Abstract
Long non-coding RNA PSMG3-AS1 is known to play critical roles in several types of cancer, while its role in prostate carcinoma (PC) is unknown. This study aimed to explore the involvement of PSMG3-AS1 in PC. In this study, RT-qPCR analysis showed that PSMG3-AS1 was upregulated, while miR-106b was downregulated in PC. PSMG3-AS1 and miR-106b were inversely and significantly correlated across PC tissue samples. In addition, in PC cells, overexpression of PSMG3-AS1 increased the DNA methylation of miR-106b and decreased the expression levels of miR-106b. In contrast, no significant alteration in the expression of PSMG3-AS1 was observed in cells transfected with miR-106b mimic. Cell proliferation analysis showed that PSMG3-AS1 reduced the inhibitory effects of miR-106b overexpression on cell proliferation. Taken together, our data suggested that PSMG3-AS1 could downregulate miR-106b through DNA methylation to suppress PC cell proliferation.
Introduction
Prostate carcinoma (PC) is a common type of male malignancy that affects about one out of nine males in their lifetime (Siegel et al. Citation2019). PC affected 1,276,106 new cases in 2018, which accounts for 7.1% of all new cancer cases during that year (Bray et al. Citation2018; Siegel et al. Citation2019). In the same year, 358,989 patients (3.8% of all cancer deaths) died of PC (Bray et al. Citation2018; Siegel et al. Citation2019). PC mainly affects elderly, while the incidence of PC shows an increasing trend among young males in recent years (Bleyer et al. Citation2020). PC patients are frequently diagnosed with local tumor. With proper treatment, the 5-year survival rate can reach 100% (Rusthoven et al. Citation2016; Xie et al. Citation2017). However, once distant metastasis has occurred, the 5-year survival rate drops to 30% (Halabi et al. Citation2003). Therefore, novel anti-cancer therapies are still needed to improve their survival conditions.
Long non-coding RNAs (lncRNAs) are a type of ncRNAs that lack protein-coding capacity and play vital roles in the development of malignancies such as tumor cell proliferation, apoptosis, migration, and invasion (Tan et al. Citation2021). For instance, GAS5 was found to be associated with PC cell proliferation by regulating the AKT/mTOR signaling pathway and sponging miR-103 (Xue et al. Citation2016). LEF1-AS1 facilitated metastasis of PC via the Wnt/β-catenin pathway (Li et al. Citation2020). PSMG3-AS1 has been reported to be upregulated in multiple cancers. For example, silencing of PSMG3-AS1 inhibited breast cancer cell proliferation and migration via sponging miR-143-3p (Cui et al. Citation2020). Overexpression of PSMG3-AS1 led to increased proliferation rates via regulating miR-143-3p (Zhang et al. Citation2020). However, the role of PSMG3-AS1 in PC has not been elucidated.
MicroRNAs (miRNAs) are ncRNAs with <200 nucleotides in length (Meng et al. Citation2020). It has been well demonstrated that miRNAs play critical roles in the occurrence and development of cancers (Wang et al. Citation2016). For instance, miR-106b was reported to be downregulated in colon cancer tissues and inhibited tumor growth and metastasis (Zhang et al. Citation2019). Moreover, miR-106b suppressed breast cancer cell invasion and motility in association with overexpression of MMP2 (Ni et al. Citation2014). Until now, no comprehensive study has been conducted on the function of miR-106b in PC.
In this study, altered expression of PSMG3-AS1 in PC and its inverse correlation with miR-106b was observed in our preliminary microarray data (data not shown). MiR-106b is also a critical player in cancer biology (Yue et al. Citation2020). We speculated that PSMG3-AS1 could interact with miR-106b to regulate PC. This study was therefore carried out to explore the interaction between PSMG3-AS1 and miR-106b in PC.
