Abstract
CD1d is a receptor on antigen-presenting cells involved in triggering cell populations, particularly natural killer T (NKT) cells, to release high levels of cytokines. NKT cells are implicated in asthma pathology and blockade of the CD1d/NKT cell pathway may have therapeutic potential. We developed a potent anti-human CD1d antibody (NIB.2) that possesses high affinity for human and cynomolgus macaque CD1d (KD ∼100 pM) and strong neutralizing activity in human primary cell-based assays (IC50 typically <100 pM). By epitope mapping experiments, we showed that NIB.2 binds to CD1d in close proximity to the interface of CD1d and the Type 1 NKT cell receptor β-chain. Together with data showing that NIB.2 inhibited stimulation via CD1d loaded with different glycolipids, this supports a mechanism whereby NIB.2 inhibits NKT cell activation by inhibiting Type 1 NKT cell receptor β-chain interactions with CD1d, independent of the lipid antigen in the CD1d antigen-binding cleft. The strong in vitro potency of NIB.2 was reflected in vivo in an Ascaris suum cynomolgus macaque asthma model. Compared with vehicle control, NIB.2 treatment significantly reduced bronchoalveolar lavage (BAL) levels of Ascaris-induced cytokines IL-5, IL-8 and IL-1 receptor antagonist, and significantly reduced baseline levels of GM-CSF, IL-6, IL-15, IL-12/23p40, MIP-1α, MIP-1β, and VEGF. At a cellular population level NIB.2 also reduced numbers of BAL lymphocytes and macrophages, and blood eosinophils and basophils. We demonstrate that anti-CD1d antibody blockade of the CD1d/NKT pathway modulates inflammatory parameters in vivo in a primate inflammation model, with therapeutic potential for diseases where the local cytokine milieu is critical.
Abbreviations
| AHR | = | airway hyper-reactivity |
| APC | = | antigen-presenting cell |
| AUC | = | area under the curve |
| BAL | = | broncho-alveolar lavage |
| BSA | = | bovine serum albumin |
| CHO | = | Chinese hamster ovary |
| ELISA | = | enzyme-linked immunosorbent assay |
| G-CSF | = | granulocyte colony stimulating factor |
| GM-CSF | = | granulocyte-macrophage colony stimulating factor |
| HEK | = | human embryonic kidney |
| HPLC | = | high performance liquid chromatography |
| IFN | = | interferon |
| IL | = | interleukin |
| MCh | = | methacholine |
| MHC | = | major histocompatibility complex |
| MIP | = | macrophage inflammatory protein |
| NKT | = | natural killer T |
| OVA | = | ovalbumin |
| PBMC | = | peripheral blood mononuclear cell |
| PBS | = | phosphate-buffered saline |
| SDS-PAGE | = | sodium dodecyl sulfate- polyacrylamide gel electrophoresis |
| SPR | = | surface plasmon resonance |
| TNF | = | tumor necrosis factor |
| VEGF | = | vascular endothelial growth factor |
Introduction
CD1d proteins are displayed on multiple antigen-presenting cell (APC) subsets, including monocytes/macrophages, dendritic cells and activated B cells, and also on certain tissue epithelia, such as the lung. CD1d presents lipid and glycolipid antigens to cognate T cells, inducing cytokine release and triggering immune responses. The major CD1d-restricted cells are natural killer T (NKT) cells, a subset of T cells that express an αβ T cell receptor and surface markers typically associated with NK cells, such as CD161 and NKG2D.Citation1 CD1d/NKT cell receptor interactions can rapidly induce a broad range of Th1- or Th2- cytokines, such as interferon (IFN)-γ, tumor necrosis factor (TNF), and interleukin (IL)-4, IL-5 and IL-13. Blockade of the CD1d/NKT cell pathway in vivo may modulate the levels of these cytokines in inflammatory settings of a systemic nature, such as systemic lupus erythematosus,Citation2,3 and local mucosal inflammation, as occurs in asthma.
Asthma is a chronic inflammatory pulmonary disorder characterized by reversible airway obstruction arising from chronic local inflammation, mucus obstruction, and bronchospasm in response to nonspecific stimuli.Citation4 The CD1d/NKT pathway appears to contribute to asthma pathology. In mouse ovalbumin (OVA)-induced models of asthma, challenged wild-type mice showed increased airway-hyper-reactivity (AHR) to inhaled methacholine (MCh) and increased airway infiltration of leukocytes, particularly monocytes/macrophages, lymphocytes and eosinophils as observed in the bronchoalveolar lavage (BAL). In contrast, OVA-induced AHR did not develop in CD1d−/- mice, and the extent of BAL cell infiltration in these CD1d−/- mice was lower than in wild-type mice.Citation5 Similarly, anti-mouse CD1d antibody treatment in the OVA model reduced BAL cellular infiltrate and levels of the Th2 cytokines IL-4 and IL-5.Citation6 Anti-CD1d antibody blockade reduced AHR and pulmonary cell influx in an ozone challenge model.Citation7 Asperamide B, a glycosphingolipid derived from Aspergillus fumigatus, a fungus associated with severe asthma in humans, directly activated mouse NKT cells and induced AHR in mice in a CD1d-restricted fashion.Citation8 Lipids from house dust or house dust mite antigens presented by CD1d were sufficient to mobilize NKT cells to the mouse lung and induce AHR.Citation9,10 Two photon intravital microscopy studies in mice showed that NKT cells were located in the intravascular compartment of the lung, poised to rapidly respond to airborne lipid antigens in a CD1d-dependent manner.Citation11 These studies highlight the potential role of the CD1d/NKT pathway in potentiating asthma-like pathology in a spectrum of mouse models.
In humans and non-human primates, the data implicating NKT cells in asthma is less developed than in mouse models. This may be associated with the lower percentages of circulating NKT cells in humans/non-human primates than mice.Citation12-14 Nevertheless, environmental lipids (from e.g., pollens, fungi) and self-lipids presented on CD1d activate human NKT cells in vitro.Citation8,15-18 KRN7000/α-galactosylceramide (α-GalCer) is a strong CD1d-restricted lipid antigen for NKT cells. Loading of autologous dendritic cells (DCs) with α-GalCer and intravenous administration to patients increased circulating blood eosinophils, suggesting a link in humans between the activation of NKT cells and the systemic mobilization of eosinophils from the bone marrow.Citation19 NKT cell numbers were elevated in the broncho-alveolar lavage (BAL) and sputum of patients with severe asthma in some studies.Citation20,21
On balance, one may hypothesize that environmental or endogenous lipids presented via CD1d could trigger activation of NKT cells, leading to the local release of cytokines, which would drive systemic mobilization of immune cells and local airway inflammation. The differences in NKT cell biology between mouse and primate systems justify investigation of a non-human primate model of asthma. Accordingly, we developed and then tested a potent anti-CD1d antibody, NIB.2, in an Ascaris suum aerosol challenge model of asthma in cynomolgus macaques. We found that anti-CD1d antibody treatment modulated several parameters of inflammation, such as lung cytokines, particularly IL-5 and IL-8, as well as BAL lymphocytes and macrophages and blood basophils. These results demonstrate the importance of the CD1d/NKT cell pathway in driving local cytokine release and increasing pulmonary inflammation, and the therapeutic potential for CD1d blockade.
Results
NIB.2 binds human and cynomolgus CD1d with high affinity
Anti-CD1d antibody NIB.2 was isolated from a phage display library and converted to human IgG4 (see Methods). The affinity of NIB.2 for human CD1d was determined by surface plasmon resonance (SPR). NIB.2 demonstrated strong binding to recombinant human CD1d (KD value 122 pM, ) and was not cross-reactive with related human proteins CD1a, CD1b, CD1c, CD1e, MR1, and HLA-B37, a representative isoform of MHC Class I (). These proteins were correctly folded, as shown by ELISA experiments using commercially-supplied antibodies targeted against these related proteins (Fig. S1). NIB.2 did not bind β2-microglobulin (β2M), the protein subunit with which CD1d forms a complex (not shown). NIB.2 also bound cynomolgus CD1d with high affinity (KD value 115 pM, ). NIB.2 did not bind recombinant murine CD1d in ELISA () or SPR experiments (not shown).
