ABSTRACT
Ricin is a toxin that could potentially be used as a bioweapon. We identified anti-ricin A chain antibodies by sequencing the antibody repertoire from immunized mice and by selecting high affinity antibodies using yeast surface display. These methods led to the isolation of multiple antibodies with high (sub-nanomolar) affinity. Interestingly, the antibodies identified by the 2 independent approaches are from the same clonal lineages, indicating for the first time that yeast surface display can identify native antibodies. The new antibodies represent well-characterized reagents for biodefense diagnostics and therapeutics development.
Introduction
Ricin is a toxin derived from the castor bean Ricinus communis, and it is classified as a Category B Agent by the Centers for Disease Control and Prevention in part because of its high lethality (LD50 about 22 μg/kg body weight for human) and ease of production.Citation1 Ricin is composed of an enzymatic A chain, which deactivates eukaryotic ribosomes by depurinating adenine 4324 in the 28S rRNA of the 60S ribosomal subunit, and a lectin B chain, which binds carbohydrates on the cell surface.Citation1,2 As a result, ricin is a potent biological weapon, and past cases of malicious exposure to ricin highlight the need for both an increase in the sensitivity of diagnostics and for treatment after poisoning.Citation3,4 Antibodies to both the A chain and B chain of ricin have been produced using hybridoma-based technology,Citation5-8 and by phage display screening of immune libraries reconstituted from the bone marrow of immunized cynomolgus macaques,Citation9 resulting in antibodies with affinities ranging from 40 pM to 5 nM. However, there are currently no US. Food and Drug Administration-approved treatments for ricin poisoning, and the diagnostic potential of these reported antibodies has yet to be tested.
In vitro screening of large combinatorial libraries using display technologies that rely on phage, bacteria, yeast, mammalian cells or even in vitro transcription/translation systems are widely employed for antibody discovery.Citation10-12 Combinatorial antibody libraries are constructed either by mining the natural diversity of immunoglobulin genes in immunized or antigen-naïve animals,Citation13-15 or by diversifying the complementarity-determining regions (CDRs) within one or more “scaffold” antibodies.Citation16 Variable heavy (VH) and variable light (VL) chain genes are then joined combinatorially, yielding, at least in theory, libraries that contain combinations of heavy chains joined with all possible light chains.Citation13-16 Typically, immune libraries constructed from mRNA obtained from the spleen, bone marrow or from peripheral blood mononuclear cells (PBMCs, primarily in the case of human donors) are more likely to encode a significant fraction of antigen-specific antibodies and thus represent the most reliable route to high affinity antibodies (provided that the antigen is immunogenic).Citation13,17 Isolation of high affinity antibodies from immune and other libraries is most readily accomplished by taking advantage of the quantitative nature of fluorescence-activated cell sorting (FACS)-based library screening methods, and yeast display has been established as the dominant technology for screening libraries by FACS.Citation18
The power of combinatorial library screening has been validated by the identification and development of therapeutic antibodies that are now entering clinical trials.Citation19 However, certain antibodies isolated from combinatorial libraries may express at low yields in mammalian cells and display poor biophysical properties in vitro,Citation20-24 which can hamper their development into therapeutics. The random pairing of heavy and light chains in combinatorial libraries results in antibodies with non-natively paired VH and VL genes, and this is potentially one cause of poor antibody expression and stability.Citation25 Moreover, a recent report has suggested that several mouse and human VH germ-line genes exhibit strong preferential pairing with specific VL chains,Citation26 and the random pairing of non-preferential VH and VL chains could lead to conformational incompatibilities, again affecting expression and stability. In the past decade, enormous efforts have been expended on advancing screening techniques to remove antibodies with mediocre biophysical properties and investigating methods to improve them.Citation21-23,27,28
Our lab has pioneered methods for the discovery of high affinity antigen-specific antibodies directly via mining of the immunoglobulin repertoire by capitalizing on next-generation sequencing technologies without the need for screening.Citation29-32 Specifically, we developed methods for high-throughput determination of the natively paired VH:VL repertoire from single B cells.Citation30,31 More recently, we have shown that antibody secreting B cells (CD138+ plasmablasts) within the draining lymph node are overwhelmingly antigen-specific, and that antibodies derived from these cells exhibit high binding affinity.Citation32
In this report, we sought to compare the isolation of high-affinity anti-ricin antibodies via mining of the draining lymph node repertoire and via yeast display of immune combinatorial libraries constructed from antibody mRNAs obtained from spleen or bone marrow cells. Overall, both approaches yielded strong ricin A chain binders (the lowest Kd values were 0.97 and 0.58 nM, respectively, for these 2 methods). Interestingly, we found that antibodies isolated by yeast display from combinatorial libraries in which the VH and VL from spleen or bone marrow had been randomly paired were clonal relatives of antibodies identified via mining of the draining lymph node repertoire, and comprised authentic, natively paired, VH and VL sequences. Thus, in hyperimmune animals where antigen-specific antibodies comprise a significant portion of the repertoire,Citation29,32-35 flow cytometric screening of libraries constructed via the random pairing of VH and VL genes can nonetheless result in the isolation of native antibodies.
Results
Overview of the experimental approach
As we have previously shown, mouse footpad immunization results in a strong immune response in the popliteal draining lymph node (DLN), characterized by the robust expansion of antibody secreting B cells (CD138+ plasmablasts).Citation32 Analysis of the DLN CD138+ B cell antibody repertoire overwhelmingly revealed that the most highly represented sequences, i.e., those present in the largest number of reads, are antigen-specific.Citation32 Therefore, we sought to explore the antibody repertoire in DLN antibody-secreting B cells, and to compare antigen-specific antibodies with those selected from bone marrow and spleen combinatorial libraries, which are commonly used in antibody discovery efforts following intraperitoneal or subcutaneous immunization.Citation9,13,36 In contrast, samples from the DLN could only support high-throughput VH:VL sequencing and repertoire analysis, as we were limited by the number of cells. Four mice were immunized by footpad injection with ricin A chain in TiterMax Gold adjuvant (), followed by 2 booster immunizations after 14 and 28 days, after which all 4 mice generated significant titers (>1 :10,000) against ricin A chain (Fig. S1A). One more booster immunization was performed, after which even higher titers were reached (Fig. S1B). Mice were sacrificed 6 d after the final booster immunization, at which point the draining lymph node was observed to be enlarged compared to the contralateral lymph node. Spleen and bone marrow were collected to screen for antigen-specific antibodies (), and the draining lymph node was collected as well for VH:VL mining ().
