 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.abstract
Fruit flies are important model organisms for functional testing of candidate genes in multiple disciplines, including the study of human diseases. Here we use a high-throughput locomotor activity assay to test the response on activity behavior of gene disruption in Drosophila melanogaster. The aim was to investigate the impact of disruption of 14 candidate genes for human attention-deficit/hyperactivity disorder (ADHD) on fly behavior. By obtaining a range of correlated measures describing the space of variables for behavioral activity we show, that some mutants display similar phenotypic responses, and furthermore, that the genes disrupted in those mutants had common molecular functions; namely processes related to cGMP activity, cation channels and serotonin receptors. All but one of the candidate genes resulted in aberrant behavioral activity, suggesting involvement of these genes in behavioral activity in fruit flies. Results provide additional support for the investigated genes being risk candidate genes for ADHD in humans.
Introduction
Model organisms are important tools for studying biological processes, and in particular the fruit fly, Drosophila melanogaster, has been used extensively. Within psychiatric genetics, D. melanogaster has shown potential to contribute with knowledge on the molecular basis of human psychiatric disorders; e.g., attention-deficit/hyperactivity disorder (ADHD)Citation1,2 and fragile X syndrome.Citation3,4 Here, we use D. melanogaster to provide further information about the genetic architecture of ADHD.
ADHD is a common psychiatric disorder with a prevalence of 3-6% in childrenCitation5-7 and 2-4% in the adult population.Citation8-10 Characteristics of ADHD patients are persistent patterns of inattention and/or hyperactivity-impulsivity behaviors interfering with daily functioning.Citation11 Evidence suggests that a large proportion of variation in the risk of developing ADHD is due to genetic variation among individuals and the heritability has been estimated to 0.76.Citation12 This has sparked numerous genetic studies of ADHD including hypothesis based candidate gene studies and hypothesis free genome-wide association studies (GWAS) resulting in identification of genes, which might be involved in the disease risk (see references in ). However, until now no genome-wide significant associated loci have been identified and significant findings have shown inconsistent results in replication studies.
Table 1. List of Mi{ET1} genotypes, Flybase ID (FB ID), the affected Drosophila genes and their human orthologs. The DIOPT score indicates the similarity between a human and a Drosophila gene. The DIOPT indicates the number of available tools that support a given orthologous gene-pair relationship. The maximum DIOPT score is 10.Citation36
Previously, D. melanogaster has been used to investigate hyperactivity and attention-like behaviors.Citation1,2,13 Several approaches have been used to assess locomotor activity in these studies. Locomotor activity profiling can be achieved using Drosophila Activity Monitors (TriKinetics, Waltham, MA, USA) counting the number of times individual flies cross an infrared light beam at one or multiple well defined regions of a test tube. Measures of activity can also be achieved using video tracking of individual flies.Citation14 Activity measures obtained from video tracking can provide more detailed information than the beam crossing methods.Citation15 Therefore, video tracking might be advantageous when studying complex phenotypes, i.e., traits related to human psychiatric disorders.
The purpose of the study presented here was to investigate the phenotypic consequences of gene disruption of 14 candidate genes for ADHD in D. melanogaster. Disruption of gene function was achieved using the Minos insertional mutants, Mi{ET1}, in which the transposable Minos element was inserted into a gene thereby disrupting the function of that gene.Citation16 To acquire detailed information of behavioral activity in these mutants we used a high-throughput behavioral activity assay. The assay relies on EthoVision XT (Noldus, Wageningen, The Netherlands) automatic computer tracking of individual animals from which a wide range of different behaviors can be extracted. The multiple correlated activity traits can then be decomposed into several distinct variables describing the space of variables defining behavioral activity in D. melanogaster. Using this setup we investigated the phenotypic consequences of disruption of individual genes within the fruit fly selected based on their orthology to human ADHD candidate genes.
We measured a range of correlated activity traits, and reduced the dimensionality of the data to new composite variables. Results provide evidence that the genes investigated are potential candidate genes for behavioral activity in D. melanogaster, and potential risk genes for ADHD. Three subgroups of mutants were identified based on the behavioral data. Mutants within each phenotypic subgroup turned to be disrupted in genes with similar molecular function; genes related to the messenger molecule cGMP, genes involved in cation channel activity, and serotogenic genes.