Results
The expression of PSMG3-AS1 and miR-106b were altered in PC and mechanisms that mediates their interaction
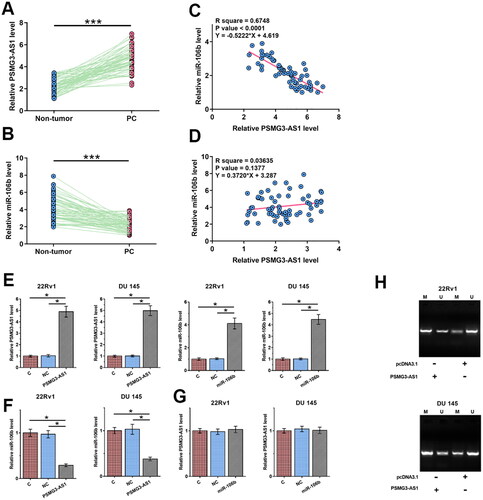
To detect the differential expression of PSMG3-AS1 and miR-106b in PC, the expression levels of PSMG3-AS1 and miR-106b in paired PC and non-tumor tissues were determined by RT-qPCR. Compared to non-tumor tissues, PSMG3-AS1 was significantly upregulated in PC tissues (, p < 0.001, df = 122), while miR-106b was significantly downregulated in PC (, p < 0.001, df = 122). Linear regression was performed to analyze the correlations between PSMG3-AS1 and miR-106b across both PC tissues (, f = 124.5, df = 60) and non-tumor tissues (, f = 2.263, df = 60). The results showed that PSMG3-AS1 and miR-106b were inversely and significantly correlated across PC tissues but not across non-tumor tissues. The inverse correlation between PSMG3-AS1 and miR-106b indicated the crosstalk between them. To further explore the interactions between PSMG3-AS1 and miR-106b, 22Rv1 and DU 145 cells were transfected with PSMG3-AS1 expression vector or miR-106b mimic. Transfections were confirmed by RT-qPCR (, p < 0.05, f = 188.2, df = 8; f = 240.4, df = 8; f = 116.2, df = 8; f = 178.1, df = 8). Overexpression of PSMG3-AS1 decreased the expression levels of miR-106b (, p < 0.05, f = 105.9, df = 8; f = 65.1, df = 8), while no significant alteration in the expression of PSMG3-AS1 was observed after overexpression of miR-106b (, f = 0.5182, df = 8; f = 0.3545, df = 8). Methylation-specific PCR (MSP) was performed to analyze the DNA methylation of miR-106b gene in cells transfected with PSMG3-AS1 expression vector. Compared to the transfection with empty pcDNA3.1 vector, transfection with PSMG3-AS1 expression vector enhanced the DNA methylation of miR-106b gene ().
Figure 1. The expression of PSMG3-AS1 and miR-106b were altered in PC and mechanisms that mediates their interaction. To analyze the expression of PSMG3-AS1 and miR-106b in PC, the expression levels of PSMG3-AS1 (A) and miR-106b (B) in paired PC tissues were determined by RT-qPCR. Linear regression was performed to analyze the correlations between PSMG3-AS1 and miR-106b across both PC tissues (C) and non-tumor tissues (D). To further explore the interactions between PSMG3-AS1 and miR-106b, 22Rv1 and DU 145 cells were subjected to PSMG3-AS1 expression vector or miR-106b mimic transfection, and transfections were confirmed by RT-qPCR (E). The expression of miR-106b in cells with PSMG3-AS1 expression vector transfection (F) and the expression of PSMG3-AS1 in cells with miR-106b mimic transfection (G) were also analyzed by RT-qPCR. MiR-106b gene methylation in cells with PSMG3-AS1 vector transfection was analyzed by MSP (H). *, p < 0.05; ***, p < 0.001.