Figure 1. Overview of the experimental approach. (A) Mouse is immunized at footpad with ricin A chain, and peripheral lymphoid organs, including bone marrow, spleen, and draining lymph node are isolated after 3 booster immunizations. (B) Total cells in bone marrow and spleen are collected, and VH and VL mRNA are reverse transcribed and amplified, which are used to construct scFv libraries, respectively. These are selected against ricin A chain using yeast surface display to isolate high-affinity binders. VH and VL cDNAs are also sequenced on an Illumina platform. (C) CD138+ antibody secreting cells are isolated from draining lymph node, and processed through our high-throughput VH:VL pairing platform.
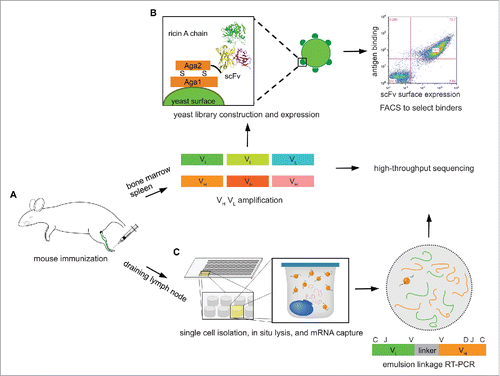
Isolation of high affinity ricin A chain antibodies by yeast display of combinatorial libraries
Single-chain variable fragment (scFv) libraries were constructed using mRNA from ∼5 × 106 total splenocytes, and separately from femur bone marrow cells (). Briefly, VH and VL genes were amplified separately, paired combinatorially by overlap extension-polymerase chain reaction that also introduced a (Gly4Ser)3 linker,Citation13 and then subcloned into the yeast display vector pCTCON2 via homologous recombination.Citation37
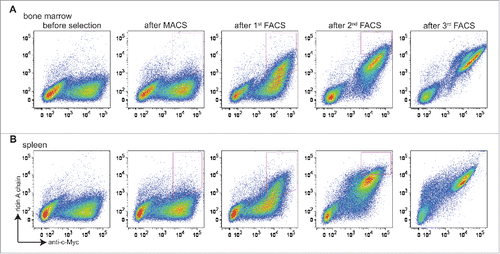
Two libraries of ∼2 × 106 transformants each (bone marrow-derived and splenocyte-derived) were obtained. Screening was performed by carrying out one round of magnetic-activated cell sorting (MACS), and then 3 rounds of FACS with progressively more stringent gates that collected cells positive both for expression (using Alexa Fluor 488-labeled anti-c-Myc) and for ricin A chain binding (using Alexa Fluor 633-labeled ricin A chain) (). For the first 2 rounds of selection, ricin A chain was used at a 1 μM concentration. Prior to selection, both libraries showed little binding to ricin A chain (, before selection). Significant enrichment of cells that stained positively both with fluorescent anti-c-Myc antibody and ricin A chain was observed from both libraries after the second round (, after first FACS). Subsequently, ricin A chain concentrations of 200 nM and 40 nM were used for the third and fourth rounds of sorting, respectively. After the fourth round, almost all of the cells that expressed scFv also showed binding to ricin A chain (, after third FACS).
Figure 2. Selection of libraries constructed from bone marrow and spleen antibody repertoires. Cells are doubly stained with chicken anti-c-Myc IgY/GaC-488 for scFv surface expression and biotinylated ricin A chain/SA-633 for antigen binding. (A) Bivariate plots are shown for bone marrow antibody library stained with 100 nM ricin A chain before selection, after MACS, after 1st FACS, after 2nd FACS, and after 3rd FACS, with x axis being surface expression, and y axis being antigen binding. (B) Bivariate plots are shown for spleen antibody library stained with 100 nM ricin A chain before and after each round of selection. For both libraries, cells in the upper-right quadrant (both express scFv on the surface and bind ricin A chain) are sought. Cells falling within strict FACS sort gates designed to ensure enrichment of clones showing increased binding to ricin A chain are collected for the next round of selection.
Twenty colonies from each library were sequenced, yielding a total of 5 unique clones (3 from the bone marrow-derived library and 2 from the splenocyte-derived library), indicating that screening had led to convergence of the libraries to a small number of scFv sequences (). Apparent equilibrium binding constants (Kd) for these 5 clones were estimated by incubating yeast cells displaying the respective antibodies with different concentrations of ricin A chain, ranging from 0.09 to 600 nM, and then measuring the mean fluorescence intensity of binding at each concentration by FACS. Binding constants were calculated using a Langmuir 1:1 binding model.Citation37 All 5 antibodies demonstrated low nanomolar affinities binding to ricin A chain (, Fig. S2). These 5 antibodies were then expressed in E.coli as single-chain antibodies (scAbs, scFv fused with human kappa light chain at the C-terminusCitation13) and purified to >95% homogeneity by Ni-NTA chromatography (as determined by SDS-PAGE). The antibodies were further purified by size-exclusion chromatography, which also revealed that all antibodies were monomeric. Surface plasmon resonance (SPR) analyses indicated that, consistent with the flow cytometric analyses,Citation37 all 5 antibodies displayed high affinity binding to ricin A chain (, Fig. S3).
Table 1. List of the antibodies isolated from the bone marrow and spleen combinatorial libraries by yeast display and abundance of their VH and VL sequences in the corresponding repertoires assessed by next-generation sequencing. Note that BM3 and BM17 have the same CDRH3 sequences, but differ in sequence outside of CDRH3 ().
Table 2. The binding kinetics of the antibodies isolated from bone marrow and spleen as measured by yeast surface titration and surface plasmon resonance.