Results and Discussion
Using automatic computer tracking several correlated activity traits were obtained. The activity assay consisted of a transparent arena with 6 × 6 individual wells (). The arena was placed in a box illuminated with light from below and the video recordings were obtained from the top of the box (). A total of eight locomotor activity traits were obtained; distance covered in 5 min. (), proportion of time spent in the center of the test arena (), the number of clockwise and counter-clockwise movements in the arena (defined as when the fly had moved 100 degrees in one direction in the arena, ), the number of shifts between active and inactive periods (or vice versa, ), the total time spent active or inactive, and finally, the longest period being inactive or active (). These eight traits were used to create an overall behavioral score for individual flies using Principal Component Analysis (PCA). The results are summarized in (ANOVA tables can be found in Tables S1-S11).
Figure 1. Schematic representation of the experimental setup. (A) Illustration of the setup used to obtain behavioral activity phenotypes of multiple flies. (B) The behavioral arena consisted of 36 individual arenas, each being 6 mm high and 16 mm in diameter. The center zone was defined as 50% of the total area. (C) From the tracking software EthoVision XT (Noldus) individual movement tracks were obtained from which several sub-phenotypes were computed. (D) Directional movement was computed and defined as when a fly had moved 100 degrees consecutively. When reaching a cumulative sum of 100 degrees one directional movement was counted (1 CW) and a new cumulative sum would start. If the fly changed direction before reaching 100 degrees (gray line) no directional movement was recorded, but directional movement in the opposite direction would be recorded (1 CCW). (E) From raw tracking files data of movement behaviors were computed based on a velocity threshold of 0.1 cm/s (dashed horizontal line). In the illustration the number of shifts between the two behavioral states (I: inactive, A: active) was five, and the asterisk indicates the longest periods in the two states. The final behavior was the average time in the two states.
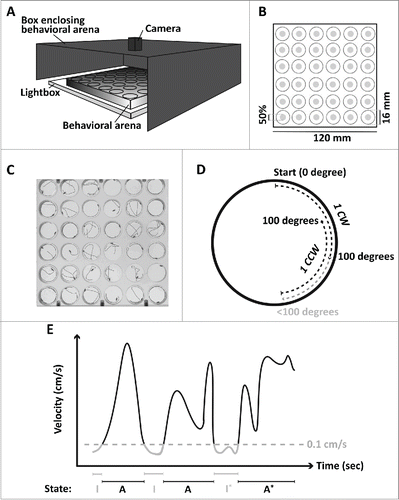
Figure 2. Summary of nine activity-behaviors. All panels show the phenotypic line means with standard errors indicated as error bars for Mi{ET1} mutants (circles) and the w1118 control line (square), sorted by decreasing distance moved. Closed circles indicate a significant difference between Mi{ET1} and w1118 after correcting for multiple testing (ANOVA tables can be found in Table S1). (A) Distance moved in 5 min(cm); (B) proportion of the time spent in the center of the arena (%); (C) Number of movements in clockwise (CW) direction; (D) number of shifts between an active and inactive period (or vice versa); (E) time in seconds (s) in the inactive state; (F) time in seconds (s) in the active state; (G) longest inactive period in seconds (s); (H) longest active period in seconds (s).
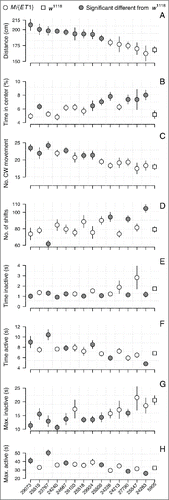
Nine of the 14 Minos mutants moved significantly longer than the control line, w1118 (). Four of the five mutants that covered the same distance as w1118, spent significantly more time in the center of the arena compared to the control (). Six mutants showed a directional movement bias (), i.e., they preferred to move in the clockwise direction in the arena. None of the mutants differed significantly from the control in the counter-clockwise direction trait (Table S4).
Three of the lines that covered the same distance as w1118 had a significantly different movement pattern (), indicated by more shifts between the active and inactive state. Only one of the mutants that moved significantly longer than w1118, namely Mi{ET1}23767, also had significantly fewer movement shifts than the control ().
Eight of the 14 Mi{ET1} mutants had significantly shorter periods of inactivity compared to the control (), and among those, the longest period of inactivity was significantly shorter for seven of the mutants (). The mutants that moved longest tended to be more in the active state than w1118 (), and the maximum time spent active was also significantly longer in these lines ().