Table 1. High affinity of NIB.2 binding to recombinant human and cynomolgus CD1d as measured by SPR
Figure 1. NIB.2 binds human and cynomolgus cell-based CD1d and demonstrates high affinity for recombinant CD1d antigens. (A) Representative SPR trace showing that NIB.2 binds strongly to human CD1d and not to related human proteins CD1a, CD1b, CD1c, CD1e, HLA-B37, and MR1. (B) NIB.2 binds recombinant human and cynomolgus CD1d/β2M expressed on transfected HEK293E cells (top panels, black histograms) and to native CD1d on primary human and cynomolgus PBMCs (bottom panels, black histograms). Negative control isotype-matched antibody showed no binding (white histograms).
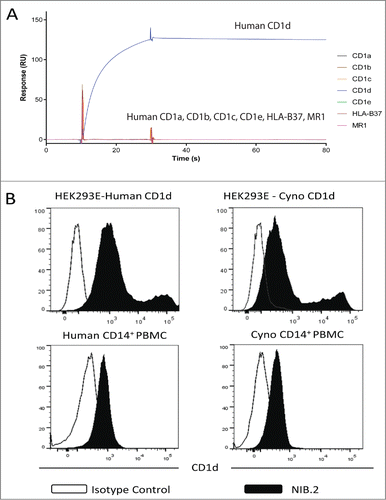
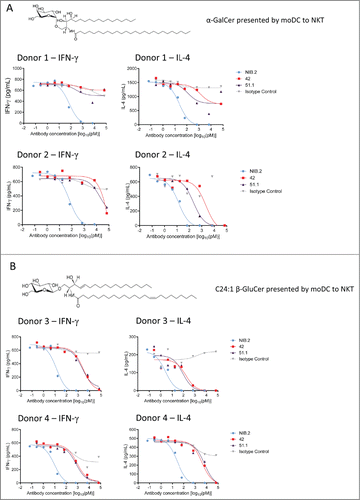
Figure 2. NIB.2 demonstrates strong neutralizing activity across 2 structurally distinct glycolipids in primary human cell-based potency assays. NIB.2 was titrated from ∼7 nM and demonstrated improved inhibition of IFN-γ and IL-4 by NKT cells after 24 h, as determined by ELISA, compared with anti-CD1d antibodies 42 and 51.1 titrated from ∼70 nM, in an assay using α-GalCer-expanded NKT cells and α-GalCer or C24:1 β-GluCer-loaded CD14+ monocyte-derived dendritic cells as CD1d-positive cells. Graphical representation of IFN-γ and IL-4 results are presented where α-GalCer (A) or C24:1 β-GluCer (B) was used as the glycolipid antigen. In all assays, the isotype human IgG4 control antibody did not markedly inhibit cytokine release compared with the reference antibodies. IC50 values from representative experiments as shown in (A) and (B) are presented in . The presented data from 2 separate donors are representative of up to n = 10 experiments with a minimum of n = 2 donors per experiment.
NIB.2 clearly bound human CD1d transfected HEK293E cells (pTT5 system) (, top left panel) in flow cytometry experiments. Similarly, NIB.2 was cross-reactive with cynomolgus CD1d transfected HEK293E cells (, top right panel). Given that cell surface CD1d in cell lines can present a number of different lipid antigens,Citation23 these results suggest that NIB.2 can recognize cell-surface CD1d molecules when a variety of different antigens may be presented. To confirm binding of NIB.2 to native CD1d, peripheral blood mononuclear cells (PBMCs) from human or cynomolgus blood were isolated and NIB.2 binding assessed by flow cytometry. NIB.2 primarily bound CD14+ cells, indicating a monocyte-like or myeloid lineage population (, bottom panels), consistent with previously reported data on cell types expressing CD1d.Citation24
NIB.2 demonstrates strong neutralizing activity across 2 structurally distinct glycolipids in primary human cell-based potency assays
To determine whether NIB.2 blocks the CD1d/NKT cell interaction in a functional manner, we utilized a primary human cell assay comprising dendritic cells and α-GalCer-expanded NKT cells. In one set of experiments, we used α-GalCer (structure in ) to stimulate NKT cell cytokine release. When α-GalCer was used, NIB.2 strongly inhibited IFN-γ, IL-4, IL-5, IL-13 and TNF cytokine release by NKT cells, with IC50 values lower than 100 pM (IFN-γ and IL-4 graphs in ; IC50 values for IFN-γ, IL-4, IL-5, IL-13 and TNF cytokines in ). In our assays, the degree of cytokine blockade by NIB.2 was higher (around 100-fold greater potency) than the reference anti-CD1d antibodies 42 and 51.1,Citation22 which showed some inhibitory activity at the highest concentration tested (∼70 nM), but a reduced level of inhibition compared with NIB.2 at lower concentrations.
Table 2. IC50 values (picomolar) from primary moDC/iNKT cell-based assays using α-GalCer. Mean IC50 values from 2 representative donors from a total of n = 10 experiments are presented. Did Not Inhibit: where inhibition was typically less than 50% of the maximal response by human NKT cells at an antibody concentration of approximately 7 nM
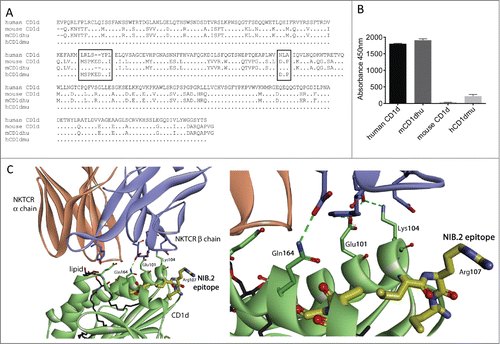
Figure 3. Characterization of NIB.2 epitope on human CD1d. (A) Amino acid sequence alignment of human CD1d with mouse CD1d and human/mouse hybrid constructs. Boxed regions denote potential residues of the NIB.2 epitope. (B) Graphical representation of ELISA results showing that NIB.2 binds human CD1d and “mCD1dhu,” a construct of mouse CD1d in which residues 106–114 (MSPKEDYPI) and 162–164 (DLP) were replaced with human CD1d sequence residues 106–112 (LRLSYPL) and 160–162 (NLA), respectively. In contrast, NIB.2 does not bind mouse CD1d or “hCD1dmu,” a human CD1d construct in which residues 106–112 (LRLSYPL) and 160–162 (NLA) were replaced with murine CD1d sequence residues 106–114 (MSPKEDYPI) and 162–164 (DLP), respectively. (C) Crystal structure of the interaction between human CD1d (green ribbon) and the Type 1 NKT cell receptor (brown ribbon – α chain; blue ribbon – β chain) with α-GalCer as a representative glycolipid (black) labeled. The partial epitope of NIB.2 on CD1d (yellow highlighted) is in close proximity to the binding site of the NKT cell receptor β chain to CD1d, suggesting that NIB.2 acts by inhibiting this interaction (crystal structure 3HUJCitation26).
A C24:1 N-acyl variant of an endogenous lipid, β-D-glucopyranosylceramide (C24:1 β-GluCer, structure in ), which differs from α-GalCer in the orientation (β- versus α-anomeric linkage) and structure (glucosyl vs. galactosyl) of the polar head group, is described as having weak activity for human NKT cells.Citation18 In our assay, C24:1 β-GluCer proved to be a weaker CD1d-restricted antigen than α-GalCer, since a higher concentration of C24:1 β-GluCer was required to achieve sufficient dynamic range in the assay. Anti-CD1d antibody NIB.2 showed orders of magnitude greater potency than antibodies 42 and 51.1 when C24:1 β-GluCer was used (IFN-γ and IL-4 graphs in ; IC50 values for IFN-γ, IL-4, IL-5, IL-13 and TNF cytokines in ). These results demonstrate that NIB.2 is able to potently inhibit CD1d-mediated lipid presentation and subsequent activation of NKT cells in the context of 2 structurally distinct glycolipids.