The question of whether in vitro screening technologies can identify native antibodies in a repertoire has generally not been addressed; the two methods often return different antigen-specific antibodies. Different screening platforms lead to the identification of different antibodies, but some platforms seem to return antibodies that are of low abundance or apparently not present at all.Citation36,38 The identification of antibodies via yeast display presented an opportunity to compare the results with the natural repertoire, and thus we sequenced both the bone marrow and spleen VH and VL repertoires using an Illumina 2 × 250 Miseq platform. From the same 5 × 106 cells of bone marrow and spleen from a given mouse that were used for yeast library construction, about 106 and 7 × 105 reads, respectively, were obtained for VH repertoires, from which 8735 and 7057 unique clonotypes (defined as the group of VH sequences that share the same germ-line V and J segments, and have >90% amino acid identity in their CDRH3sCitation39) were identified (). Similar to previous reports,Citation29,40 both repertoires were observed to have skewed germ-line V gene usage. For example, both repertoires showed biases for IGHV1, 5, 14, and IGKV1, 4, 6 families (), with IGKV4 as the most commonly used light chain family in the bone marrow, and IGKV6 as the most commonly used light chain in the spleen. We noticed the preferential usage of the IGKV1 family in bone marrow relative to spleen, and of the IGKV6 family in spleen relative to bone marrow. It will be interesting to determine if similar repertoire biases toward specific germline light chain families are observed with other antigens and adjuvant combinations. Both repertoires showed a CDRH3 length distribution comparable to those previously reported from animals immunized with different antigens,Citation29 with a median CDRH3 length of 13 amino acids in the bone marrow repertoire and 11 amino acids in the spleen repertoire ().
Figure 3. Characteristics of bone marrow and spleen antibody repertoires from the same mouse immunized with ricin A chain. (A) Germ-line VH gene usage in bone marrow and spleen repertoires. (B) Germ-line VL gene usage in the 2 repertoires. (C) CDRH3 length distribution in the 2 repertoires. Green denotes bone marrow repertoire, and blue denotes spleen repertoire.
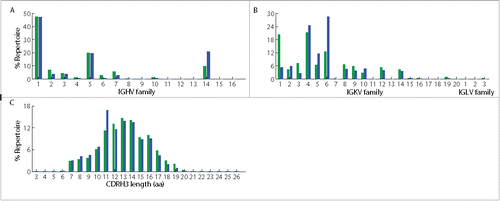
Table 3. Next-generation sequencing statistics of bone marrow and spleen repertoires.
Importantly, we now find that the antibodies identified by yeast surface display are among the most abundant antibodies in the sequenced repertoire (). This indicates for the first time that yeast surface display of combinatorial libraries can identify native antibodies in the repertoire.
Isolation of high affinity anti-ricin A chain antibodies by next-generation sequencing of DLN B cells
CD138+ antibody secreting B cells were isolated from the draining lymph node and the paired VH:VL repertoire was determined, as previously described.Citation32 Briefly, a total of 90,000 CD138+ antibody secreting cells were isolated by magnetic sorting by first depleting CD45R+, CD49b+, and CD19+ cells, and then enriching for CD138+ cells. Approximately 40,000 antibody secreting cells were deposited into microfabricated nanowell plates such that >98% of wells contained a single cell, after which poly(dT) magnetic beads were added to capture mRNAs, the cells were lysed, and the poly(dT) beads were collected. The beads were emulsified, and overlap extension RT-PCR was performed to synthesize linked VH:VL amplicons ().Citation30 The region of the linked amplicons comprising CDRH3 and CDRL3 was then sequenced in an Illumina MiSeq 2 × 250 run. Full-length VH and VL sequences were identified in separate MiSeq 2 × 250 runs. After bioinformatics analysis,Citation30 212 unique VH:VL pairs were identified in the draining lymph node repertoire, and these showed great polarization, indicative of clonal expansion. Specifically, the top 10 most abundant VH:VL pairs constituted 65.8% of the total reads (). The virtual absence of antibodies using IGLV families in the repertoire was expected, as only 5% of all mouse antibodies use the lambda light chain.Citation40
Figure 4. Characteristics of CD138+ antibody-secreting cells repertoire in draining lymph node from mouse immunized with ricin A chain. (A) The frequency of each unique antibody clone in the repertoire, which is calculated as the percentage of its read counts in the read counts of all clones, is shown with its rank, which is ordered by the number of read counts of each unique clone. Clones with only 1 read are removed, and CDRH3 nucleotide sequences are clustered to 96% identity. Inset shows the distribution of the top 10 most abundant clones in the repertoire. (B) Germ-line V gene family usage in the same repertoire is shown. (C) CDRH3 length (calculated as amino acid length) distribution of antibodies in the repertoire is shown as the percentage of antibodies that have the denoted CDRH3 length in the whole repertoire.
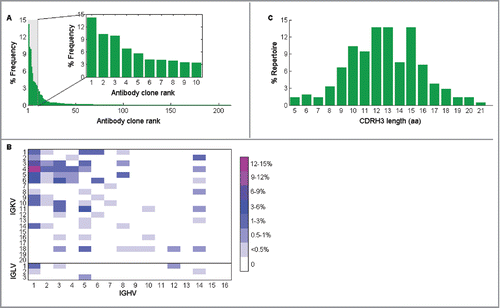
Comparisons with mouse antibody repertoires generated by immunization with other antigens showed similar germ-line gene usage between the anti-ricin A chain antibody repertoire and other repertoires (, compare to the repertoires in ref 29 and 32 generated against other antigens).Citation29,32 The CDRH3 length distribution in CD138+ antibody secreting cells from the draining lymph node showed 3 peaks at 12, 13, and 15 amino acids ().
Synthetic genes for the top 10 highest frequency paired VH:VL sequences from the DLN repertoire were constructed and the respective mouse-human chimeric antibodies (mouse VH fused with the human IgG1 constant domain, and mouse VL fused with the human kappa constant domain) were expressed in Expi293 cells and purified. Four/10 recombinant antibodies displayed binding by ELISA (, Fig. S4). SPR analysis was used to obtain antigen-binding kinetics for these 4 clones (, Fig. S3). All but one antibody showed high affinity binding to the ricin A chain. Interestingly, the affinities of the antibodies that bound ricin A chain correlated with their abundance in the VH:VL repertoire, with the antibody displaying the lowest Kd (RAM1.2) being the most abundant.
Table 4. List of the antibodies isolated from CD138+ antibody-secreting cells in the draining lymph node.
Table 5. The binding kinetics of the antibodies isolated from the draining lymph node as measured by ELISA and surface plasmon resonance.