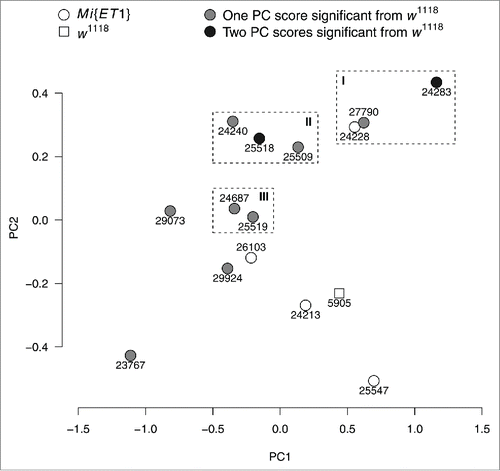
We obtained several correlated traits related to locomotor activity. Therefore, we used the unsupervised learning method, PCA, to summarize the set of variables with a smaller number of representative variables that collectively explain most of the variability in the data. The first and second principal component (PC) accounted for 43.3% and 18.3% of the total variation. Therefore, the scores from the first and second principal component were good behavioral scores encompassing the general behavioral activity for individual flies. Ten of the 14 tested mutants differed significantly from w1118 in the score from principal components 1 or 2 (). Two of the mutants, Mi{ET1}24283 and Mi{ET1}25518, differed significantly in both scores ( and Tables S10-S11).
Figure 3. Principal components (PC) 1 and 2. Each symbol shows the mean score of PC1 and PC2 for Mi{ET1} (circles) and w1118 (square). Symbols filled with light gray indicate that one score was (either the score from PC1 or PC2) significantly different from w1118, and symbols with black indicate that scores from both PC1 and PC2 were significantly different from w1118. The three rectangles indicate mutants with similar scores along both major axes. Lines within each subgroup share molecular functions; I contains genes related to cGMP, II contains genes related to serotonin and serotonin receptors, and III contains genes related to cations.
The main advantage of using video tracking was the level of information acquired. This enabled us to distinguish between the different mutants and to create subphenotypes. Mi{ET1}23767 and Mi{ET1}25519 covered the same distance (), however, Mi{ET1}23767 had significantly fewer shifts between the active and inactive state (). Similarly, Mi{ET1}29073 and Mi{ET1}25519 also covered the same distance in 5minutes (), but Mi{ET1}29073 spent more time being active, and had longer bouts of activity (). Likewise, there was no significant difference in the distance that Mi{ET1}24283 and w1118 covered (), but Mi{ET1}24283 spent significantly more time in the center of the arena (), had significantly more shifts between the active and inactive state (), and the time spent as active was significantly lower ().
All mutants and the control line were indistinguishable in the counter-clockwise movement metric (Table S4), whereas six mutants showed a clockwise movement preference (). Recent studies suggest, that at the population level there is no genetic variation for left or right movement preference when tested in a Y-maze, but at an individual-to-individual level, i.e., intragenotypic, such variation does exist.Citation17,18 The Y-maze assay and our assay cannot be directly compared, however, it was surprising that six mutants showed a clockwise movement bias. This could indicate genetic variation for orientation, which is beyond the scope of this study.
The mutant lines tested in this study were all selected based on being candidate genes for ADHD and high similarity between candidate genes and Drosophila genes (). Thus, it could be hypothesized that the molecular functions of the genes were similar. Based on the PCA, three subgroups of mutants were identified (). The grouping was based on mutants with similar phenotypic score in the two major principal component axes. Consulting the literature showed, that the genes disrupted within the subgroups had a shared molecular function; i.e. that these genes, within grouping, were associated to the same cellular molecular function (see below).
The first group, Group I (), contained mutants with mutagenesis in the genes DIP-θ (Mi{ET1}27790), Pkg21D (Mi{ET1}24228) and Nos (Mi{ET1}24283). Nos codes for a nitric oxide synthase.Citation19 Nitric oxide is known to activate guanylyl cyclaseCitation20 and thereby increases cyclic guanosine monophosphate (cGMP); a cellular messenger. Pkg21D is a gene regulating cGMP-dependent protein kinase activity.Citation21 The function of DIP-θ is currently unknown, however, based on the phenotypic similarity to the other mutants, the function could be related to cGMP as well. The molecular functions of the human orthologs of Pkg21D and Nos, PRKG1 and NOS1, are similar to the fly genes; namely cGMP-dependent kinase and nitric oxide synthase.Citation22 Thus, the genes in group I are related to cGMP. The second group, group II (), contained lines with the Minos insertion in the genes 5-HT1B (Mi{ET1}24240), Alk (Mi{ET1}25509) and Fur1 (Mi{ET1}25518). 5-HT1B is related to serotonin receptor activity.Citation23 The gene Alk is a receptor signaling protein for tyrosine-kinase activity,Citation24 attaching phosphate groups to serine and threonine.Citation25 The minor amino acid, phosphoserine, is a good predictor for levels of pyridoxal-5-phosphate, which is critical for the conversion of tryptophan to serotonin.Citation26 Fur1 is a serine protease.Citation27 The human orthologs in this group are; HTR1B, a serotonin receptor; ALK, a tyrosine-kinase; and FURIN which is a protease.Citation22 Thus, the second group contained lines with genes disrupted in genes related to serotonin receptors and serotonin production. The final group, group III (), contained two genes both related to cations; CG5027 (Mi{ET1}24787) and nAchRα6 (Mi{ET1}25519). Based on prediction (by FlyBaseCitation28) CG5027 is thought to be involved in calcium ion binding, whereas nAchRα6 is involved in an acetylcholine-activated cation-selective channel.Citation29 The human ortholog of nAchRα6, CHRNA7, is likewise a acetylcholine cation channel,Citation22 whereas the ortholog of CG5027, TMX3, is a disulfide isomerase.Citation22 To summarize, using PCA the dimensionality of the data was reduced, and the first two axes explained more than 60% of the variability in the data. Based on this, three phenotypic subgroups, among the 14 mutants, were identified. Overall, the genes disrupted in these groups were involved in cGMP processes, serotonin receptors, and cation channels.