Table 3. IC50 values (picomolar) from primary moDC/iNKT cell-based assays using C24:1 β-GluCer. Mean IC50 values from 2 representative donors from a total of n = 3 experiments are presented. Did Not Inhibit: where inhibition was typically less than 50% of the maximal response by human NKT cells at an antibody concentration of approximately 7 nM
In similar potency assays where THP-1 monocyte-like cells were loaded with α-GalCer or C24:1 β-GluCer and co-cultured with α-GalCer-expanded human NKT cells, NIB.2 demonstrated significantly improved inhibition of IFN-γ, IL-4, and IL-13 cytokine release by NKT cells, with IC50 values typically lower than 100 pM (Fig. S2). The extent of inhibitory activity by NIB.2 was significantly higher than 42 and 51.1, which clearly showed inhibition of cytokine release, albeit at higher antibody concentrations.
Characterization of the NIB.2 epitope on human CD1d
We considered the possibility that NIB.2 might recognize CD1d in complex specifically with a particular lipid. However, a number of lines of evidence suggest that the epitope bound by NIB.2 is recognized when a variety of lipid antigens are present in complex with CD1d: (1) NIB.2 binds to CD1d on transfected HEK cells and native CD1d on human PBMCs () – these cells would be expected to present a diverse array of lipids;Citation16,23 (2) NIB.2 neutralizes NKT cell cytokine release activity mediated by CD1d+ cells loaded with 2 different glycolipids (); and (3) epitope mapping, as described below.
To elucidate the epitope of NIB.2 on human CD1d, we performed hydrogen/deuterium experiments similar to those reported previously,Citation25 using a Fab fragment from NIB.1, which is a closely related precursor molecule of NIB.2. These experiments revealed regions between residues 108–113 and 145–161 that were protected (, where % Difference is less than −10%. With these results, the sequence of human CD1d was aligned with murine CD1d () and a series of recombinant CD1d constructs produced. “hCD1dmu” was human CD1d in which amino acids located between positions 106 and 112 (LRLSYPL) and 160–162 (NLA) were replaced with the murine CD1d sequence. “mCD1dhu” was murine CD1d in which the amino acids located between positions 106 and 114 (MSPKEDYPI) and 160–162 (DLP) were replaced with corresponding amino acids from the human CD1d sequence (). Given that NIB.2 does not bind to mouse CD1d (), this epitope reconstitution strategy was directed toward identifying human CD1d sequences critical for NIB.2 binding. All CD1d constructs were recombinantly expressed in HEK293E cells and proteins purified, then detected by polyclonal antibodies against human or mouse CD1d in an ELISA format (not shown). NIB.2 bound to human CD1d, but not mouse CD1d (). NIB.2 bound to mCD1dhu, a mouse CD1d into which residues 106–112 and 160–162 of human CD1d had been introduced, suggesting human CD1d sequence amino acids 106–112 and 160–162 are involved in the binding of NIB.2 to human CD1d (). Likewise, when these amino acids on human CD1d were replaced by mouse sequence at the corresponding locations (hCD1dmu), NIB.2 showed substantially reduced binding (). Taken together these results indicate that the sequence of human CD1d between residues 106–112 and 160–162 form part of the epitope on CD1d to which NIB.2 binds.
Table 4. Hydrogen/deuterium exchange experiments to determine anti-CD1d antibody epitope. Analysis of the effects of a Fab from NIB.1 (a closely related precursor molecule of NIB.2) on the rate of deuterium/hydrogen exchange on human CD1d located a region between residues 108–113 and 145–161 that was protected. % Difference refers to the difference in deuteration levels in each segment of CD1d after on/off exchange experiments at 23°C, pH 7
When these regions were analyzed on an X-ray crystal structure of human CD1d in complex with a Type 1 NKT cell receptor (crystal structure 3HUJ),Citation26 they were found in close proximity to each other in the tertiary structure, even though these individual regions are distant in primary sequence (). Both LRLSYPL (amino acids 106–112) and NLA (amino acids 160–162) are located on or in close proximity to the α-helices of CD1d and in close proximity to the binding site of the Type 1 NKT cell receptor β-chain on human CD1d (). The significance of this observation is that the potential epitope of NIB.2 is distinct from the lipid binding groove of CD1d. This supports the concept that NIB.2 inhibits the interaction of the NKT cell receptor β-chain with CD1d, and thus prevents downstream activation of the NKT cell.
NIB.2 decreases lung cytokine release in a cynomolgus macaque Ascaris suum challenge model of asthma
We investigated the effect of NIB.2 on various physiological endpoints in a well-established lung challenge model in Ascaris suum-sensitive cynomolgus macaques (Macaca fascicularis) to test whether blockade of the CD1d/NKT interaction by NIB.2 translates to modulation of inflammation in vivo. Prolonged and repeated inhalation challenges induce allergic responses in cynomolgus macaques in this model.Citation27-31 A sequential study design was used. A. suum-sensitive macaques (n = 10) were dosed in a blinded manner in 2 stages: first with vehicle (buffer only on days -8, -5 and -1), followed by A. suum challenge and analysis of endpoints (), including pulmonary function, enumeration of BAL and blood leukocyte subsets, and cytokine expression in BAL cell-free supernatants. After a 17-day washout period, the study was repeated with the same macaques dosed with NIB.2 (3 intravenous doses at 20 mg/kg) instead of vehicle.
Figure 4. NIB.2 decreased lung cytokine release in a cynomolgus macaque Ascaris suum challenge model of asthma. (A) Model schematic, showing sequential study design and endpoints (n = 10 macaques). (B) Pulmonary A. suum challenge induced IL-5, IL-8, IL-1 receptor antagonist and G-CSF in the lungs of challenged cynomolgus macaques at day 1 post challenge. Compared with vehicle, NIB.2 treatment significantly reduced BAL levels of IL-5, IL-8 and IL-1 receptor antagonist to baseline (dotted lines in graphs). (C) Compared with vehicle, NIB.2 significantly reduced baseline levels of GM-CSF, IL-6, IL-15, IL-12/23p40, MIP-1α, MIP-1β and VEGF, with trend to reduction of TNF (not significant). *ρ < 0.05, **ρ < 0.01.
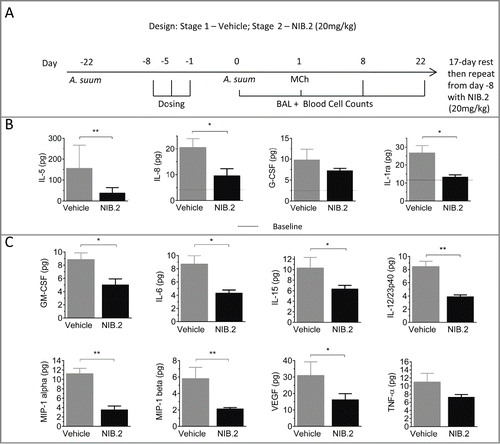
We assessed cell-free supernatants from BAL taken from the right and left lung lobes at 1, 8 and 22 d following pulmonary A. suum challenge. The peak cytokine response was at day 1 after A. suum challenge, with IL-5, IL-8, IL-1 receptor antagonist and G-CSF expression above baseline measurements taken at day 0 (, mean at baseline indicated by dotted lines versus day 1 measurements after vehicle treatment, gray bars). NIB.2 treatment led to a blockade of these cytokines, especially IL-5 and IL-8 at day 1 (). Furthermore, although there was no induction of GM-CSF, IL-6, IL-15, IL-12/23(p40), MIP-1α, MIP-1β and VEGF at day 1 following A. suum challenge, NIB.2 treatment led to statistically significant reduction of these cytokines (; baseline levels in vehicle stage in gray; NIB.2 in black). There was also a trend to reduction with BAL TNF following NIB.2 treatment, although this result did not reach statistical significance (ρ = 0.0645, ). Overall, NIB.2 treatment reduced BAL levels of particular cytokines implicated in airway inflammation, such as IL-5 and IL-8.