We compared the native antibodies recovered from the DLN to yeast display-selected antibodies from the bone marrow and spleen combinatorial libraries. Interestingly, BM1, BM3, BM17 and SP19 were found to share germ-line V and J segments with RAM1.5, and SP1 had the same germ-line V and J segments as RAM1.4. The two groups of antibodies also had the same VH:VL pairing. Furthermore, antibodies recovered from the DLN without selection had very similar CDRH3s with those isolated by yeast display from bone marrow and spleen (), as well as additional amino acid substitutions, indicating that they correspond to somatic variants derived from the same antibody lineage during clonal expansion ().
Figure 5. Sequence comparison between yeast surface display and next-generation sequencing discovered antibodies. (A), (B) VH alignments of BM1, BM3, BM17, SP19 and RAM1.5 and of SP1 and RAM1.4 showed mutations through the complete VH sequences, indicating they are somatic variants. (C), (D) VL alignments of BM1/BM3/BM17, SP19 and RAM1.5 and of SP1 and RAM1.4 also showed mutations. Mutations are indicated by different colors.

Discussion
In vitro screening of antibody libraries constructed from different sources (synthetic, naïve, or immune) is one of the most commonly used techniques for antibody discovery.Citation11,12 In this work, we identified anti-ricin A chain antibodies by eliciting an immune response in mice and then exploring the resultant antibody repertoires via both yeast display of combinatorial libraries from the bone marrow and spleen and paired VH:VL sequencing from the draining lymph node.
While both approaches generated high affinity clones (Kd ranging from 0.55 nM to 6.12 nM), as has previously been observed,Citation38 unexpectedly the antibodies generated by these 2 very different approaches were found to be clonal relatives. Different amino acids encoded by different nucleotides in the isolated antibodies from different lymphoid organs indicated that BM1, BM3, BM17 (derived from bone marrow using yeast display), SP19 (derived from the spleen using yeast display) and RAM1.5 (derived from the draining lymph node using paired VH:VL sequencing), and separately SP1 and RAM1.4 were clonal variants (). A previous report has shown that while naïve mouse spleen and lymph node repertoires share some common CDRH3s, the scFvs isolated from these lymphoid organs following intraperitoneal and subcutaneous immunization were unique.Citation41 In contrast, when we probed different lymphoid organs, clonal variants were obtained. These differing conclusions may reflect differences in the immunization routes used, the numbers of boost immunizations, and significant differences introduced by the expression biases in phage compared to yeast display. The clonal variants we isolated from different lymphoid organs indicate that these antibodies were likely encoded by the same ancestral B cells that homed to different lymphoid organs and experienced clonal expansion there.
We have previously shown that by dissecting the draining lymph node antibody repertoire with paired VH:VL sequencing, the highly abundant clones were usually antigen specific as a result of clonal expansion.Citation32 Here, we again showed that the highly abundant clones in the draining lymph node probed with paired VH:VL sequencing encoded high-affinity, antigen-specific antibodies. Comparison of the antibodies isolated using these 2 approaches supports our hypothesis that yeast display can recover native antibodies in a repertoire, as antibodies isolated by yeast display are clonal variants of those isolated by paired VH:VL sequencing, which identifies native VH:VL pairings. Given the relative abundance of the VH and VL sequences of antibodies in the bone marrow and spleen repertoires, the calculated probability of obtaining these native antibodies by combinatorially random pairing would be about 0.2%. Thus, in hyperimmunized animals the proportion of randomly paired VH and VL genes is sufficiently high that natively paired heavy and light chains occur. Moreover, high affinity native pairs can be isolated following yeast surface expression and FACS. To our knowledge this is the first report that yeast display selections can identify native antibodies from the immune repertoire.
The fact that an in vitro combinatorial library screening method can identify native V gene pairs is relevant for antibody discovery. Numerous successes in developing antibody drugs from combinatorial libraries generally validate this approach.Citation19 However, some high-affinity antibodies identified by library screening have mediocre biophysical properties, such as low yield and poor stability.Citation20-24 Such deficiencies are thought to emanate from random or non-native pairings of VH and VL that lead to unfavorable conformations that in turn promote poor biophysical properties.Citation25,26 Another contributor may be that expression in a non-native host (i.e., mouse antibody libraries screened by phage display) leads to selection at the expression level against certain clones.Citation38 In contrast, antibodies with natively paired heavy and light chains, such as those isolated from hybridomas, express better, and in general show much better stability.Citation25,26,42 Our methods can therefore potentially streamline the huge amount of effort put into the discovery process.Citation20-22,27,28
Our results support the utility of next-generation sequencing in combinatorial library screening.Citation10,29,32 Hyperimmunization leads to a high degree of polarization of the B-cell repertoire in a particular compartment, such that the dominant antibodies are highly represented and combinatorial screening by a technique with low expression bias, such as yeast display, can further yield high-affinity native antibodies. The high-affinity anti-ricin A chain antibodies we identified may provide a greater diversity of reagents for both diagnostics and therapeutics.
Materials and methods
Cell line and media
The yeast strain EBY100 (MATa AGA1::GAL1-AGA1::URA3 ura3-52 trp1 leu2Δ200 his3Δ200 pep4::HIS3 prb11.6R can1 GAL) was used for library construction and screening. Yeast cells were maintained in YPD medium (20 g/l dextrose, 20 g/l peptone, and 10 g/l yeast extract); after library transformation, they were maintained in SDCAA medium (20 g/l dextrose, 6.7 g/l yeast nitrogen base, 5 g/l casamino acids, 8.56 g/l NaH2PO4.H2O, and 10.2 g/l Na2HPO4.7H2O). SGCAA medium (identical to SDCAA except 20 g/l galactose is used instead of dextrose) was used for library induction. E. coli strain DH10β was used for subcloning, and E.coli strain Jude-1 was used for soluble scAb expression. Expi293 cells (Invitrogen) were used for IgG expression, and were maintained in Expi293 expression medium (Invitrogen).
Antigen and antibodies
Ricin A chain was purchased from Sigma-Aldrich (cat# L9514). It was biotinylated using an EZ-Link Sulfo-NHS-LC-Biotin kit (Thermo Scientific). Chicken anti-c-Myc IgY, Alexa Fluor 488-goat anti-chicken IgG (GaC-488), and streptavidin-Alexa Fluor 633 (SA-633) were obtained from Invitrogen (cat# A-21281, A-11039, and S21375, respectively). Anti-biotin microbeads were purchased from Miltenyi Biotec (cat# 130-090-485).
