.org/10.1016/j.ydbio.2003.10.040
- Curtis BJ, Zraly CB, Marenda DR, Dingwall AK. Histone lysine demethylases function as co-repressors of SWI/SNF remodeling activities during Drosophila wing development. Dev Biol 2011; 350:534-47; PMID:21146519; http://dx.doi.org/10.1016/j.ydbio.2010.12.001
- Lai AY, Wade PA. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat Rev Cancer 2011; 11:588-96; PMID:21734722; http://dx.doi.org/10.1038/nrc3091
- Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene 2007; 26:5433-5438; PMID:17694084; http://dx.doi.org/10.1038/sj.onc.1210611
- Singh AP, Archer TK. Analysis of the SWI/SNF chromatin-remodeling complex during early heart development and BAP250a repression cardiac gene transcription during P19 cell differentiation. Nucleic Acids Res 2014; 42:2958-75; PMID:24335282; http://dx.doi.org/10.1093/nar/gkt1232
- Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 1997; 89:357-64; PMID:9150135; http://dx.doi.org/10.1016/S0092-8674(00)80216-0
- Alenghat T, Meyers K, Mullican SE, Leitner K, Adeniji-Adele A, Avila J, Bucan M, Ahima RS, Kaestner KH, Lazar MA. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature 2008; 456:997-1000; PMID:19037247; http://dx.doi.org/10.1038/nature07541
- Sif S, Saurin AJ, Imbalzano AN, Kingston RE. Purification and characterization of mSin3A-containing BRG1 and hBRM chromatin remodeling complexes. Genes Dev 2001; 15:603-18; PMID:11238380; http://dx.doi.org/10.1101/gad.872801
- Kuzmichev A, Zhang Y, Erdjument-Bromage H, Tempst P, Reinberg D. Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1). Mol Cell Biol 2002; 22:835-48; PMID:11784859; http://dx.doi.org/10.1128/MCB.22.3.835-848.2002
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen Receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 2003; 115:751-763; PMID:14675539; http://dx.doi.org/10.1016/S0092-8674(03)00934-6
- Underhill C, Qutob MS, Yee SP, Torchia J. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J Biol Chem 2000; 275:40463-70; PMID:11013263; http://dx.doi.org/10.1074/jbc.M007864200
- Yudkovsky N, Logie C, Hahn S, Peterson CL. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev 1999; 13:2369-2374; PMID:10500094; http://dx.doi.org/10.1101/gad.13.18.2369
- Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 1999; 97:299-311; PMID:10319811; http://dx.doi.org/10.1016/S0092-8674(00)80740-0
- Varga-Weisz P. ATP-dependent chromatin remodeling factors: nucleosome shufflers with many missions. Oncogene 2001; 20:3076-85; PMID:11420723; http://dx.doi.org/10.1038/sj.onc.1204332
- Jang AR, Moravcevic K, Saez L, Young MW, Sehgal A. Drosophila TIM binds importin α1, and acts as an adapter to transport PER to the nucleus. PLoS Genet 2015; 11:e1005205; PMID:25915778; http://dx.doi.org/10.1371/journal.pgen.1004974
- Menet JS, Pescatore S, Rosbash M. CLOCK:BMAL1 is a pioneer-like transcription factor. Genes Dev 2014; 28:8-13; PMID:24395244; http://dx.doi.org/10.1101/gad.228536.113
- Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell 2006; 125:497-508; PMID:16678094; http://dx.doi.org/10.1016/j.cell.2006.03.033
- Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y, Sassone-Corsi P. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature 2007; 450:1086-90; PMID:18075593; http://dx.doi.org/10.1038/nature06394
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature 1999; 399:491-6; PMID:10365964; http://dx.doi.org/10.1038/20974
- Mahesh G, Jeong E, Ng FS, Liu Y, Gunawardhana K, Houl JH, Yildirim E, Amunugama R, Jones R, Allen DL et al. Phosphorylation of the transcription activator CLOCK regulates progression through a ˜24-h feedback loop to influence the circadian period in Drosophila. J Biol Chem 2014; 289:19681-93; PMID:24872414; http://dx.doi.org/10.1074/jbc.M114.568493
- Blau J, Young MW. Cycling vrille expression is required for a functional Drosophila clock. Cell 1999; 99:661-71; PMID:10612401; http://dx.doi.org/10.1016/S0092-8674(00)81554-8
