Mechanosensitive channels are the molecules closely matching the definition of “machines” in our macroworld—they convert external mechanical forces into motion of the parts to open or close a water-filled pore, with no chemical energy inputs involved. Bacterial mechanosensitive channel MscL from E. coli is the first known and probably the best understood representative of this group.Citation1 A minimalistic bundle of 5 pairs of interlocked α-helices in the membrane requires no external protein connections. The barrel responds directly to the lipid bilayer tension by tilting of the helices and iris-like expansion that opens a large (∼3 nm) pore.Citation1 Robustness of MscL makes it a promising template for engineering nano-devices operated by force or chemical modification.Citation2 Known crystal structures and models enabled molecular simulations which visualized lipid interactions and forces acting on specific channel segments.Citation3 The experimentally estimated timescale of the opening, is in the order of microsecond,Citation4 whereas the process of subsequent equilibration within the subset of open states may take up to several seconds.Citation5 Simulations striving for proper representation of these stochastic events must be at least as long. Atomistic simulations on this time scale are currently unattainable thus calling for radical simplifications. Over the last decade, a helpful approximation for MscL was developed using solid-state mechanical engineering tools that present the channel/membrane system as interacting meshworks of finite elastic elements.Citation6
Despite the apparent simplicity, MscL is surrounded by solvent and a highly anisotropic bilayer that both change their interactions with the protein during the gating. This poses several challenges for the finite-element approach. One is the necessity to account for large changes of the solvent-accessible area and associated hydration contribution. In part, it was recently addressed by explicitly introducing terms for the hydration energetics.Citation6 The second oversimplification of the finite-element approach was that, while effective elasticities of different elements can be derived from atomistic calculations, they were assumed to be constant and it is unclear how they change under varying solvation or sequestration inside the lipid. The paper by Bavi et al. published in the current issueCitation7 addresses these questions. Their elasticity tests of isolated α-helices of MscL in steered molecular dynamics simulations gave different moduli estimations for different domains. As one can expect, the elastic modulus values changed to some extent upon switching from M. tuberculosis MscL to its modeled E. coli homolog. Most importantly, the α-helices became considerably softer when the helix was hydrated, compared with simulations in vacuo.Citation7 That finding might be crucial for the mechanics of MscL in continuum-based models because the hydration of the pore-lining helices increases dramatically upon channel opening.Citation8 Moreover, as the lipid-facing helices tilt in the stretching (and thinning) bilayer, their hydration is likely to change too. This calls for advancement of the finite-element model to the one where elements are sensitive to the changing environment and adjust their stiffness through the simulation. The change in elasticity can be essential for the gating time course and may potentially contribute to the well-known “hysteresis” where the tension required for opening is higher than the one for closing.
Building a proper mechanical model for this challenging system is iterative and far from completion. The future steps in quantifying elasticity of MscL helices might include 1) simulations of helices in aligned pairs (as they are in the crystal structures of M. Tuberculosis MscL) where they reinforce and partially shield each other from the medium; 2) more realistic heterogeneous membrane environment (not a simple water box); 3) inclusion of bending and twisting deformations, which are more likely helix force modalities than pure axial stretch. These developments are straightforward to implement with theflexible constant-force steering protocol that the authors of the current paper have advanced.Citation7 Moreover, grounding their work on MD simulations allows for a clear comparison of different force fields, water models, mediums, channel conformations and homologs. The approach may also enable a feedback from the finite-element models to refine MD simulations settings. The opportunities for understanding and designing new custom-tailored nano-machines are truly exciting.
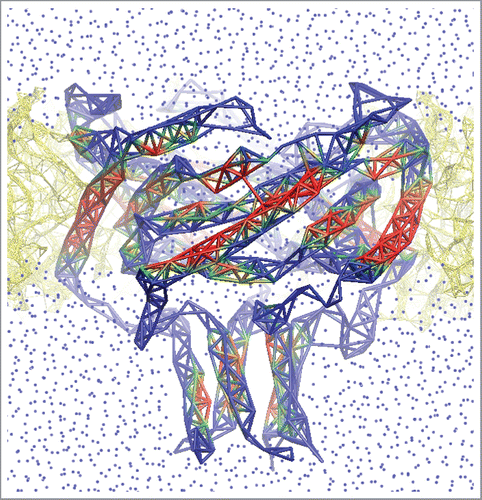
Figure 1. Finite-element representation of an open E.coli MscL model. The conformation was taken from explicit-medium MD simulation.Citation8 Lipids (yellow) are shown as bonds between the heavy atoms within 4 Å cut-off. Protein is shown as a meshwork of thick bonds between the α-carbons within 7 Å (colored by solvation: red-hidden, blue-solvated). Water is pictured as blue dots.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Kung C, Martinac B, Sukharev S. Mechanosensitive channels in microbes. Ann Rev Microbiol 2010; 64:313-29; PMID: 20825352; https://doi.org/10.1146/annurev.micro.112408.134106
- Kocer A. Mechanisms of mechanosensing—mechanosensitive channels, function and re-engineering. Curr Opin Chem Biol 2015; 29:120-7; PMID: 26610201; https://doi.org/10.1016/j.cbpa.2015.10.006
- Vanegas JM, Arroyo M. Force transduction and lipid binding in MscL: a continuum-molecular approach. PLoS One 2014; 9:e113947; PMID: 25437007; https://doi.org/10.1371/journal.pone.0113947
- Shapovalov G, Lester H. Gating transitions in bacterial ion channels measured at 3 μs resolution. J Gen Physiol 2004; 124:151-61; PMID: 15277576; https://doi.org/10.1085/jgp.200409087
- Chiang C-SS, Anishkin A, Sukharev S. Gating of the large mechanosensitive channel in situ: estimation of the spatial scale of the transition from channel population responses. Biophys J 2004; 86:2846-61; PMID: 15111402; https://doi.org/10.1016/S0006-3495(04)74337-4
- Zhu L, Wu J, Liu L, Liu Y, Yan Y, Cui Q, Chen X. Gating mechanism of mechanosensitive channel of large conductance: a coupled continuum mechanical-continuum solvation approach. Biomech Model Mechanobiol 2016; 15:1557-76; PMID: 27009075; https://doi.org/10.1007/s10237-016-0783-4
- Bavi N, Bavi O, Vossoughi M, Naghdabadi R, Hill AP, Martinac B, Jamali Y. Nanomechanical properties of MscL alpha helices: A steered molecular dynamics study. Channels 2017; 11(3):209-223; PMID: 27753526; https://doi.org/10.1080/19336950.2016.1249077
- Anishkin A, Akitake B, Kamaraju K, Chiang C-SS, Sukharev S. Hydration properties of mechanosensitive channel pores define the energetics of gating. J Phys Condens Matter 2010; 22:454120; PMID: 21339607; https://doi.org/10.1088/0953-8984/22/45/454120