PSMG3-AS1 increased proliferation of 22Rv1 and DU 145 cells through miR-106b
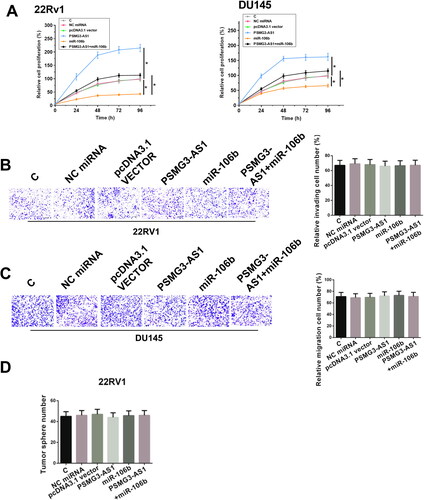
The roles of PSMG3-AS1 and miR-106b in regulating the proliferation of 22Rv1 and DU 145 cells were assessed by CCK-8 assay. Overexpression of miR-106 decreased the cell proliferation rate, while overexpression of PSMG3-AS1 increased the cell proliferation rate. In addition, co-transfection analysis showed that overexpression of PSMG3-AS1 suppressed the role of miR-106b in cell proliferation (, df = 60). Moreover, Transwell assay and cell stemness assay results showed that there was no significant change in cell invasion, migration and stemness with the overexpression of PSMG3-AS1 in 22Rv1 cells () and DU 145 cells (data not shown). Therefore, PSMG3-AS1 may participate in PC mainly by regulating cell proliferation.
Figure 2. PSMG3-AS1 promoted the proliferation of 22Rv1 and DU 145 cells through miR-106b. CCK-8 assay was performed following cell transfection assays to explore the functions of PSMG3-AS1 and miR-106b in 22Rv1 and DU 145 cell proliferation (A). Besides cell proliferation assay, invasion, migration and stemness assays were also used to detect the role of PSMG3-AS1 in 22Rv1 cells (B–D). Experiments were performed in three independent replicates and mean±SD values were presented and compared. Representative images of three biological replicates of invasion (B) and migration (C) assays were presented. *, p < 0.05.
PSMG3-AS1 inhibition impairs PC tumor growth
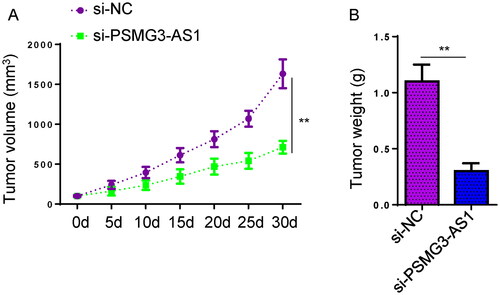
To explore the functions of PSMG3-AS1 in PC in vivo, xenograft tumor assay was performed. 22Rv1 cells transfected with si-NC or si-PSMG3-AS1 were injected into the mice subcutaneously. As shown in ( (A, p < 0.01, df = 57; B, p < 0.01, df = 4), mice in PSMG3-AS1-deficient group exhibited a remarkable decrease in tumor volume and tumor weight in comparison to that in the control group. Therefore, PSMG3-AS1 plays an essential role in promoting PC tumor growth.
Figure 3. PSMG3-AS1 inhibition impairs PC tumor growth. To explore the role of PSMG3-AS1 in the in vivo growth of PC tumors, 22Rv1 cells transfected with si-NC or si-PSMG3-AS1 were injected into BALB/c nude mice to construct xenograft tumor models. Tumor volume was measured every 5 days until 30 days to monitor tumor growth (A). The xenograft tumors were weighed and compared among groups at the end of experiments. (B). The xenograft tumors were weighed and compared among groups at the end of experiments Experiments were performed in three independent replicates and mean±SD values were presented and compared. *, p < 0.05.
Discussion
The expression of PSMG3-AS1 in PC and its interaction with miR-106b in PC were explored in this study. Our results revealed altered expression of PSMG3-AS1 and miR-106b in PC. Moreover, PSMG3-AS1 may decrease the expression levels of miR-106b through methylation to increase the proliferation of PC cells.
A recent study reported the role of PSMG3-AS1 in breast cancer (Cui et al. Citation2020). Overexpression of PSMG3-AS1 absorbs miR-143-3p to increase cell proliferation and migration in breast cancer cells (Cui et al. Citation2020). Besides, miR-449b-5p and miR-143-3p target PSMG3-AS1 to suppress cell proliferation in lung adenocarcinoma and hepatocellular carcinoma, respectively (Yue et al. Citation2020; Zhang et al. Citation2020). Although the function of PSMG3-AS1 in glioblastomas is unknown, overexpression of PSMG3-AS1 separates glioblastomas from sarcoidosis, which is frequently misdiagnosed as glioblastomas in clinical practice (Chen et al. Citation2020). The expression pattern and functions of PSMG3-AS1 in other types of cancer remain unclear. To the best of our knowledge, the present study is the first to report the upregulation of PSMG3-AS1 in PC. In addition, overexpression of PSMG3-AS1 increased cancer cell proliferation, suggesting the oncogenic role of PSMG3-AS1 in PC.