Mouse immunizations
This study was approved by the University of Texas Institutional Animal Care and Use Committee under protocol# AUP-2013-00009. Ricin A chain was mixed with TiterMax Gold adjuvant (Sigma-Aldrich, cat# T2684) at a 1:1 ratio and pipetted several times to obtain stable emulsions for immunization. On day 1, 4 female BALB/c mice at 6 weeks of age (Jackson Laboratory) were injected subcutaneously at the left hind footpad with 5 μg of ricin A chain in 25 μl antigen adjuvant mixture. A booster immunization was performed on day 14. Seven days after the booster immunization, the serum antibody titers against ricin A chain were determined. About 30 μl of blood was collected from each mouse at a small tail vein incision made with a scalpel blade, and coagulated at room temperature (RT) for 30 min. Following centrifugation at 13000 rpm for 15 min, the supernatant (serum) was used for ELISAs. The serum was first serially diluted with phosphate-buffered saline (PBS) +2% milk (PBSB) at a 1:3 ratio from 1:100 to 1:218,700. The diluted serum was applied in triplicate onto ELISA plates (Corning) that had been coated with 50 μl of 4 μg/ml of ricin A chain overnight (O/N) at 4°C and then blocked with PBSB at RT for 2 hours. After incubation at RT for 1 hour, plates were washed with PBS+0.05% Tween-20 (PBST), followed by adding 50 μl of a 1:5000 diluted horseradish peroxidase (HRP)-conjugated goat anti-mouse antibody (Jackson ImmunoResearch, cat# 115-035-166). After 1 hr incubation, plates were washed again with PBST, after which 50 μl of TMB Ultra ELISA substrate (Thermo Scientific, cat# 34028) was added. The reaction was quenched after 10 min with 50 μl of 2 M H2SO4. Absorbance at 450 nm was determined using a Tecan M200 plate reader, and the serum titer was calculated as the dilution at which the absorbance was 3 times higher than background. The second booster was performed on day 28, after which significant titers (>1 :10,000) were generated in all mice. The final booster was performed on day 42, and 6 d later, mice were sacrificed for lymphoid organ collection.
Lymphoid organ collection
Bone marrow, spleen, and draining lymph node tissues were collected separately. Thirty min before sacrifice, 10 μl 2% Evans Blue in PBS (Sigma-Aldrich, w/v, cat# E2129) was injected into the left hind footpad. After CO2 asphyxiation and cervical dislocation, the blue-stained popliteal lymph node behind the knee was collected. The spleen was also collected. For bone marrow collection, after clipping the ends of the tibias and femurs, bone marrow was flushed out with PBS+0.1% bovine serum albumin (BSA)+2 mM EDTA (PBSM) using a syringe. Each lymphoid organ tissue sample was first mechanically disrupted using a needle, then passed through a 70 μm cell strainer (Corning) to collect single cells. The single cell suspension of each lymphoid organ was washed with 20 ml of PBSM buffer and resuspended in 2 ml red blood cell lysis buffer (155 mM NH4Cl, 12 mM NaHCO3, and 0.1 mM EDTA). After incubation at RT for 5 min, cells were diluted with 20 ml of PBS and then spun down. Cells were washed twice more with PBSM buffer, then cells from the bone marrow and spleen were used for yeast library construction, and cells from draining lymph node were used for high-throughput VH:VL pairing.
Yeast library construction for bone marrow and spleen repertoires
Cells from the bone marrow and spleen were resuspended in TRI Reagent (Invitrogen), and RNA was extracted from each sample. cDNA was generated using 200 ng of RNA as a template with the SuperScript III First-Strand synthesis kit (Invitrogen). VH and VL sequences were amplified using mouse V gene specific primers (Table S1). For scFv library construction, 50 ng of VH and VL DNA were used as templates for overlap extension PCR, and amplified with primers that contained 50 nucleotides at each end that were in common with the display vector, which should be sufficient to promote homologous recombination upon yeast transformation. The display vector pCTCON2 was linearized by NheI and BamHI digestionCitation37 and the digested backbone was purified and co-transformed with 5-fold mass excess of scFv fragments into electrocompetent yeast cells.Citation43 The libraries were cultured in SDCAA medium at 30°C for 2 d.
Yeast library screening
Cells were induced in SGCAA medium at 20°C for 2 d. For the 1st round of selection, 2 × 107 cells (10-fold coverage relative to the number of transformants) were incubated with 1 μM biotinylated ricin A chain at RT for 1 hr and washed thoroughly with PBS+0.5% BSA+2 mM EDTA (PBSA) to remove any unbound ricin A chain protein. Cells were then mixed with 400 μl of anti-biotin microbeads and incubated at 4°C for 30 min, and washed again. Cells that bound ricin A chain were selected using MidiMACS system (Miltenyi Biotec), and recovered in SDCAA medium at 30°C. For the 2nd round of selection, 107 cells in 200 μl PBS+0.1% BSA (PBSF) were labeled with 1 μM biotinylated ricin A chain and 4 μg/ml chicken anti-c-Myc IgY at RT for 1hr. After washing with PBSF, cells were incubated with 5 μg/ml SA-633 and 5 μg/ml GaC-488 at 4°C for 30 min. After washing, cells were resuspended in PBSF buffer and sorted using FACS Aria (BD Bioscience). Gated cells were collected from the double positive quadrant as shown in . The subsequent 2 rounds of selection were performed in a similar way, except that the concentration of ricin A chain used was decreased to 200 nM and 40 nM, respectively. After 4 rounds of selection, 20 random colonies from each library were picked, and plasmids were isolated using Zymoprep Yeast Plasmid Miniprep II kit (Zymo Research). Plasmids were amplified in E. coli DH10β and sequenced. Three and 2 unique clones were thus isolated from the bone marrow and spleen libraries, respectively.