A recent study by van der Voet et al.Citation2 investigated an ADHD-associated dopamine transporter knockdown in D. melanogaster (DAT; corresponds to our Mi{ET1}25547) and found, compared to control flies, an increased activity pattern of this mutant at dark conditions. Such evidence is not directly comparable to our study, because they used conditional knockdown of DAT and a light beam crossing method. No difference in activity was observed at daytime, which corresponds to the data reported here (). However, we found that Mi{ET1}25547 spent significantly more time in the center of the arena (). Thus, despite two different assays were used, and that the molecular method of gene disruption was also different, both studies provide evidence for aberrant activity behavior of D. melanogaster when the function of DAT was disrupted.
Locomotor activity is a trait, which has been used in experimental Drosophila genetics previously; ranging from finding the underlying genetic architecture of the trait,Citation30,31 characterizing the effects of inbreeding,Citation32 responses to stressors,Citation33 and to study human related diseases, such as ADHD and Alzheimer.Citation2,34 In these studies Drosophila activity is typically assessed using an activity monitor system (e.g., TriKinetics, Waltham, MA, USA), which scores the number of times individual flies interrupt one or more infrared light beams in a glass vial. However, as shown in this study, obtaining a single metric can lead to incomplete description of the behavior. This was illustrated by comparing mutants with similar responses in one sub-phenotype to another sub-phenotype; for example, Mi{ET1}24283 and w1118 covered the same distance in 5 minutes, but Mi{ET1}24283 showed a significant positional preference for the center of the arena. But more importantly, using the complete set of sub-phenotypes and reducing the dimensionality to the major axes of variation, mutants were grouped by phenotypic similarity (). Furthermore, studies using activity monitors to investigate sleep patterns in Drosophila have compared light crossing methods to video tracking and report high similarity between the methods,Citation15 but beam crossing methods overestimate sleep and video tracking methods are more accurate.Citation14,35 Thus, it is reasonable to transfer such results to patterns of behavioral activity.
Using animal models to study complex human diseases requires accurate and well-defined phenotypes, and an assay that reliably measures such traits. We believe that the type of assay used here and the proposed data analysis provide a more accurate estimate of Drosophila behavioral activity compared to the traditional type of locomotor activity assay used for Drosophila. The main limitation of the results presented here, concerns the duration of the recordings, which was limited to 5 minutes. However, we compared the results to those based on the whole 10 min. of recordings (see Methods), and very high correlations between the results were observed (Fig. S1), thus, the conclusions would have been the same independent on duration of recordings. However, obtaining behavioral activity for a limited time period will only give an immediate impression of the activity state of a given individual. Despite this, clear phenotypic separations of the lines were possible. The experimental design can easily be extended to encompass circadian rhythms by prolonging the test duration, and in addition; recordings in the infrared spectrum will allow differential light settings, without compromising the detailed information level.
By obtaining data on multiple correlated behavioral activity traits for insertional gene disrupted mutants, the behavioral phenotypic consequences of gene disruption of potential candidate genes for ADHD were investigated. Among the mutant lines investigated all but one resulted in an aberrant behavior compared to the control line, suggesting a potential role of those genes in ADHD. In addition, we show that the mutant lines could be grouped based on behavioral characteristics, and we show, that the disrupted genes within those groups had shared molecular function; both in D. melanogaster and humans. The first group contained genes related to cGMP production and cGMP-dependent kinases, the second group contained genes with relation to serotonin receptors, and the last group contained genes involved in cation channels.