MiR-106b plays different roles in different types of cancer (Cai et al. Citation2011; Shen et al. Citation2013; Zheng et al. Citation2015; Yen et al. Citation2016). For instance, miR-106b is upregulated in hepatocellular carcinoma and promotes cancer development by interacting with hepatitis B virus (Yen et al. Citation2016). MiR-106b is also upregulated in laryngeal carcinoma and it targets RB to promote cell proliferation (Cai et al. Citation2011). In contrast, miR-106b is downregulated and plays a tumor suppressive role by targeting Prrx1 to suppress epithelial-mesenchymal transition in colorectal cancer. We showed that miR-106b was downregulated in PC and overexpression of miR-106b decreased proliferation of PC cells. Therefore, miR-106b plays a tumor suppressive role in PC.
LncRNAs may regulate the expression of miRNAs and protein coding genes through methylation (Liu et al. Citation2016; Yu et al. Citation2022). For instance, KIF9-AS1 can recruit methylation factor DNMT1 to increase RAI2 DNA methylation, thereby accelerating the growth of hepatocellular carcinoma (Yu et al. Citation2022). We showed that overexpression of PSMG3-AS1 decreased the expression levels of miR-106b in PC cells by enhancing the DNA methylation miR-106b gene. Interestingly, significant correlation between PSMG3-AS1 and miR-106 was only observed across PC tissues but not non-tumor tissues. The data suggested that certain pathological factors may mediate the crosstalk between PSMG3-AS1 and methylation factors involved in the DNA methylation of miR-106b gene. Future studies are needed to identify these factors. It is worth noting that our data only showed that the function of PSMG3-AS1 in PC was at least partially mediated by miR-106b. PSMG3-AS1 may also interact with other factors to regulate cancer cell behaviors. The mechanism of the function of PSMG3-AS1 in PC remains to be further elucidated. Moreover, the close correlation between PSMG3-AS1 and miR-106b was not observed in normal tissues, suggesting that the regulation of DNA methylation of miR-106b by PSMG3-AS1 is mediated by certain pathological factors.
In conclusion, PSMG3-AS1 is upregulated in PC and it may downregulate miR-106b by enhancing the DNA methylation of miR-106 to increase PC cell proliferation.
Materials and methods
PC patients and tissue collection
A total of 62 male PC patients (stage I (n = 11), II (n = 19), III (n = 15) and IV (n = 17)) admitted at Soochow University Affiliated Wuxi Ninth Hospital between May 2017 and May 2019 were enrolled in this study. Ethics Committee of the aforementioned hospital approved this study before the admission of patients. During biopsy, PC and paired non-tumor tissues were obtained from each patient. F Information about the patients with PC tissues was listed in .
Table 1. Correlation between PSMG3-AS1 and clinicopathological characteristics of prostate carcinoma.
PC cells and transfections
Human PC cell lines 22Rv1 and DU 145 were purchased from the American Type Culture Collection (ATCC; Manassas, VA) and cultivated following manufacturer’s instruction. Transfection of PSMG3-AS1 vector and miR-106b mimic were performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA).
RNA isolation and RT-qPCR
Total RNAs were isolated using RNeasy Mini Kit (Qiagen, Germantown, MD). SSRT III system (Thermo Fisher Scientific, Waltham, MA) was used to prepare cDNA, and expression levels of PSMG3-S1 and miR-106b were determined by qPCRs with 18S rRNA and U6 as the endogenous control. Expression was normalized using the 2−ΔΔCT method.
Methylation-specific PCR
Genomic DNAs were extracted from transfected cells using Genomic DNA Extraction Kit (TianGen, Beijing, China). romoter sequence (2000 bp) of miR-106b was identified using UCSC Genome Browser (http://genome.ucsc.edu/). MethPrimer (http://www.urogene.org/methprimer) was then applied to predict CpG islands. Primers used in MSP and routine PCR were also designed using MethPrimer.
