ABSTRACT
Raising functional antibodies against G protein-coupled receptors (GPCRs) is challenging due to their low density expression, instability in the absence of the cell membrane's lipid bilayer and frequently short extracellular domains that can serve as antigens. In addition, a particular therapeutic concept may require an antibody to not just bind the receptor, but also act as a functional receptor agonist or antagonist. Antagonizing the glucose-dependent insulinotropic polypeptide (GIP) receptor may open up new therapeutic modalities in the treatment of diabetes and obesity. As such, a panel of monoclonal antagonistic antibodies would be a useful tool for in vitro and in vivo proof of concept studies. The receptor is highly conserved between rodents and humans, which has contributed to previous mouse and rat immunization campaigns generating very few usable antibodies. Switching the immunization host to chicken, which is phylogenetically distant from mammals, enabled the generation of a large and diverse panel of monoclonal antibodies containing 172 unique sequences. Three-quarters of all chicken-derived antibodies were functional antagonists, exhibited high-affinities to the receptor extracellular domain and sampled a broad epitope repertoire. For difficult targets, including GPCRs such as GIPR, chickens are emerging as valuable immunization hosts for therapeutic antibody discovery.
Abbreviations
| BLI | = | biolayer interferometry |
| ECD | = | extracellular domain |
| GEM | = | gel encapsulated microenvironment |
| GIP | = | glucose dependent insulinotropic peptide |
| GIP | = | glucose dependent insulinotropic peptide receptor |
| mAb | = | monoclonal antibody |
| PK | = | protein kinase |
Introduction
Glucose-dependent insulinotropic polypeptide, also named gastric inhibitory polypeptide (or GIP), has long been known as one of the incretins stimulating insulin secretion in response to food intake.Citation1, 2 However, in the context of diabetes the insulinotropic action of GIP is markedly diminished.Citation3 In contrast, the effects of GIP on fat deposition and lipid metabolism in adipose tissue are not impaired, thus promoting the development of insulin resistance and obesity. Furthermore, GIP stimulates the secretion of glucagon, which might contribute to the lack of postprandial glucagon suppression and hyperglycemia seen in patients with type 2 diabetes.Citation4-7
GIP, a 42-amino acid peptide, is released into circulation from K cells in the duodenum and small intestine upon nutrient ingestion. GIP exerts activity via its receptor, GIPR. GIPR is expressed primarily in the pancreas, adipose tissue, stomach, small intestine, bone and central nervous system.Citation2, 8 The GIP receptor is a member of the class B (Secretin) family of G protein-coupled receptors (GPCRs) and activation results in the stimulation of adenylyl cyclase and Ca(2+)-independent phospholipase A(2) and activation of protein kinase (PK) A and PKB. GIPR is coupled to GαS, and activation of the receptor leads to an increase of the second messenger cAMP. GIPR is characterized by a large extracellular loop (extracellular domain - ECD) that serves as the site of specific interaction with its ligand, binding with low affinity (µM range). The ECD confers the selectivity of the receptor to its ligand, and upon binding a conformational change leads to receptor activation with potency in the pM range.Citation9-11
Various groups have shown that GIPR antagonism has a beneficiary impact on disease phenotype in rodent models. Under a high fat diet, GIPR knockout mice show an increased insulin-sensitivity, a resistance against diet-induced obesity, suppression of liver steatosis, and reduced plasma cholesterol and triglyceride levels.Citation12, 13 Similar effects are seen with a variety of antagonistic peptides,Citation14-16 and recently with antagonistic antibodies raised in a phage display campaign.Citation17
GPCRs, however, are difficult targets for antibody campaigns. Often, GPCRs occur in low density on the cell surface and are very unstable when purified from the cellular membrane, presenting a challenge in obtaining sufficient amounts of immunogen in which native epitopes are maintained for antibody recognition. Furthermore, a particular therapeutic concept may require an antibody that does not just bind the GPCR but acts as an agonist or antagonist, which may necessitate the recognition of particular, potentially ligand-sensitive, epitopes. These additional requirements may further reduce the effective hit rate in antibody generation campaigns.Citation18
In addition to the challenges of preparing native GCPR protein for use as an immunogen, the receptor structure itself offers relatively few antigenic determinants at the cell surface that are potentially available for antibody binding. This paucity, when combined with the further restriction imposed by immunological tolerance, can lead to very poor immunogenicity for sequence-conserved GPCR targets. Human and murine GIPR have 81% identity at the protein level, and indeed our previous antibody campaigns using mice and rats as hosts resulted in a very small panel of functional antibody clones with limited epitope coverage. For the campaign described here, we selected chicken as an alternative host to generate a more diverse panel of antibodies. The rationale for using chicken is based upon the greater evolutionary distance between humans and chickens compared with humans and other mammals such as mice. This evolutionary distance allows chickens to produce a more vigorous and diverse immune response when challenged with human proteins.Citation19 In the specific case of GIPR, the chicken and human orthologs have only 37% sequence identity (based upon an incomplete chicken sequence). Another major advantage of chicken immunization is the generation of antibodies that recognize “pan-mammalian” epitopes; such antibodies are difficult or impossible to generate in mammalian hosts. An additional advantage of chicken antibodies comes with the broad species cross-reactivity, obviating the need to generate surrogate antibodies for the purpose of experimentation in various disease models.