Methods
Drosophila genetics
In order to identify genes that most consistently have demonstrated association to ADHD in human candidate gene studies and GWAS a list of candidate genes for ADHD was constructed () based on literature mining Using the online tool DIOPTCitation36 Drosophila orthologs were predicted. To test the phenotypic consequence of gene disruption of the candidate genes, Minos insertional mutants (Mi{ET1}) were used. These mutants were originally generated by mutagenesis using single insertion of the Minos transposon, thus, interfering with gene function.Citation16,37 For each of the 14 candidate genes Mi{ET1} insertional mutants, and the corresponding control line (w1118), were obtained from Bloomington Drosophila Stock Center (NIH P40OD018537) (). All Minos mutants were originally generated in the isogenic w1118 background.Citation37
Flies were reared on oatmeal-sugar-yeast-agar medium and maintained at 22.5°C at 12 hlight/dark cycles. Male flies were tested in the behavioral activity assay at the age of 2-3 d.
Behavioral activity assay
The intention of the behavioral activity assay was to create a high-throughput system for obtaining activity related behavioral traits for individual flies. The platform used consisted of a 120 mm × 120 mm plate (6 mm thick) containing 36 individual circular arenas with a diameter of 16 mm (, Video S1). The plate was created in transparent polycarbonate to allow illumination of each arena with light from below and video recordings from above. The light source was a light box (LP400, Dörr, Chesterfield, UK) to ensure a contrast between the flies and the background. To avoid external disturbances the whole system was placed in a box allowing airflow to avoid rising temperatures. Video recordings were obtained using an iPad Air2 (Apple, Cupertino, California, USA).
Videos were analyzed using the tracking software EthoVision XT from Noldus (Wageningen, The Netherlands). The software recorded 30 x-y coordinates per second, from which different behaviors could be computed (see section below).
All recordings were performed between 08:00 – 11:00 AM to reduce the influence of circadian rhythm. Each bout of recordings was 10, min however, the first 5 minwere discarded to allow the flies to acclimate to their new environment. The Mi{ET1} mutants were divided into three experimental blocks, with all mutants within block represented on one plate together with the control line, w1118.Citation38 Recordings of 16 plates per block were obtained resulting in a total of 96 observations per Mi{ET1} mutant and 288 observations for w1118.
Decomposition of locomotor activity
A key factor for developing an alternative strategy for acquiring locomotor activity measures for Drosophila is the ability to decompose the trait into distinct features. By obtaining movement tracks of individual flies, such decomposition is possible. Another important key concept is to avoid making assumptions that potentially limit the outcome. Selecting parameters based on assumptions of which could be important for the trait might bias the results. Therefore, we sought to avoid this by obtaining a large set of features related to activity (see below).
From the tracking software, EthoVision XT, several features were obtained: Total distance traveled (cm), time spent in the inner zone (defined as an area of 50% of the total area, ) and a measure of preferred moving direction, measured as the number of times a fly had moved 100 degrees (either clockwise or counter-clockwise) from the starting position; i.e., when the flies had moved 100 degrees a new starting position was set, and the cumulative sum of degrees moved was reset ().
From the raw tracking files patterns of movement were computed by defining periods of activity and inactivity according to a velocity threshold, here set to <0.01 cm/s (). The velocity (v) for each ith data point was computed as a moving average of five consecutive data points;
where di is the distance traveled from time i−1 to i. The mean and maximum periods of activity and inactivity, and the number of shifts between the two states were computed ().
Statistical analyses
The phenotypic consequences of each disrupted gene were assessed by comparing each Mi{ET1} insertional mutant line to the control line using analysis of variance (ANOVA) by fitting . The model specifications were equal for all analyses, such that
was the response variable and
was a vector containing the estimated effects of the two lines and an experimental block effect. Model assumptions were checked and data were transformed when needed (i.e., the proportion of time spent in the center was transformed by the arcsine function, the number of shifts in behavior were log-transformed, the mean time of moving/not moving was log transformed, and the maximum time moving/not moving was square-root transformed). All p values were adjusted for multiple testing using a false discovery rate of 0.05.
To create an overall score for the combined activity behavior for individual flies, all eight traits (i.e., distance, time spent in the center, number of rotations (clockwise and counter-clockwise), number of shifts between active and inactive periods (or vice versa), total time spent active and inactive and longest period being inactive or active) for all 14 Mi{ET1} mutants and the control line, w1118, were used in one PCA. Prior to the analysis the individual traits were centered and scaled to a mean of zero and standard deviation of one. The new trait for individual flies constituted the score from the first principal component.
All analyses were conducted in the free programming language R.Citation39
Abbreviations
| ADHD | = | Attention-deficit/hyperactivity disorder |
| GWAS | = | Genome-wide association study |
| PC | = | Principal component |
| PCA | = | Principal component analysis |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KFLY_A_1158365_Supplementals.zip
Download Zip (3.4 MB)Acknowledgments
We thank Pernille Merete Sarup for many useful discussions related to the analysis of the data and Helle Blendstrup and Henriette Casper Jensen for technical assistance. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study.