Cell proliferation assay, sphere-formation assay, transwell and assay and animal experiment
Proliferation ability of 22Rv1 and DU 145 cells was analyzed using cell counting kit-8 (CCK-8) assay every 24 h for four times.
22Rv1 cells were seeded into 6-well culture plates (Corning, NY). Cells (2 × 105) were cultured in serum-free DMEM supplemented with EGF, hFGF (Peprotech, USA), insulin, and penicillin/streptomycin (Gibco Paisley, Scotland). Spheroids clones were fixed and stained with crystal violet, and counted under a light stereomicroscope (Olympus, Tokyo, Japan).
The migration and invasion of cells were evaluated by transwell assay using 24-well plates with 8-μm pore polycarbonate membranes (BD Biosciences, USA).
Male BALB/c nude mice (5 weeks old) were used for the establishment of PC Xenograft tumor models. The volume of Xenografted tumor was calculated every 5 days. All mice were sacrificed 30 days later to harvest tumors. Then, tumors were weighed.
Data analysis
Three independent replicates were included in all experiments. All data are presented as the mean ± SD and were analyzed using the SPSS 18.0 software (SPSS, Inc.). Normal distribution of data was evaluated using the Shapiro-Wilk normality test. Student’s t-test (both paired and unpaired) were used for comparisons between two groups. One-way ANOVA followed by Tukey’s post hoc test was performed to compare differences among multiple groups. Linear regression was used to analyze correlations. p < 0.05 was considered statistically significant.
Ethics approval
Ethics Committee of Soochow University Affiliated Wuxi Ninth Hospital approved this study, and the study was conducted in accordance with the Declaration of Helsinki published by the World Medical Association. Informed consent was also obtained from all patients.
Authors’ contributions
Conception: QX; Experimental studies: LZ, YC; Data analysis: LZ, ZW; Statistical analysis: LZ; Manuscript writing: LZ; Manuscript editing and revision: QX. All authors have read and approve the submission of the manuscript.
| Abbreviations | ||
| PC | = | Prostate carcinoma |
| lncRNAs | = | Long noncoding RNAs |
| ncRNA | = | Non-coding RNA |
| miRNAs | = | MicroRNAs |
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All data generated or analyzed during this study are available from the corresponding author upon reasonable request.
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
References
- Bleyer A, Spreafico F, Barr R. 2020. Prostate cancer in young men: an emerging young adult and older adolescent challenge. Cancer. 126(1):46–57.
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. 2018. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 68(6):394–424.
- Cai K, Wang Y, Bao X. 2011. MiR-106b promotes cell proliferation via targeting RB in laryngeal carcinoma. J Exp Clin Cancer Res. 30(1):73.
- Chen L, Wang G, Xu Z, Lin K, Mu S, Pan Y, Shan M. 2020. Overexpression of LncRNA PSMG3-AS1 distinguishes glioblastomas from sarcoidosis. J Mol Neurosci. 70(12):2015–2019.
- Cui Y, Fan Y, Zhao G, Zhang Q, Bao Y, Cui Y, Ye Z, Chen G, Piao X, Guo F, et al. 2020. Novel lncRNA PSMG3AS1 functions as a miR1433p sponge to increase the proliferation and migration of breast cancer cells. Oncol Rep. 43(1):229–239.
- Halabi S, Small EJ, Kantoff PW, Kattan MW, Kaplan EB, Dawson NA, Levine EG, Blumenstein BA, Vogelzang NJ. 2003. Prognostic model for predicting survival in men with hormone-refractory metastatic prostate cancer. J Clin Oncol. 21(7):1232–1237.
- Li W, Yang G, Yang D, Li D, Sun Q. 2020. LncRNA LEF1-AS1 promotes metastasis of prostatic carcinoma via the Wnt/beta-catenin pathway. Cancer Cell Int. 20(1):543.
- Liu HT, Fang L, Cheng YX, Sun Q. 2016. LncRNA PVT1 regulates prostate cancer cell growth by inducing the methylation of miR-146a. Cancer Med. 5(12):3512–3519.