Plasmids encoding the 5 unique antibody clones above were re-transformed into strain EBY100. Cells were cultured and induced for scFv expression as described above. 106 cells were labeled with 1:3 serially diluted ricin A chain, ranging from 0.09 nM to 600 nM. The labeling process was the same as that used for library sorting. After labeling, cells were analyzed on BD FACS Aria and the mean fluorescence intensity (MFI) values were used to calculate apparent equilibrium binding constant (Kd), as described.Citation37
scAb expression and characterization
For soluble antibody purification, antibody genes were subcloned into pMopac16 vector and expressed as scAb fragments in E.coli Jude-1 cellsCitation13 and then purified by Ni-NTA chromatography according to the manufacturer's instructions (Thermo Scientific). After elution, proteins were dialyzed in PBS, then loaded onto a Hiload 16/600 Superdex 75 pg column (GE Healthcare) and antibody-containing fractions were pooled and concentrated to 2 mg/ml.
For affinity validation, SPR was performed for each antibody clone using a BIAcore 3000 biosenor (Biacore). About 400 RU (response units) of scAbs were immobilized on the CM5 sensor chip (GE Healthcare) using amine coupling chemistry. All binding experiments were done in HBS-EP buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 3.4 mM EDTA, and 0.005% P20 surfactant) (GE Healthcare). Ricin A chain was injected at concentrations of 25, 50, 100, 200, and 400 nM with a flow rate of 30 μL/min for 2 min and a dissociation time of 10 min. The chip was regenerated after each injection by 100 mM citric acid, pH 3.0. The response generated by flowing ricin A chain over a BSA-coupled surface was used as control and consequently subtracted. Experiments were carried out in triplicates. All kinetic parameters were determined in BIAevaluation 3.0 software using a 1:1 Langmuir model and were reported as the average of the 3 technical replicates.
V gene repertoire sequencing and analysis
VH and VL cDNA for bone marrow and spleen samples (the same cDNAs used for yeast library construction) were prepared and sequenced using the Illumina 2 × 250 paired end Miseq platform as described previously.Citation30 Briefly, raw sequences were first filtered for sequences containing at least half of the nucleotides at a minimum Phred quality score of 20 to obtain high quality sequences, then assembled to obtain full length VH and VL sequences, respectively. Sequences represented by at least 2 reads were submitted to IMGT (the international immunogenetics information system, www.imgt.org) High V-Quest analysisCitation44 and those with productive VH or VL, and in-frame V(D)J junctions (CDRH3 or CDRL3) were analyzed further.
Paired VH: VL sequencing of antibody secreting cells from the draining lymph node
From single cell suspension of the draining lymph node, CD138+ antibody secreting B cells were isolated using a mouse CD138+ Plasma Cell Isolation kit (Miltenyi Biotec).Citation32 The paired VH:VL gene repertoire was determined as described.Citation30 Briefly, isolated single cells were deposited into 125 pL wells in PDMS slides together with poly(dT) magnetic beads (Invitrogen). After cell lysis in situ, beads with captured mRNA from the same cell were collected, washed, and emulsified. Mouse V gene specific primers (Supplementary Table 2) were used to link endogenous VH and VL in the subsequent emulsion overlap extension RT-PCR. A second nested PCR was used to amplify the linked VH:VL pairs. Sequencing was performed by Illumina 2 × 250 paired end Miseq. The raw Miseq reads were filtered for reads where at least half of the nucleotides had a minimum Phred quality score of 20 to remove low-quality reads, and were then submitted to IMGT for V(D)J germ-line alignment.Citation44 Sequence data were again filtered for productive VH or VL, and for in-frame V(D)J junctions (CDRH3 or CDRL3), and were then matched using their Illumina read IDs. CDRH3 nucleotide sequences were clustered to 96% identity, and CDRH3:CDRL3 pairs represented by 2 or more reads were rank-ordered. In two separate sequencing runs the complete VH and VL sequences encoded within the linked VH:VL amplicons were determined. The sequences for the VH:VL junction and the separate VH and VL sequences were assembled to yield natively paired full length VH and VL sequences, as described.Citation30
Antibody expression and characterization
Consensus VH and VL sequences of the top 10 most abundant antibodies from the draining lymph node were synthesized using gblocks (IDT) and subcloned into pcDNA3.4 vectors (Invitrogen) as fusions with human IgG1 and kappa constant domains, respectively. After sequence verification, the heavy and light chain plasmids were mixed at 1:3 ratio, and transfected into Expi293 cells for antibody expression. Six days after transfection, the supernatant was collected and antibodies were purified using protein A chromatography (Thermo Scientific).
To verify the specificity of these antibodies, ELISAs were carried out with both ricin A chain and BSA as a control. ELISA plates were coated with 50 μl of 4 μg/ml ricin A chain or BSA at 4°C O/N. On the next day, plates were blocked with PBSB at RT for 2 hr. After that, 1:5 serially diluted antibodies with concentrations ranging from 300 nM to 3.84 pM were added to the plates and the plates were incubated at RT for 1 hr. After washing with PBST, 50 μl of 1:5000 diluted HRP-conjugated donkey anti-human antibody (Jackson ImmunoResearch, cat# 709-035-149) was added and the plates were again incubated at RT for 1hr. After washing with PBST, 50 μl of TMB substrate was added and the reaction was stopped after 10 min by adding 50 μl of 2 M H2SO4. The absorbance at 450 nm was read and EC50 was calculated. For each antibody that showed binding to ricin A chain, affinity determinations were carried out via SPR analyses, as described above for scAbs.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental_Data.docx
Download MS Word (586.7 KB)Acknowledgments
We thank Dr. K. D. Wittrup for providing pCTCON2 vector and EBY100 strain, K. H. Hoi for assistance with bioinformatics analysis, and C. Das for help with antibody expression.
Funding
This work was funded by the Defense Threat Reduction Agency under grant No. HDTRA-1-13-1-0031.
References
- Lord JM, Roberts LM, Robertus JD. Ricin: structure, mode of action, and some current applications. J Fed Am Societies Exp Biol 1994; 8:201-8; PMID:8119491.