Funding
This work was funded by the Carlsberg Foundation (grant number 2013_01_0949) and a Sapere Aude grant from the Danish Research Council to TNK (DFF – 4002-00036).
References
- van Swinderen B, Brembs B. Attention-like deficit and hyperactivity in a Drosophila memory mutant. J Neurosci 2010; 30:1003-14; PMID:20089909; http://dx.doi.org/10.1523/JNEUROSCI.4516-09.2010
- van der Voet M, Harich B, Franke B, Schenck A. ADHD-associated dopamine transporter, latrophilin and neurofibromin share a dopamine-related locomotor signature in Drosophila. Mol Psychiatry 2015; 10:1-9
- Schenck A, Bardoni B, Langmann C, Harden N, Mandel JL, Giangrande A. CYFIP/Sra-1 controls neuronal connectivity in Drosophila and links the Rac1 GTPase pathway to the fragile X protein. Neuron 2003; 38:887-98; PMID:12818175; http://dx.doi.org/10.1016/S0896-6273(03)00354-4
- Wan L, Dockendorff TC, Jongens TA, Dreyfuss G. Characterization of dFMR1, a Drosophila melanogaster homolog of the Fragile X Mental Retardation protein. Mol Cell Biol 2000; 20:8536-47; PMID:11046149; http://dx.doi.org/10.1128/MCB.20.22.8536-8547.2000
- Polanczyk G, De Lima MS, Horta BL, Biederman J, Rohde LA. The worldwide prevalence of ADHD: A systematic review and metaregression analysis. Am J Psychiatry 2007; 164:942-8; PMID:17541055; http://dx.doi.org/10.1176/ajp.2007.164.6.942
- Thomas R, Sanders S, Doust J, Beller E, Glasziou P. Prevalence of attention-deficit/hyperactivity disorder: A systematic review and meta-analysis. Pediatrics 2015; 135:e994-1001; PMID:25733754; http://dx.doi.org/10.1542/peds.2014-3482
- Faraone SV, Sergeant J, Gillberg C, Biederman J. The worldwide prevalence of ADHD: is it an American condition? World Psychiatry 2003; 2:104-13; PMID:16946911
- Simon V, Czobor P, Balint S, Meszaros A, Bitter I. Prevalence and correlates of adult attention-deficit hyperactivity disorder: Meta-analysis. Br J Psychiatry 2009; 194:204-11; PMID:19252145; http://dx.doi.org/10.1192/bjp.bp.107.048827
- Ramos-Quiroga JA, Montoya A, Kutzelnigg A, Deberdt W, Sobanski E. Attention deficit hyperactivity disorder in the European adult population: prevalence, disease awareness, and treatment guidelines. Curr Med Res Opin 2013; 29:1093-104; PMID:23742051; http://dx.doi.org/10.1185/03007995.2013.812961
- Kessler R, Avenevoli S, Costello JE, Green JG, Gruber MJ, Heeringa S, Merikangas KR, Pennell BE, Sampson NA, Zaslavsky AM. Validity of the World Health Organization Adult ADHD Self-Report Scale (ASRS) Screener in a representative sample of health plan members. Int J Methods Psychiatr Res 2009; 18:69-83; PMID:19507169; http://dx.doi.org/10.1002/mpr.279
- American Psychiatric Association. American Psychiatric Association Diagnostic and Statistical Manual of Mental Disorders. 4th ed. Arlington, VA; 2000
- Faraone SV, Perlis RH, Doyle AE, Smoller JW, Goralnick JJ, Holmgren MA, Sklar P. Molecular genetics of attention-deficit hyperactivity disorder. Biol Psychiatry 2005; 57:1313-23; PMID:15950004; http://dx.doi.org/10.1016/j.biopsych.2004.11.024
- van Swinderen B, Flores KA. Attention-like processes underlying optomotor performance in a Drosophila choise maze. Dev Neurobiol 2006; 2:129-45
- Gilestro GF. Video tracking and analysis of sleep in Drosophila melanogaster. Nat Protoc 2012; 7:995-1007; PMID:22538850; http://dx.doi.org/10.1038/nprot.2012.041
- Garbe DS, Bollinger WL, Vigderman A, Masek P, Gertowski J, Sehgal A, Keene AC. Context-specific comparison of sleep acquisition systems in Drosophila. Biol Open 2015; 4:1558-68; PMID:26519516; http://dx.doi.org/10.1242/bio.013011
- Metaxakis A, Oehler S, Klinakis A, Savakis C. Minos as a genetic and genomic tool in Drosophila melanogaster. Genetics 2005; 171:571-81; PMID:15972463; http://dx.doi.org/10.1534/genetics.105.041848