NIB.2 modulates BAL and blood inflammatory cell composition and airway hyper-reactivity in the A. suum model
Aerosol challenge with A. suum increased BAL macrophages, eosinophils and neutrophils, with peak responses at day 1 post challenge. NIB.2 showed a trend to reduction in the total number of BAL cells compared with vehicle, although this result was not statistically significant (). A. suum challenge induced eosinophils at both day 1 () and day 8 (not shown) post challenge. At day 1 post challenge, the A. suum-induced increase in eosinophils was greater in the NIB.2-treated group than the vehicle (), and eosinophil levels were similar between groups at day 8 post challenge (not shown). There was no overall reduction in either total numbers or percentage of neutrophils with NIB.2 treatment compared with vehicle (). Although statistical significance was not reached, NIB.2 showed a trend to reduction of BAL macrophage numbers compared with vehicle (). At day 1 post A. suum challenge, there was no overall induction of BAL lymphocytes. However, BAL lymphocyte numbers and percentages were significantly reduced after NIB.2 treatment compared with vehicle (). There were no marked changes to BAL mast cells after A. suum challenge or NIB.2 treatment ().
Figure 5. Effect of NIB.2 on lung and systemic cellular changes and AHR in a cynomolgus macaque Ascaris suum challenge model of asthma (n = 10). Changes in BAL leukocyte numbers (A), percentages of leukocyte subsets in BAL (B) and blood leukocyte numbers (C) at day 1 post challenge with A. suum. NIB.2 treatment reduced blood basophils, reaching significance at days 8 and 22 post challenge (D). Compared with vehicle, NIB.2 treatment showed trend to reduced airway hyper-reactivity (E), but this did not reach statistical significance (F), as shown by measuring the area under the curve (AUC) from 0–10 mg/mL methacholine (MCh) from E. *ρ < 0.05, **ρ < 0.01.
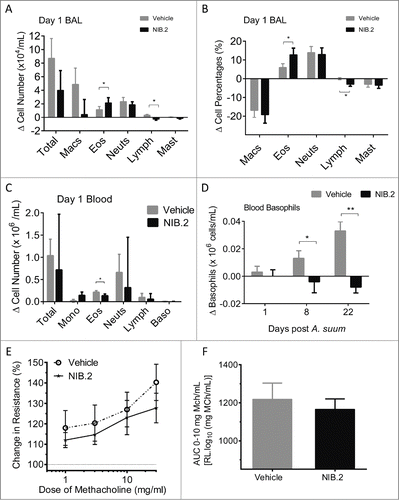
At day 1 post A. suum challenge, there was an increase in the total number of peripheral blood leukocytes, particularly eosinophils, lymphocytes and basophils (). NIB.2 reduced by approximately 25% the mean number of total blood leukocytes, though this result was not statistically significant. NIB.2 treatment significantly reduced blood eosinophils (). NIB.2 reduced by approximately 50% the Ascaris-induced blood neutrophils, although statistical significance was not reached (). Further, NIB.2 significantly reduced the Ascaris-induced increases in blood basophils at 8 and 22 d after challenge compared with vehicle ().
Dynamic lung resistance in response to MCh challenge was measured approximately 24 h after A. suum exposure in both treatment stages. To assess the effect of NIB.2 vs. vehicle on modulation of lung function, the area under the curve (AUC) for the dose response curves for the dose range 0 to 10 mg/mL MCh was calculated. The MCh-induced airway resistance measured 24 h post A. suum challenge showed a trend toward reduction in NIB.2 versus vehicle, as indicated by reduced dynamic airways resistance values at varying doses of MCh (). However, when measured as the AUC in the dose range of 0–10 mg/mL MCh this apparent reduced airways resistance was not statistically significant (ρ = 0.6250, ).
Discussion
In this report, we describe a monoclonal anti-CD1d antibody NIB.2 that shows potent neutralizing activity in vitro and in vivo blockade of lung inflammation in a cynomolgus macaque AHR model. In vitro NIB.2 shows improved potency relative to previously described anti-human CD1d antibodies, 42 and 51.1.Citation22 This enhanced potency could be explicable in terms of increased affinity or by epitope specificity. The epitope of NIB.2 was characterized herein and is distinct from 42 and 51.1 based on the lack of competition of NIB.2 binding to human CD1d in a competition ELISA (data not shown). To characterize the epitope of the antibody on CD1d, we used hydrogen-deuterium exchange to identify key binding regions on CD1d. We then introduced these regions into mouse CD1d (to which NIB.2 does not bind) and recovered binding of NIB.2 confirming these regions on CD1d as critical for NIB.2 binding. NIB.2 binds regions on or in close proximity to the α-helices of CD1d in a manner that inhibits Type I NKT cell receptor β-chain interactions with CD1d. The majority of human CD1d-restricted NKT cells (known as Type 1 NKT) express a semi-invariant T cell receptor comprising Vα24Jα18 paired with Vβ11.Citation1 There is also a smaller, more diverse NKT cell population (Type 2 NKT) that do not express the canonical semi-invariant T-cell receptor rearrangement. These cells are less frequent than Type 1 NKT cells and their biological significance in asthma is not as well characterized. The X-ray crystal structures of some Type 2 NKT cell receptors in complex with CD1d have been solved (crystal structures 3HE6, 4EN3Citation26,32,33), and it is now known that there are distinct differences between the docking modes of Type 1 vs. Type 2 NKT cell receptors with human CD1d. The predicted epitope of NIB.2 suggests inhibition of Type 1, but perhaps not Type 2 NKT cell interactions with CD1d. This makes sense given that NIB.2 was identified by screening assays that used Type 1 NKT cells. Inhibition of Type 1 NKT cell activity may be sufficient in human asthma, especially since studies implicating NKT cells in asthma generally refer to semi-invariant (Type 1) NKT cells.Citation20,21
NIB.2 has been shown to block the CD1d/NKT interaction across structurally distinct glycolipids presented by CD1d. This is important from a translational perspective where a desirable attribute of an effective antibody therapeutic is binding of CD1d independent of the glycolipid antigen presented. To demonstrate this, we showed that NIB.2 bound human and cynomolgus CD1d on the surface of transfected HEK293E cells, and on native CD1d in human and cynomolgus PBMCs. We also demonstrated in vitro potency of NIB.2 in cell-based assays using 2 distinct glycolipids, α-GalCer and C24:1 β-GluCer. Because CD1d can present a range of lipids naturally,Citation23 these results support the concept that NIB.2 should confer neutralizing activity in a lipid antigen-independent manner.
The high potency of NIB.2 in vitro suggests strong neutralization by NIB.2 of NKT cell signaling and cytokine release through blockade of the CD1d/Type 1 NKT interaction (). This in vitro activity is also exhibited in vivo, as shown by blockade of certain A. suum-induced cytokines, such as IL-5 and IL-8 (), which are known to influence the recruitment and function of eosinophils and neutrophils, respectively. Further, although A. suum did not increase levels of the inflammatory mediators GM-CSF, IL-6, IL-15, IL-12/23p40, MIP-1α, MIP-1β, VEGF or TNF, the levels of these mediators were reduced after NIB.2 treatment when compared with vehicle treatment. In addition, there was evidence of NIB.2-mediated reduction of peripheral blood eosinophils and basophils and BAL lymphocytes.
IL-13 has been implicated as an important cytokine in AHR induction, and as such, was included in the panel of cytokines monitored. At the timepoints measured, IL-13 was detected at low levels in BAL samples from placebo and treated groups, with no modulation of IL-13 levels by NIB.2 observed (data not shown). Peak A. suum challenge-induced IL-13 expression in BAL has been reported as 6h post-challenge.Citation28 Based on animal welfare considerations, the earliest BAL assessment in the present study was at 24h, and given the low levels of IL-13 detected, this is likely to be beyond the optimal time window to observe suppression by NIB.2.