- Meng Q, Liang C, Hua J, Zhang B, Liu J, Zhang Y, Wei M, Yu X, Xu J, Shi S. 2020. A miR-146a-5p/TRAF6/NF-kB p65 axis regulates pancreatic cancer chemoresistance: functional validation and clinical significance. Theranostics. 10(9):3967–3979.
- Ni X, Xia T, Zhao Y, Zhou W, Wu N, Liu X, Ding Q, Zha X, Sha J, Wang S. 2014. Downregulation of miR-106b induced breast cancer cell invasion and motility in association with overexpression of matrix metalloproteinase 2. Cancer Sci. 105(1):18–25.
- Rusthoven CG, Jones BL, Flaig TW, Crawford ED, Koshy M, Sher DJ, Mahmood U, Chen RC, Chapin BF, Kavanagh BD, et al. 2016. Improved survival with prostate radiation in addition to androgen deprivation therapy for men with newly diagnosed metastatic prostate cancer. J Clin Oncol. 34(24):2835–2842.
- Shen G, Jia H, Tai Q, Li Y, Chen D. 2013. miR-106b downregulates adenomatous polyposis coli and promotes cell proliferation in human hepatocellular carcinoma. Carcinogenesis. 34(1):211–219.
- Siegel RL, Miller KD, Jemal A. 2019. Cancer statistics, 2019. CA Cancer J Clin. 69(1):7–34.
- Tan YT, Lin JF, Li T, Li JJ, Xu RH, Ju HQ. 2021. LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun (Lond). 41(2):109–120.
- Wang W, Zhu Y, Li S, Chen X, Jiang G, Shen Z, Qiao Y, Wang L, Zheng P, Zhang Y. 2016. Long noncoding RNA MALAT1 promotes malignant development of esophageal squamous cell carcinoma by targeting beta-catenin via Ezh2. Oncotarget. 7(18):25668–25682.
- Xie W, Regan MM, Buyse M, Halabi S, Kantoff PW, Sartor O, Soule H, Clarke NW, Collette L, Dignam JJ, et al. 2017. Metastasis-free survival is a strong surrogate of overall survival in localized prostate cancer. J Clin Oncol. 35(27):3097–3104.
- Xue D, Zhou C, Lu H, Xu R, Xu X, He X. 2016. LncRNA GAS5 inhibits proliferation and progression of prostate cancer by targeting miR-103 through AKT/mTOR signaling pathway. Tumor Biol. 37(12):16187–16197.
- Yen CS, Su ZR, Lee YP, Liu IT, Yen CJ. 2016. miR-106b promotes cancer progression in hepatitis B virus-associated hepatocellular carcinoma. World J Gastroenterol. 22(22):5183–5192.
- Yu Y, Lu X, Yan Y, Wang Y, Meng J, Tian S, Mu J. 2022. The lncRNA KIF9-AS1 accelerates hepatocellular carcinoma growth by recruiting DNMT1 to promote RAI2 DNA methylation. J Oncol. 2022:3888798.
- Yue N, Ye M, Zhang R, Guo Y. 2020. MiR-449b-5p targets lncRNA PSMG3-AS1 to suppress cancer cell proliferation in lung adenocarcinoma. BMC Pulm Med. 20(1):152.
- Zhang J, Huang J, Chen W, Hu Z, Wang X. 2020. miR-143-3p targets lncRNA PSMG3-AS1 to inhibit the proliferation of hepatocellular carcinoma cells. Cancer Manag Res. 12:6303–6309.
- Zhang J, Liu H, Zhao P, Zhou H, Mao T. 2019. Has_circ_0055625 from circRNA profile increases colon cancer cell growth by sponging miR-106b-5p. J Cell Biochem. 120(3):3027–3037.
- Zheng L, Zhang Y, Lin S, Sun A, Chen R, Ding Y, Ding Y. 2015. Down-regulation of miR-106b induces epithelial-mesenchymal transition but suppresses metastatic colonization by targeting Prrx1 in colorectal cancer. Int J Clin Exp Pathol. 8(9):10534–10544.