- Audi J, Belson M, Patel M, Schier J, Osterloh J. Ricin Poisoning A Comprehensive Review. J Am Med Assoc 2005; 294:2342-51; PMID:16278363; http://dx.doi.org/10.1001/jama.294.18.2342
- Knight B. Ricin-a potent homicidal poison. Br Med J 1979; 1:350-1; PMID:421122
- Mayor S. UK doctors warned after ricin poison found in police raid. Br Med J 2003; 326:126; PMID:12531838; http://dx.doi.org/10.1136/bmj.326.7381.126
- Maddaloni M, Cooke C, Wilkinson R, Stout AV, Eng L, Pincus SH. Immunological Characteristics Associated with the Protective Efficacy of Antibodies to Ricin. J Immunol 2004; 172:6221-8; PMID:15128810; http://dx.doi.org/10.4049/jimmunol.172.10.6221
- Mantis NJ, McGuinness CR, Sonuyi O, Edwards G, Farrant SA. Immunoglobulin A antibodies against ricin A and B subunits protect epithelial cells from ricin intoxication. Infect Immun 2006; 74:3455-62; PMID:16714576; http://dx.doi.org/10.1128/iai.02088-05
- Neal LM, O'Hara J, Brey RN, Mantis NJ. A monoclonal immunoglobulin G antibody directed against an immunodominant linear epitope on the ricin A chain confers systemic and mucosal immunity to ricin. Infect Immun 2010; 78:552-61; PMID:16714576; http://dx.doi.org/10.1128/iai.02088-05
- Prigent J, Panigai L, Lamourette P, Sauvaire D, Devilliers K, Plaisance M, Volland H, Créminon C, Simon S. Neutralising antibodies against ricin toxin. PLoS One 2011; 6:e20166; PMID:21633505; http://dx.doi.org/10.1371/journal.pone.0020166
- Pelat T, Hust M, Hale M, Lefranc M-P, Dübel S, Thullier P. Isolation of a human-like antibody fragment (scFv) that neutralizes ricin biological activity. BMC Biotechnol 2009; 9:1-13; PMID:19563687; http://dx.doi.org/10.1186/1472-6750-9-60
- Georgiou G, Ippolito GC, Beausang J, Busse CE, Wardemann H, Quake SR. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat Biotechnol 2014; 32:158-68; PMID:24441474; http://dx.doi.org/10.1038/nbt.2782
- Bradbury ARM, Sidhu S, Dübel S, McCafferty J. Beyond natural antibodies: the power of in vitro display technologies. Nat Biotechnol 2011; 29:245-54; PMID:21390033; http://dx.doi.org/10.1038/nbt.1791
- Hoogenboom HR. Selecting and screening recombinant antibody libraries. Nat Biotechnol 2005; 23:1105-16; PMID:16151404; http://dx.doi.org/10.1038/nbt1126
- Hayhurst A, Happe S, Mabry R, Koch Z, Iverson BL, Georgiou G. Isolation and expression of recombinant antibody fragments to the biological warfare pathogen Brucella melitensis. J Immunol Methods 2003; 276:185-96; PMID:12738372; http://dx.doi.org/10.1016/s0022-1759(03)00100-5
- Sheets MD, Amersdorfer P, Finnern R, Sargent P, Lindquist E, Schier R, Hemingsen G, Wong C, Gerhart JC, Marks JD. Efficient construction of a large nonimmune phage antibody library: the production of high-affinity human single-chain antibodies to protein antigens. Proc Natl Acad Sci U S A 1998; 95:6157-62; PMID:9600934; http://dx.doi.org/10.1073/pnas.95.11.6157
- Feldhaus MJ, Siegel RW, Opresko LK, Coleman JR, Feldhaus JMW, Yeung YA, Cochran JR, Heinzelman P, Colby D, Swers J, et al. Flow-cytometric isolation of human antibodies from a nonimmune Saccharomyces cerevisiae surface display library. Nat Biotechnol 2003; 21:163-70; PMID:12536217; http://dx.doi.org/10.1038/nbt785
- Knappik A, Ge L, Honegger A, Pack P, Fischer M, Wellnhofer G, Hoess A, Wölle J, Plückthun A, Virnekäs B. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J Mol Biol 2000; 296:57-86; PMID:10656818; http://dx.doi.org/10.1006/jmbi.1999.3444
- Maruyama T, Rodriguez LL, Jahrling PB, Sanchez A, Khan AS, Nichol ST, Peters CJ, Parren PWHI, Burton DR. Ebola Virus Can Be Effectively Neutralized by Antibody Produced in Natural Human Infection. J Virol 1999; 73:6024-30; PMID:10364354
- Angelini A, Chen TF, De Picciotto S, Yang NJ, Tzeng A, Santos MS, Van Deventer JA, Traxlmayr MW, Wittrup KD. Protein Engineering and Selection Using Yeast Surface Display. Methods Mol Biol 2015; 1319:3-36; PMID:26060067; http://dx.doi.org/10.1007/978-1-4939-2748-7_1
- Brekke OH, Sandlie I. Therapeutic Antibodies for Human Diseases at the Dawn of the Twenty-first Century. Nat Rev Drug Discov 2003; 2:52-62; PMID:12509759; https://dx.doi.org/10.1038/nrd984
- Wörn A, Plückthun A. Stability engineering of antibody single-chain Fv fragments. J Mol Biol 2001; 305:989-1010; PMID:11162109; http://dx.doi.org/10.1006/jmbi.2000.4265
- Liu Y, Caffry I, Wu J, Geng SB, Jain T, Sun T, Reid F, Cao Y, Estep P, Yu Y, et al. High-throughput screening for developability during early-stage antibody discovery using self-interaction nanoparticle spectroscopy. MAbs 2014; 6:483-92; PMID:24492294; http://dx.doi.org/10.4161/mabs.27431
- Xu Y, Roach W, Sun T, Jain T, Prinz B, Yu TY, Torrey J, Thomas J, Bobrowicz P, Vásquez M, et al. Addressing polyspecificity of antibodies selected from an in vitro yeast presentation system: A FACS-based, high-throughput selection and analytical tool. Protein Eng Des Sel 2013; 26:663-70; PMID:24046438; http://dx.doi.org/10.1093/protein/gzt047
- Wang S, Liu M, Zeng D, Qiu W, Ma P, Yu Y, Chang H, Sun ZW. Increasing stability of antibody via antibody engineering: Stability engineering on an anti-hVEGF. Proteins Struct Funct Bioinforma 2014; 82:2620-30; PMID:24916692; http://dx.doi.org/10.1002/prot.24626