In the Ascaris suum challenge model in cynomolgus macaques, increased dynamic airways resistance to inhaled MCh is a well-established hallmark of AHR. NIB.2 treatment showed a trend toward reduction in airways resistance, which is notable because only a few biologics have been reported to reduce AHR in this model.Citation28 Other biological therapeutics were either reported not to improve airways resistance in cynomolgus macaque AHR models,Citation30 or AHR results were not reported.Citation31 It is also notable that, although eosinophils are a distinguishing feature of the Ascaris suum cynomolgus model, in human asthma, eosinophils are only one component of a multi-faceted, chronic allergic inflammatory state that also features other leukocyte subsets including lymphocyte-driven activation.
Therapeutic agents that directly target eosinophil activity/function reduce eosinophil numbers in Ascaris challenge models.Citation30,31 We show here that the anti-CD1d antibody modulates cellular parameters, such as blood basophils, blood eosinophils and BAL lymphocytes, suggesting that inhibition of local pulmonary cytokine release has concomitantly effected reduction in parameters of systemic and pulmonary cellular inflammation.
The Ascaris suum challenge model is known to drive acute eosinophilia, and this is observed at day 1 post aerosol challenge, a reasonably early timepoint. The Ascaris suum extract may induce eosinophils in an acute manner, as well as through a T cell-dependent response. The eosinophil-centric responses at 24 h post-challenge may represent a skewed acute innate immune response rather than a T cell-dependent adaptive response. The mechanism of action of NIB.2 is predicted to have indirect effects on eosinophilia through the modulation of cytokines, and therefore sustained dosing may be required to modulate BAL eosinophils. The modulation of NIB.2 on blood eosinophil and basophil numbers but not lung eosinophils suggests that local blockade of cytokines translates into reduced systemic levels of cells (perhaps egressing from bone marrow), but that it may take longer to see similar effects on local infiltrating eosinophils with CD1d blockade.
In summary, NIB.2 demonstrated strong in vitro blockade of NKT cell activation and this translated into potent in vivo modulation of multiple inflammatory endpoints through the modulation of lung cytokines and cellular subsets infiltrating into the lung and in circulation. It is possible that with an extended dosing regimen typical of clinical administration of biologics, NIB.2 treatment has the potential to significantly improve the regulation of cellular infiltration and AHR seen in this model of allergic airway inflammation.
Methods
Generation of Antibody NIB.2
Antibody NIB.2 was isolated from a naïve human phagemid library. Antigen-binding fragments (Fabs) that bound to both human and cynomolgus CD1d/β2M were converted to full antibodies of the human IgG4 isotype.
Cynomolgus CD1d
Cynomolgus macaque (Macaca fascicularis) spleen (Biochain) was obtained and primers (F1 – GTGCCTGCTGTTTCTGCTG; R1 – TGCCCTGATAGGAAGTTTGC) based on rhesus CD1d mRNA (PubMed Accession number: NM_001033114) were used to amplify cynomolgus CD1d cDNA. A 1 kb DNA product was amplified by PCR, ligated into pGEM-T Easy (Promega) and sequenced using M13 forward and reverse primers.
Recombinant protein expression
Recombinant proteins such as CD1d/β2M constructs were produced in a mammalian HEK293E/pTT5 expression system.Citation34 Human CD1d/β2M was expressed in HEK293E cells using a DNA expression construct encoding the extracellular domain of CD1d (residues 1–301, from UniProt Accession number P15813) with a C-terminal protease cleavage site and a polyhistidine tag, co-transfected with a DNA expression construct encoding β2M. The CD1d/β2M protein complex was purified from supernatant by nickel affinity chromatography using a HisTrapTM FF column (GE Healthcare Life Sciences, 17–5255–01). Proteins were then concentrated and buffer exchanged using Amicon Ultra centrifugal filters (Millipore, Cat # UFC801024). Similar methods were used for expression and purification of other recombinant proteins with a C-terminal polyhistidine tag, such as human CD1a, CD1b, CD1c, CD1e, MR1 and HLA-B37, mouse CD1d and cynomolgus CD1d. To ensure that the purified proteins were folded correctly, proteins were detected using an ELISA-based approach involving capture of the proteins on a high protein-bind plate (ThermoFisher, 442404) and recognition using commercial mouse IgG antibodies specific for the various targets and optimized for ELISA assays (anti-CD1a: Biolegend, 300102; anti-CD1b: Biolegend, 329102; anti-CD1c: Biolegend, 331502; anti-HLA-B37 detected with anti-HLA-A,B,C: Biolegend, 311402; anti-MR1: Abnova, H000003140-M03). Mouse IgGs were then detected in ELISA format with a HRP-conjugated anti-mouse IgG (Dako, P0447). For flow cytometry experiments to detect CD1d/β2M on the cell surface of HEK293E cells, pTT5 constructs of human CD1d were co-transfected with β2M and harvested 48 h after transfection. To express recombinant NIB.2 antibody, a DNA vector containing NIB.2 heavy and light chains was stably expressed in Chinese hamster ovary cells and cell-free supernatant from fed-batch overgrows in a bioreactor was assessed for protein yield by Protein-A HPLC. NIB.2 was purified by a chromatography schema including Protein A.
Flow cytometry of transfectants and peripheral blood mononuclear cells
HEK293E cells were transfected with human or cynomolgus CD1d/β2M constructs for 24 h, washed several times in cold PBS with 2% w/v bovine serum albumin (BSA), incubated at 2–8°C for 30 min with NIB.2 (∼70 nM) or human IgG4 isotype control (Sigma Aldrich, I4639) and counter-stained with fluorescein isothiocyanate-conjugated anti-human IgG4 (Sigma-Aldrich, F9890). Human or cynomolgus PBMCs were isolated from buffy coats or whole blood by centrifugation over a density gradient (Axis-Shield, LymphoprepTM) with the brake off. The PBMC layer was washed several times in cold PBS with 2% w/v BSA and stained with NIB.2 or human IgG4 isotype control as described above. Cells were co-stained with Pacific BlueTM-conjugated anti-human CD14 (BD Biosciences, 558121), phycoerythrin-conjugated anti-human CD11c (Biolegend, 337216), allophycocyanin-conjugated anti-human CD3 (BD, 340440) and allophycocyanin/Cy7-conjugated anti-human CD20 (Biolegend, 302314) to identify leukocyte populations. NKT cell populations were analyzed by flow cytometry using phycoerythrin-conjugated α-GalCer-loaded CD1d tetramers (ProImmune, D001–2), peridinin chlorophyll protein-conjugated anti-CD3 (BD Biosciences, 347344) and allophycocyanin-conjugated anti-Vα24Jα18 (Miltenyi, 130–094–839). The isotype control human IgG4 antibody (Sigma Aldrich, I4639) was negative for binding in all experiments.
Characterization of NIB.2 by surface plasmon resonance
A Biacore T200 biosensor (GE Healthcare) was used to determine the affinity of NIB.2 for human CD1d and related proteins. The surface of a CM5 Series S sensor chip (GE Healthcare, BR-1005–30) was immobilized with protein A (Pierce, 21181) to 3000 resonance units using standard amine coupling chemistry. Interactions were measured at 25°C with 10 Hz data collection. Running buffer was 10 mM HEPES-Na pH 7.5, 150 mM NaCl, 3 mM EDTA, 0.005% Tween 20. NIB.2 was captured to a density of approximately 350 resonance units on flow cell 2. Flow cell 1 was used as a blank reference. The human CD1d and related protein analytes were injected over a concentration range of 6.8 to 217 nM, and a buffer reference was also included. Contact times for the association and dissociation phases were 30 seconds and 600 seconds, respectively. The data was processed by subtracting the uncoated surface signal from a blank injection of running buffer and the corrected signal subtracted from the sample binding curves (double referencing). Kinetic binding analysis was performed using Biacore T200 evaluation software and data was fitted using a 1:1 Langmuir binding model to generate ka, kd and KD values.