- Ayroles JF, Buchanan SM, O’Leary C, Skutt-Kakaria K, Grenier JK, Clark AG, Hartl DL, de Bivort BL. Behavioral idiosyncrasy reveals genetic control of phenotypic variability. Proc Natl Acad Sci 2015; 112:201503830; http://dx.doi.org/10.1073/pnas.1503830112
- Buchanan SM, Kain JS, de Bivort BL. Neuronal control of locomotor handedness in Drosophila. Proc Natl Acad Sci 2015; 112:201500804; http://dx.doi.org/10.1073/pnas.1500804112
- Regulski M, Tully T. Molecular and biochemical characterization of dNOS: a Drosophila Ca2+/calmodulin-dependent nitric oxide synthase. Proc Natl Acad Sci U S A 1995; 92:9072-6; PMID:7568075; http://dx.doi.org/10.1073/pnas.92.20.9072
- Derbyshire ER, Marletta MA. Biochemistry of soluble guanylate cyclase. Handb Exp Pharmacol 2009; 191:17-31; PMID:19089323; http://dx.doi.org/10.1007/978-3-540-68964-5_2
- MacPherson MR, Lohmann SM, Davies SA. Analysis of Drosophila cGMP-dependent protein kinases and assessment of their in vivo roles by targeted expression in a renal transporting epithelium. J Biol Chem 2004; 279:40026-34; PMID:15218025; http://dx.doi.org/10.1074/jbc.M405619200
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res 2002; 12:996-1006; PMID:12045153; http://dx.doi.org/10.1101/gr.229102
- Gasque G, Conway S, Huang J, Rao Y, Vosshall LB. Small molecule drug screening in Drosophila identifies the 5HT2A receptor as a feeding modulation target. Sci Rep 2013; 3:srep02120; PMID:23817146; http://dx.doi.org/10.1038/srep02120
- Lee HH, Norris A, Weiss JB, Frasch M. Jelly belly protein activates the receptor tyrosine kinase Alk to specify visceral muscle pioneers. Nature 2003; 425:507-12; PMID:14523446; http://dx.doi.org/10.1038/nature01916
- Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 1988; 241:42-52; PMID:3291115; http://dx.doi.org/10.1126/science.3291115
- Young SN. How to increase serotonin in the human brain without drugs. J Psychiatry Neurosci 2007; 32:394-9
- Hayflick S, Wolfgang J, Forte A. A Unique Kex24-like endoprotease from Drosophila melanogaster is expressed in the central nervous system during early embryogenesis. J Neurosci 1992; 12:705-17; PMID:1545235
- Attrill H, Falls K, Goodman JL, Millburn GH, Antonazzo G, Rey AJ, Marygold SJ. FlyBase: Establishing a gene group resource for Drosophila melanogaster. Nucleic Acids Res 2015; 1:1-7
- Lansdell SJ, Collins T, Goodchild J, Millar NS. The Drosophila nicotinic acetylcholine receptor subunits Dα5 and Dα7 form functional homomeric and heteromeric ion channels. BMC Neurosci 2012; 13:73; PMID:22727315; http://dx.doi.org/10.1186/1471-2202-13-73
- Ayroles JF, Carbone MA, Stone EA, Jordan KW, Lyman RF, Magwire MM, Rollmann SM, Duncan LH, Lawrence F, Anholt RRH, et al. Systems genetics of complex traits in Drosophila melanogaster. Nat Genet 2009; 41:299-307; PMID:19234471; http://dx.doi.org/10.1038/ng.332
- Harbison ST, McCoy LJ, Mackay TFC. Genome-wide association study of sleep in Drosophila melanogaster. BMC Genomics 2013; 14:281; PMID:23617951; http://dx.doi.org/10.1186/1471-2164-14-281
- Manenti T, Pertoldi C, Moghadam NN, Schou MF, Kjærsgaard A, Cavicchi S, Loeschcke V. Inbreeding affects locomotor activity in Drosophila melanogaster at different ages. Behav Genet 2015; 45:127-34; PMID:25252771; http://dx.doi.org/10.1007/s10519-014-9683-5
- Kjærsgaard A, Demontis D, Kristensen TN, Le N, Faurby S, Pertoldi C, Sørensen JG, Loeschcke V. Locomotor activity of Drosophila melanogaster in high temperature environments: Plastic and evolutionary responses. Clim Res 2010; 43:127-34; http://dx.doi.org/10.3354/cr00870
- Fernandez-Funez P, Zhang Y, Sanchez-Garcia J, de Mena L, Khare S, Golde TE, Levites Y, Rincon-Limas DE. Anti-Aβ single-chain variable fragment antibodies exert synergistic neuroprotective activities in Drosophila models of Alzheimer disease. Hum Mol Genet 2015; 24:6093-105; PMID:26253732; http://dx.doi.org/10.1093/hmg/ddv321