- Ponsel D, Neugebauer J, Ladetzki-Baehs K, Tissot K. High affinity, developability and functional size: The holy grail of combinatorial antibody library generation. Molecules 2011; 16:3675-700; PMID:21540796; http://dx.doi.org/10.3390/molecules16053675
- Tiller T, Schuster I, Deppe D, Siegers K, Strohner R, Herrmann T, Berenguer M, Poujol D, Stehle J, Stark Y, et al. A fully synthetic human Fab antibody library based on fixed VH/VL framework pairings with favorable biophysical properties. MAbs 2013; 5:445-70; PMID:23571156; http://dx.doi.org/10.4161/mabs.24218
- Jayaram N, Bhowmick P, Martin ACR. Germline VH/VL pairing in antibodies. Protein Eng Des Sel 2012; 25:523-9; PMID:22802295; http://dx.doi.org/10.1093/protein/gzs043
- Mcconnell AD, Zhang X, Macomber JL, Chau B, Joseph C, Rahmanian S, Hare E, Spasojevic V, Horlick RA, King DJ, et al. A general approach to antibody thermostabilization. MAbs 2014; 6:1274-82; PMID:25517312; http://dx.doi.org/10.4161/mabs.29680
- Rouet R, Lowe D, Christ D. Stability engineering of the human antibody repertoire. FEBS Lett 2014; 588:269-77; PMID:24291820; http://dx.doi.org/10.1016/j.febslet.2013.11.029
- Reddy ST, Ge X, Miklos AE, Hughes RA, Kang SH, Hoi KH, Chrysostomou C, Hunicke-Smith SP, Iverson BL, Tucker PW, et al. Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nat Biotechnol 2010; 28:965-9; PMID:20802495; http://dx.doi.org/10.1038/nbt.1673
- DeKosky BJ, Ippolito GC, Deschner RP, Lavinder JJ, Wine Y, Rawlings BM, Varadarajan N, Giesecke C, Dörner T, Andrews SF, et al. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat Biotechnol 2013; 31:166-9; PMID:23334449; http://dx.doi.org/10.1038/nbt.2492
- DeKosky BJ, Kojima T, Rodin A, Charab W, Ippolito GC, Ellington AD, Georgiou G. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nat Med 2015; 21:86-91; PMID:25501908; http://dx.doi.org/10.1038/nm.3743
- Wang B, Kluwe CA, Lungu OI, DeKosky BJ, Kerr SA, Johnson EL, Jung J, Rezigh AB, Carroll SM, Reyes AN, et al. Facile Discovery of a Diverse Panel of Anti-Ebola Virus Antibodies by Immune Repertoire Mining. Sci Rep 2015; 5:13926; PMID:26355042; http://dx.doi.org/10.1038/srep13926
- Becker PD, Legrand N, van Geelen CMM, Noerder M, Huntington ND, Lim A, Yasuda E, Diehl SA, Scheeren FA, Ott M, et al. Generation of human antigen-specific monoclonal IgM antibodies using vaccinated “human immune system” mice. PLoS One 2010; 5:1-10; PMID:20957227; http://dx.doi.org/10.1371/journal.pone.0013137
- Kuroiwa Y, Kasinathan P, Sathiyaseelan T, Jiao J, Matsushita H, Sathiyaseelan J, Wu H, Mellquist J, Hammitt M, Koster J, et al. Antigen-specific human polyclonal antibodies from hyperimmunized cattle. Nat Biotechnol 2009; 27:173-81; PMID:19151699; http://dx.doi.org/10.1038/nbt.1521
- Kurosawa N, Yoshioka M, Fujimoto R, Yamagishi F, Isobe M. Rapid production of antigen-specific monoclonal antibodies from a variety of animals. BMC Biol 2012; 10:1-14; PMID:23017270; http://dx.doi.org/10.1186/1741-7007-10-80
- Saggy I, Wine Y, Shefet-Carasso L, Nahary L, Georgiou G, Benhar I. Antibody isolation from immunized animals: comparison of phage display and antibody discovery via V gene repertoire mining. Protein Eng Des Sel 2012; 25:539-49; PMID:22988130; http://dx.doi.org/10.1093/protein/gzs060
- Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Isolating and engineering human antibodies using yeast surface display. Nat Protoc 2006; 1:755-68; PMID:17406305; http://dx.doi.org/10.1038/nprot.2006.94
- Bowley DR, Labrijn AF, Zwick MB, Burton DR. Antigen selection from an HIV-1 immune antibody library displayed on yeast yields many novel antibodies compared to selection from the same library displayed on phage. Protein Eng Des Sel 2007; 20:81-90; PMID:17242026; http://dx.doi.org/10.1093/protein/gzl057
- Lavinder JJ, Wine Y, Giesecke C, Ippolito GC, Horton AP, Lungu OI, Hoi KH, DeKosky BJ, Murrin EM, Wirth MM, et al. Identification and characterization of the constituent human serum antibodies elicited by vaccination. Proc Natl Acad Sci U S A 2014; 111:2259-64; PMID:24469811; http://dx.doi.org/10.1073/pnas.1317793111
- Lu J, Panavas T, Thys K, Aerssens J, Naso M, Fisher J, Rycyzyn M, Sweet RW. IgG variable region and VH CDR3 diversity in unimmunized mice analyzed by massively parallel sequencing. Mol Immunol 2014; 57:274-83; PMID:24211535; http://dx.doi.org/10.1016/j.molimm.2013.09.008
- Venet S, Kosco-Vilbois M, Fischer N. Comparing CDRH3 diversity captured from secondary lymphoid organs for the generation of recombinant human antibodies. MAbs 2013; 5:690-8; PMID:23924800; http://dx.doi.org/10.4161/mabs.25592
- Dessain SK, Adekar SP, Berry JD. Exploring the native human antibody repertoire to create antiviral therapeutics. Curr Top Microbiol Immunol 2008; 317:155-83; PMID:17990793; http://dx.doi.org/10.1007/978-3-540-72146-8_6
- Benatuil L, Perez JM, Belk J, Hsieh CM. An improved yeast transformation method for the generation of very large human antibody libraries. Protein Eng Des Sel 2010; 23:155-9; PMID:20130105; http://dx.doi.org/10.1093/protein/gzq002
- Lefranc M-P, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, Wu Y, Gemrot E, Brochet X, Lane J, et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res 2009; 37:D1006-12; PMID:18978023; http://dx.doi.org/10.1093/nar/gkn838