Isolation and expansion of NKT cells
PBMCs were enriched for Type 1 NKT cells by positive selection using MACSTM microbeads against the Vα24-Jα18 iNKT junction region (Miltenyi, 130–094–842). Monocytes used as CD1d+ stimulator cells were treated with mitomycin C (Sigma-Aldrich, M4287) and loaded with α-galactosylceramide, ((2S,3S,4R)-1-O-(D-galactosyl)-N-hexacosanoyl-2-amino-1,3,4-octadecanetriol, Fukanoshi, KRN7000) to a final concentration of 116.5 nM and co-cultured at 37°C and 5% CO2 at a 1:1 ratio with 1 × 104 NKT cells per well in 96-well round bottom plates. At 40 h post incubation, human IL-2 (R&D Systems, 202-IL-500) was added to a final concentration of 667 pM. Cells were kept in culture for 7–14 d and phenotyped by flow cytometry.
Human NKT cell potency assays
α-GalCer or the C24:1 N-acyl variant of β-D-glucopyranosylceramide, “C24:1 β-GluCer” (D-glucosyl-ß-1,1′ N-(15Z-tetracosenoyl)-D-erythro-sphingosine, Avanti, 860549) was solubilized in dimethylsulfoxide at 37°C for 2 h. NKT cells were used in assays at purity of 70% or higher as determined by flow cytometry. To perform the assay, monocyte-derived dendritic cells or THP-1 cells were cultured in 96-well flat bottom plates at 2 × 104 cells per well and loaded with α-GalCer at 116.5 nM or C24:1 β-GluCer at ∼12 μM for 1 h. Anti-CD1d, NIB.2, reference or control antibodies were added to the cultures for 1 hr, prior to addition of previously α-GalCer-expanded NKT cells in a 1:1 ratio with the dendritic cells. After approximately 16 h co-culture, cell-free supernatants were assayed for IFN-γ, IL-4, IL-5, IL-13 and TNF levels by ELISA (R&D Systems). Mouse anti-human CD1d monoclonal IgG antibodies 42 (BD Biosciences, 550254) and 51.1 (eBioscience, 14–0016–82 or Biolegend, 350304) were used as reference antibodies. Human IgG4 (Sigma-Aldrich, I4639) was used as an isotype-matched negative control. For the cell-based potency assays, data is presented in single-point form and is representative of n = 10 experiments (where α-GalCer was used) or n = 3 experiments (where C24:1 β-GluCer was used) where each experiment was done with at least n = 2 donors. A three-parameter curve of best fit (log10 antagonist versus response) was conducted using GraphPad Prism (version 6). IC50 data from the presented representative experiments is shown in . For IC50 data, antibody concentrations in μg/mL are converted to molar equivalents based on a conservative assumption that the molecular weight of IgG is ∼150 kDa.
Epitope mapping experiments
Preparation of Fabs. Prior to finalization of the NIB.2 sequence, epitope mapping was performed with a closely-related precursor antibody NIB.1 which varied from NIB.2 only by 3 residues in variable heavy chain framework 1. To prepare Fab for hydrogen/deuterium exchange experiments, NIB.1 was prepared by papain digest (Pierce, 44985) according to the manufacturer's instructions. Intact Fab was removed from Fc (fragment crystallizable) containing protein by running the sample over a HiTrap Protein A column, (GE Healthcare, 17–0402–01) equilibrated with 1x PBS pH 7.0 and collecting the flow-through. Fabs were analyzed by size exclusion chromatography (SEC) using a TSK gel G3000SWx1 column (TOSOH) at 0.5 ml/min with 1X PBS as a running buffer. The results indicated that the Fabs were >95% pure (not shown). To confirm binding of the Fabs to human CD1d, an ELISA was performed in which human CD1d was coated at 1 μg/mL in PBS onto a 96-well NuncTM MaxisorpTM ELISA plate (ThermoFisher, 442404) overnight at 4°C. The wells were then washed with 3 separate washes with 1X PBS with 0.05% Tween-20 (Sigma, P1379). The wells were blocked with 1% BSA in PBS for 1 h at room temperature. The wells were then washed with 1X PBS as described above. A titration of Fab or full-length antibody was then performed in serial dilutions from a concentration of 10 μg/mL with a PBS only control included. One hundred microliters per well of secondary antibody (Goat Anti-human Kappa F+B HRPO Conjugate, Invitrogen, 1:2000 dilution) was added for 1 h at room temperature followed by addition of 50 μL of TMB (Sigma, T0440) and reaction was stopped by adding 50 μL of 1 M HCl after color development. The absorbance was read at 450 nm (referenced at 620 nm).
Hydrogen/Deuterium Exchange. These experiments were performed in a similar manner to that described.Citation25 Briefly, human CD1d was diluted in 1 X PBS (pH 7) to a concentration of 12.8 μM and then equilibrated with Fab fragment of NIB.1 at 14.1 μM. 14 μL of this solution was mixed with 26 μL of 50 mM Phosphate pH 7 in D2O (where D is deuterium). Separate solutions were prepared and incubated for 30, 100, 300 or 1000s at 23°C. At the end of the incubation period 20 μL of 2 M urea, 1 M TCEP pH 3.0 was added to each solution. The sample was then passed over an immobilized pepsin column at 200 μL/min in 0.05% trifluoracetic acid (TFA) in H2O. Peptic fragments were loaded onto a reversed-phase trap column and desalted with 0.05% TFA in H2O for 3 mins. Peptides were separated by a C18 column with a linear gradient of 13% to 35% of buffer comprising 95% acetonitrile, 5% H2O, 0.0025% TFA) over 23 mins. The peptides were then analyzed by mass spectrometry in profile mode. Fully deuterated control samples were prepared by mixing 32.2 μL of 0.615 mg/mL CD1d with 59.8 μL of 100 mM TCEP in D2O and incubating at 60°C for 3 h.
CD1d human:mouse constructs
Numbering of residues of CD1d in this manuscript includes the signal sequence of the CD1d protein. Human CD1d in which residues 106–112 (LRLSYPL) and 160–162 (NLA) were replaced with murine CD1d sequence residues 106–114 (MSPKEDYPI) and 162–164 (DLP), respectively, was generated by gene synthesis and designated huCD1dmu. Mouse CD1d in which residues 106–114 (MSPKEDYPI) and 162–164 (DLP) were replaced with human CD1d sequence residues 106–112 (LRLSYPL) and 160–162 (NLA), respectively, was generated by gene synthesis and designated muCD1dhu. Both constructs were expressed in HEK293E/pTT5 and purified as described above.
CD1d human:mouse construct ELISA
All solutions were added at 50 μL/well. Human and mouse CD1d, hCD1dmu and muCD1dhu were coated on 96-well NuncTM MaxisorpTM ELISA Plates (ThermoFisher, 442404) at 1 μg/mL in PBS overnight at 4°C. The wells were washed with buffer (PBS + 0.05 % Tween-20) 3 times. The plates were blocked for 1 h at room temperature in blocking buffer (PBS + 1% BSA) before being washed 3 times. NIB.2 or controls in diluent (PBS + 1% BSA + 0.05% Tween-20) were added to the wells in a half log titration starting from 10 μg/mL, and no antibody (0 μg/mL) was included as a negative control. The plate was incubated at room temperature for 1 h, and then washed as described above. Secondary horseradish peroxidase-conjugated goat anti human IgG antibody (directed against both heavy and light chains) (Life Technologies, 62–7120) was added at 1:2000 dilution in antibody diluent and incubated for 1 h at room temperature. After washing the plate, 50 μL of 3,3′,5,5′-tetramethylbenzidine (Sigma-Aldrich, T0440) was added to each well. 50 μL of 1 M hydrochloric acid was added after color development to stop the reaction. The absorbance of each well of the plate was read at 450 nm (referenced at 620 nm).