- Zimmerman JE, Raizen DM, Maycock MH, Maislin G, Pack AI. A video method to study Drosophila sleep. Sleep 2008; 31:1587-98; PMID:19014079
- Hu Y, Flockhart I, Vinayagam A, Bergwitz C, Berger B, Perrimon N, Mohr SE. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics 2011; 12:357; PMID:21880147; http://dx.doi.org/10.1186/1471-2105-12-357
- Bellen HJ, Levis RW, He Y, Carlson JW, Evans-Holm M, Bae E, Kim J, Metaxakis A, Savakis C, Schulze KL, et al. The Drosophila gene disruption project: Progress using transposons with distinctive site specificities. Genetics 2011; 188:731-43
- Ryder E, Blows F, Ashburner M, Bautista-Llacer R, Coulson D, Drummond J, Webster J, Gubb D, Gunton N, Johnson G, et al. The DrosDel Collection: A set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics 2004; 167:797-813; PMID:15238529; http://dx.doi.org/10.1534/genetics.104.026658
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/. Version 3.2.0. 2015
- Neale BM, Lasky-Su J, Anney R, Franke B, Zhou K, Maller JB, Vasquez AA, Asherson P, Chen W, Banaschewski T, et al. Genome-wide association scan of attention deficit hyperactivity disorder. Am J Med Genet Part B Neuropsychiatr Genet 2008; 147B:1337-44; http://dx.doi.org/10.1002/ajmg.b.30866
- Hawi Z, Foley D, Kirley A, McCarron M, Fitzgerald M, Gill M. Dopa decarboxylase gene polymorphisms and attention deficit hyperactivity disorder (ADHD): no evidence for association in the Irish population. Mol Psychiatry 2001; 6:420-4; PMID:11443526; http://dx.doi.org/10.1038/sj.mp.4000903
- Neale BM, Medland S, Ripke S, Anney RJL, Asherson P, Buitelaar J, Franke B, Gill M, Kent L, Holmans P. Case-control genome-wide association study of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry 2010; 49:906-20; PMID:20732627; http://dx.doi.org/10.1016/j.jaac.2010.06.007
- Banaschewski T, Becker K, Scherag S, Franke B, Coghill D. Molecular genetics of attention-deficit/hyperactivity disorder: an overview. Eur Child Adolesc Psychiatry 2010; 19:237-57; PMID:20145962; http://dx.doi.org/10.1007/s00787-010-0090-z
- Yang L, Neale BM, Liu L, Lee SH, Wray NR, Ji N, Li H, Qian Q, Wang D, Li J, et al. Polygenic transmission and complex neuro developmental network for attention deficit hyperactivity disorder: Genome-wide association study of both common and rare variants. Am J Med Genet Part B Neuropsychiatr Genet 2013; 162:419-30; http://dx.doi.org/10.1002/ajmg.b.32169
- Mick E, McGough J, Loo S, Doyle AE, Wozniak J, Wilens TE, Smalley S, McCracken J, Biederman J, Faraone SV. Genome-wide association study of the child behavior checklist dysregulation profile. J Am Acad Child Adolesc Psychiatry 2011; 50:807-17; PMID:21784300; http://dx.doi.org/10.1016/j.jaac.2011.05.001
- Neale BM, Medland SE, Ripke S, Asherson P, Franke B, Lesch KP, Faraone S V, Nguyen TT, Schäfer H, Holmans P, et al. Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry 2010; 49:884-97; PMID:20732625; http://dx.doi.org/10.1016/j.jaac.2010.06.008
- Stergiakouli E, Hamshere M, Holmans P, Langley K, Zaharieva I, Hawi Z, Kent L, Gill M, Williams N, Owen MJ, et al. Investigating the contribution of common genetic variants to the risk and pathogenesis of ADHD. Am J Psychiatry 2012; 169:186-94; PMID:22420046; http://dx.doi.org/10.1176/appi.ajp.2011.11040551
- Lesch KP, Timmesfeld N, Renner TJ, Halperin R, Röser C, Nguyen TT, Craig DW, Romanos J, Heine M, Meyer J, et al. Molecular genetics of adult ADHD: converging evidence from genome-wide association and extended pedigree linkage studies. J Neural Transm 2008; 115:1573-85; PMID:18839057; http://dx.doi.org/10.1007/s00702-008-0119-3