Ascaris suum model of asthma in cynomolgus macaques
All animal procedures were conducted under protocols reviewed and approved by the Animal Ethics Committee of the testing facility. Ten male cynomolgus macaques (∼14–16 y old at start of experiments) were selected on the basis of positive bronchoconstrictor responses to A. suum. Animals were already sensitized to A. suum, but had not been challenged with the allergen for more than 12 months prior to study start. Each animal was exposed to A. suum (Greer Laboratories) by nebulization to determine their level of sensitivity 22 d prior to the first treatment. Dosing groups were blinded to the technical staff and Study Director until the conclusion of the study period. The macaques were treated by intravenous slow bolus with vehicle (buffer for antibody) in Stage 1 or NIB.2 at a dose of 20 mg/kg in Stage 2 (). For experimental procedures, fasted animals were sedated with ketamine (KetaVedTM ∼8–10 mg/kg by intramuscular injection) and anesthetized with isoflurane (Vet One 4–5% for induction, 1–2.5% for maintenance). An appropriately sized endotracheal tube was placed in the trachea of the anesthetized animal and an esophageal balloon catheter connected to a differential pressure transducer (Validyne) was used to measure pulmonary pressure. Macaques were mechanically ventilated (volume ∼15–18 mL/kg; ∼30 breaths/min) during each of the experiments. Airflow and tidal volume was measured using a pneumotach (3500 series [0–35 L/min]; Hans Rudolph) located in front of the endotracheal tube and connected to a differential pressure transducer (Validyne). Pulmonary function measurements were calculated from the simultaneous measurement of transpulmonary pressure, tidal volume and respiratory flow using a custom-designed software acquisition system (EMKA Technologies). During anesthesia animals were placed on a circulating water heating pad and covered with blankets to maintain a safe body temperature. The animals’ temperature, heart rate and blood O2 saturation were monitored while under anesthesia and during recovery.
Measurement of Airway Hyper-reactivity to Ascaris and methacholine
Ascaris suum extract (Greer Laboratories, XPB33 × 1A50) was suspended in sterile water (5 mg/mL) on the day of use and delivered via PARI LC nebulizer connected to the inspiratory line of the respirator by a 2-way breathing valve. All animals received the same dose of A. suum throughout the study (exposure duration of 30 seconds). MCh (TCI, Cat # M0073) was similarly prepared in sterile water on the day of use and delivered by inhalation using a HEART® nebulizer. Five breaths of increasing doses in half log increments were delivered until an approximately 150% increase in resistance over baseline was reached.
Blood and BAL Analysis
Blood samples were taken under anesthesia using potassium EDTA as anticoagulant and examined for hematology parameters and cell differentials (Advia). BAL was performed under anesthesia by guidance of a pediatric fiberoptic bronchoscope past the carina into the right and left upper lobe of the lungs and wedged into a fifth to seventh generation bronchus. Two lobes were lavaged at each of the time points with three 5 mL aliquots of sterile saline (pH 5.5). Supernatant was separated from the cells by centrifugation (up to 1600 rpm, 10 min, 4°C), and lavage fluid analyzed by LuminexTM multiplex assay (Millipore, MXPRCYMGPX23) for IL-4, IL-5, IL-10, IL-13, IFN-γ, TNF, IL-2, IL-15, IL-12/23p40, IL-1 receptor antagonist, IL-6, IL-8, GM-CSF, G-CSF, MIP-1α, MIP-1β, TGF-α, sCD40L, MCP-1, VEGF, IL-17A, IL-1β, IL-18. Measurements were extrapolated to total protein levels. BAL cells were prepared by cytospin and examined using a modified Wright-Giemsa stain, with differential counts on at least 200 nucleated cells per slide to enumerate neutrophils, macrophages, lymphocytes, mast cells and eosinophils. The total cell numbers were normalized to the recovery volume for individual lavage washes. Results from each stage (vehicle vs. NIB.2) were expressed as the change in cell number or percentage relative to the stage-specific day 0 timepoint (prior to A. suum challenge).
Statistical analysis
Data is presented as means ± SEM. Within the A. suum challenge study, differences in analyzed parameters following vehicle or NIB.2 treatment (normalized to change from day 0 baseline) were tested for statistical significance (ρ < 0.05) by a non-parametric paired test (Wilcoxon matched-pairs signed rank test for paired data with non-Gaussian distribution).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental_Material.zip
Download Zip (1.2 MB)Acknowledgments
The authors wish to thank the Australian Red Cross Blood Service for provision of human buffy coat samples.
Supplementary Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Rossjohn J, Pellicci DG, Patel O, Gapin L, Godfrey DI. Recognition of CD1d-restricted antigens by natural killer T cells. Nat Rev Immunol 2012; 12:845-57; PMID:23154222; http://dx.doi.org/10.1038/nri3328
- Zeng D, Liu Y, Sidobre S, Kronenberg M, Strober S. Activation of natural killer T cells in NZB/W mice induces Th1-type immune responses exacerbating lupus. J Clin Invest 2003; 112:1211-22; PMID:14561706; http://dx.doi.org/10.1172/JCI200317165
- Morshed SR, Takahashi T, Savage PB, Kambham N, Strober S. Beta-galactosylceramide alters invariant natural killer T cell function and is effective treatment for lupus. Clin Immunol 2009; 132:321-33; PMID:19564135; http://dx.doi.org/10.1016/j.clim.2009.05.018
- Murdoch JR, Lloyd CM. Chronic inflammation and asthma. Mutat Res 2010; 690:24-39; PMID:19769993; http://dx.doi.org/10.1016/j.mrfmmm.2009.09.005
- Akbari O, Stock P, Meyer E, Kronenberg M, Sidobre S, Nakayama T, Taniguchi M, Grusby MJ, DeKruyff RH, Umetsu DT. Essential role of NKT cells producing IL-4 and IL-13 in the development of allergen-induced airway hyperreactivity. Nat Med 2003; 9:582-8; PMID:12669034; http://dx.doi.org/10.1038/nm851
- Lisbonne M, Diem S, de Castro Keller A, Lefort J, Araujo LM, Hachem P, Fourneau JM, Sidobre S, Kronenberg M, Taniguchi M, et al. Cutting edge: invariant V alpha 14 NKT cells are required for allergen-induced airway inflammation and hyperreactivity in an experimental asthma model. J Immunol 2003; 171:1637-41; PMID:12902459; http://dx.doi.org/10.4049/jimmunol.171.4.1637
- Pichavant M, Goya S, Meyer EH, Johnston RA, Kim HY, Matangkasombut P, Zhu M, Iwakura Y, Savage PB, DeKruyff RH, et al. Ozone exposure in a mouse model induces airway hyperreactivity that requires the presence of natural killer T cells and IL-17. J Exp Med 2008; 205:385-93; PMID:18250191; http://dx.doi.org/10.1084/jem.20071507
- Albacker LA, Chaudhary V, Chang YJ, Kim HY, Chuang YT, Pichavant M, Dekruyff RH, Savage PB, Umetsu DT. Invariant natural killer T cells recognize a fungal glycosphingolipid that can induce airway hyperreactivity. Nat Med 2013; 19:1297-304; PMID:23995283; http://dx.doi.org/10.1038/nm.3321
- Wingender G, Rogers P, Batzer G, Lee MS, Bai D, Pei B, Khurana A, Kronenberg M, Horner AA. Invariant NKT cells are required for airway inflammation induced by environmental antigens. J Exp Med 2011; 208:1151-62; PMID:21624935; http://dx.doi.org/10.1084/jem.20102229
- Damayanti T, Kikuchi T, Zaini J, Daito H, Kanehira M, Kohu K, Ishii N, Satake M, Sugamura K, Nukiwa T. Serial OX40 engagement on CD4+ T cells and natural killer T cells causes allergic airway inflammation. Am J Respir Crit Care Med 2010; 181:688-98; PMID:20019337; http://dx.doi.org/10.1164/rccm.200910-1598OC
- Scanlon ST, Thomas SY, Ferreira CM, Bai L, Krausz T, Savage PB, Bendelac A. Airborne lipid antigens mobilize resident intravascular NKT cells to induce allergic airway inflammation. J Exp Med 2011; 208:2113-24; PMID:21930768; http://dx.doi.org/10.1084/jem.20110522
- Lee PT, Benlagha K, Teyton L, Bendelac A. Distinct functional lineages of human V(alpha)24 natural killer T cells. J Exp Med 2002; 195:637-41; PMID:11877486; http://dx.doi.org/10.1084/jem.20011908
- Scheuplein F, Thariath A, Macdonald S, Truneh A, Mashal R, Schaub R. A humanized monoclonal antibody specific for invariant Natural Killer T (iNKT) cells for in vivo depletion. PLoS One 2013; 8:e76692; PMID:24086759; http://dx.doi.org/10.1371/journal.pone.0076692
- Pellicci DG, Hammond KJ, Uldrich AP, Baxter AG, Smyth MJ, Godfrey DI. A natural killer T (NKT) cell developmental pathway iInvolving a thymus-dependent NK1.1(-)CD4(+) CD1d-dependent precursor stage. J Exp Med 2002; 195:835-44; PMID:11927628; http://dx.doi.org/10.1084/jem.20011544
- Bashir ME, Lui JH, Palnivelu R, Naclerio RM, Preuss D. Pollen lipidomics: lipid profiling exposes a notable diversity in 22 allergenic pollen and potential biomarkers of the allergic immune response. PLoS One 2013; 8:e57566; PMID:23469025; http://dx.doi.org/10.1371/journal.pone.0057566
- Abos-Gracia B, del Moral MG, Lopez-Relano J, Viana-Huete V, Castro L, Villalba M, Martinez-Naves E. Olea europaea pollen lipids activate invariant natural killer T cells by upregulating CD1d expression on dendritic cells. J Allergy Clin Immunol 2013; 131:1393-9.e5; PMID:23265858; http://dx.doi.org/10.1016/j.jaci.2012.11.014
- Agea E, Russano A, Bistoni O, Mannucci R, Nicoletti I, Corazzi L, Postle AD, De Libero G, Porcelli SA, Spinozzi F. Human CD1-restricted T cell recognition of lipids from pollens. J Exp Med 2005; 202:295-308; PMID:16009719; http://dx.doi.org/10.1084/jem.20050773
- Brennan PJ, Tatituri RV, Brigl M, Kim EY, Tuli A, Sanderson JP, Gadola SD, Hsu FF, Besra GS, Brenner MB. Invariant natural killer T cells recognize lipid self antigen induced by microbial danger signals. Nat Immunol 2011; 12:1202-11; PMID:22037601; http://dx.doi.org/10.1038/ni.2143
- Richter J, Neparidze N, Zhang L, Nair S, Monesmith T, Sundaram R, Miesowicz F, Dhodapkar KM, Dhodapkar MV. Clinical regressions and broad immune activation following combination therapy targeting human NKT cells in myeloma. Blood 2013; 121:423-30; PMID:23100308; http://dx.doi.org/10.1182/blood-2012-06-435503
- Akbari O, Faul JL, Hoyte EG, Berry GJ, Wahlstrom J, Kronenberg M, DeKruyff RH, Umetsu DT. CD4+ invariant T-cell-receptor+ natural killer T cells in bronchial asthma. N Engl J Med 2006; 354:1117-29; PMID:16540612; http://dx.doi.org/10.1056/NEJMoa053614
- Matangkasombut P, Marigowda G, Ervine A, Idris L, Pichavant M, Kim HY, Yasumi T, Wilson SB, DeKruyff RH, Faul JL, et al. Natural killer T cells in the lungs of patients with asthma. J Allergy Clin Immunol 2009; 123:1181-5; PMID:19356791; http://dx.doi.org/10.1016/j.jaci.2009.02.013
- Exley M, Garcia J, Balk SP, Porcelli S. Requirements for CD1d recognition by human invariant Valpha24+ CD4-CD8- T cells. J Exp Med 1997; 186:109-20; PMID:9207002; http://dx.doi.org/10.1084/jem.186.1.109
- Yuan W, Kang SJ, Evans JE, Cresswell P. Natural lipid ligands associated with human CD1d targeted to different subcellular compartments. J Immunol 2009; 182:4784-91; PMID:19342656; http://dx.doi.org/10.4049/jimmunol.0803981
- Yue SC, Shaulov A, Wang R, Balk SP, Exley MA. CD1d ligation on human monocytes directly signals rapid NF-kappaB activation and production of bioactive IL-12. Proc Natl Acad Sci U S A 2005; 102:11811-6; PMID:16091469; http://dx.doi.org/10.1073/pnas.0503366102
- Clarke AW, Poulton L, Wai HY, Walker SA, Victor SD, Domagala T, Mraovic D, Butt D, Shewmaker N, Jennings P, Doyle AG. A novel class of anti-IL-12p40 antibodies: potent neutralization via inhibition of IL-12-IL-12Rbeta2 and IL-23-IL-23R. MAbs 2010; 2:539-49.
- Pellicci DG, Patel O, Kjer-Nielsen L, Pang SS, Sullivan LC, Kyparissoudis K, Brooks AG, Reid HH, Gras S, Lucet IS, et al. Differential recognition of CD1d-alpha-galactosyl ceramide by the V beta 8.2 and V beta 7 semi-invariant NKT T cell receptors. Immunity 2009; 31:47-59; PMID:19592275; http://dx.doi.org/10.4161/mabs.2.5.13081
- Gundel RH, Gerritsen ME, Gleich GJ, Wegner CD. Repeated antigen inhalation results in a prolonged airway eosinophilia and airway hyperresponsiveness in primates. J Appl Physiol (1985) 1990; 68:779-86; PMID:2180898
- May RD, Monk PD, Cohen ES, Manuel D, Dempsey F, Davis NH, Dodd AJ, Corkill DJ, Woods J, Joberty-Candotti C, et al. Preclinical development of CAT-354, an IL-13 neutralizing antibody, for the treatment of severe uncontrolled asthma. Br J Pharmacol 2012; 166:177-93; PMID:21895629; http://dx.doi.org/10.1111/j.1476-5381.2011.01659.x
- Wang L, Jenkins TJ, Dai M, Yin W, Pulido JC, Lamantia-Martin E, Hodge MR, Ocain T, Kolbeck R. Antagonism of chemokine receptor CCR8 is ineffective in a primate model of asthma. Thorax 2013; 68:506-12; PMID:23457038; http://dx.doi.org/10.1136/thoraxjnl-2012-203012
- Hart TK, Cook RM, Zia-Amirhosseini P, Minthorn E, Sellers TS, Maleeff BE, Eustis S, Schwartz LW, Tsui P, Appelbaum ER, et al. Preclinical efficacy and safety of mepolizumab (SB-240563), a humanized monoclonal antibody to IL-5, in cynomolgus monkeys. J Allergy Clin Immunol 2001; 108:250-7; PMID:11496242; http://dx.doi.org/10.1067/mai.2001.116576
- Egan RW, Athwal D, Bodmer MW, Carter JM, Chapman RW, Chou CC, Cox MA, Emtage JS, Fernandez X, Genatt N, et al. Effect of Sch 55700, a humanized monoclonal antibody to human interleukin-5, on eosinophilic responses and bronchial hyperreactivity. Arzneimittelforschung 1999; 49:779-90; PMID:10514907
- Patel O, Pellicci DG, Gras S, Sandoval-Romero ML, Uldrich AP, Mallevaey T, Clarke AJ, Le Nours J, Theodossis A, Cardell SL, et al. Recognition of CD1d-sulfatide mediated by a type II natural killer T cell antigen receptor. Nat Immunol 2012; 13:857-63; PMID:22820603; http://dx.doi.org/10.1038/ni.2372
- Lopez-Sagaseta J, Kung JE, Savage PB, Gumperz J, Adams EJ. The molecular basis for recognition of CD1d/alpha-galactosylceramide by a human non-Valpha24 T cell receptor. PLoS Biol 2012; 10:e1001412; PMID:23109910; http://dx.doi.org/10.1371/journal.pbio.1001412
- Durocher Y, Perret S, Kamen A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res 2002; 30:E9; PMID:11788735; http://dx.doi.org/10.1093/nar/30.2.e9