Here, we used chicken immunization to obtain antagonistic antibodies against GIPR and compared those to antibodies previously raised in mice and rats. Our data show that for GIPR, chickens were the superior host, resulting in a larger number of antibodies, a higher frequency of functional antagonists and antibodies covering a broader epitope space.
Results
Enriching for species cross-reactive clones with immunization and screening strategies
Initially gel encapsulated microenvironment (GEM) screens were performed with lymphocytes from chicken 11272 (hGIPR-Fc immunization) using cellular reporters, parental Chinese hamster ovary (CHO) cells dyed blue with vital dye and CHO cells stably expressing mGIPR. This cellular reporter GEM screen yielded a high frequency of species cross-reactive clones (38% cross-reactive). Secondly, lymphocytes from chicken 11272 were used in GEM screens with reporter beads featuring hGIPR-Fc and hFc were coated onto white or blue beads, respectively. The reporter bead GEM screen was not as efficient as the cellular screen in obtaining species cross-reactive clones (4.8% cross-reactive). Therefore, going forward cellular reporters were used when GEM screens were set up for the remaining chicken lymphocytes. Some immunization protocols included mGIPR DNA in addition to hGIPR-Fc protein, with the goal of steering the response toward a more cross-reactive repertoire; the results were not extremely dramatic, but these immunizations did deliver a higher percentage of cross-reactive clones. Immunizations using only DNA resulted in low titer and yielded significantly fewer antibody clones overall.
Large and diverse antibody panels generated to GIPR
A total of 694 clones were generated from the chicken immunization. Of those, 462 were unique sequences. 206 were cross-reactive with human, cynomolgus, murine or rat GPCR and were moved along the screening cascade to EC50 determinations. 125 clones had an EC50 on human GPCR-expressing CHO cells that were within 4-fold of the control antibody. A total of 172 unique clones were advanced to further study.
Immunizing chickens with GIPR yields more antagonistic antibodies than classic hybridoma technology using mouse and rat as host species
The chicken immunization campaign yielded a total of 172 antibodies with unique sequences that were screened for functional activity, i.e., their ability to antagonize GIPR in a cAMP assay in vitro. In comparison, classic mouse and rat immunizations in conjunction with hybridoma technology, resulted in just 87 hybridoma supernatant samples whose activity could be assessed (, ). Notably, before obtaining those 87 hybridoma supernatants, multiple rodent immunization campaigns, which used a combination of GIPR cDNA, GIPR-Fc fusion protein and cell lines overexpressing GIPR as antigen and used both GIPR+/+ and GIPR−/− host strains, failed entirely at producing antagonistic anti-GIPR antibodies (data not shown).
Figure 1. Anti-GIPR antibodies raised in chicken vs. mouse and rat hybridoma antibodies. (A) illustrates the number and functional activity (antagonism as measured in GIPR specific cAMP assay) of anti-GIPR antibodies raised in chicken vs. previous campaigns using classic mouse and rat hybridoma technology. In (B), the chicken antibodies are broken down into those containing cysteines in the CDR3 of the heavy chain (Cys) and those without (no Cys) while also giving the fraction of antagonistic vs. not antagonistic antibodies in each population.
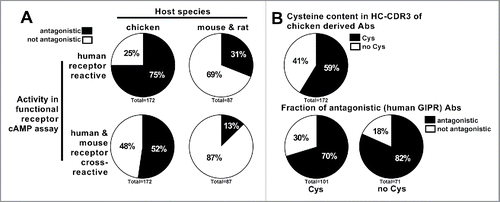
Among the chicken-derived antibodies, 129 (75%) were tested to be antagonistic in a human-GIPR specific assay, whereas 90 (52%) showed additional murine cross-reactivity by also exhibiting antagonistic behavior in a mouse-GIPR specific assay. For the population of hybridoma antibodies raised in mouse and rat, this fraction was only 31% (27) and 13% (11), respectively. During the antibody generation process, each chicken antibody was sequenced and only unique sequences were taken forward.
One feature of chicken antibodies is the presence of non-canonical cysteine residues in the complementarity-determining region (CDR)3 of the heavy chain. These cysteine residues can potentially form intra-chain disulfide bonds, and are particularly likely to occur in long CDR3s, where they are thought to play a role in stabilizing the secondary structure of the CDR3 loop.Citation20 Among the population of GIPR antibodies tested in this work, 101 (59%) contained cysteines in the HC-CDR3 (). The presence of cysteines, however, had no apparent effect on the likelihood of an antibody exhibiting antagonistic behavior in the GIPR cAMP assay. 71 (70%) and 58 (82%) of the antibodies possessed antagonistic activity among the cysteine-containing and cysteine-free antibody populations, respectively.
The antibodies used in this work were derived from 5 chickens (). The likelihood of antibody antagonism and the fraction of antibodies containing HC-CDR3 cysteines varied considerably by the host chicken, suggesting multiple chickens should be used in each immunization to obtain the best possible antibody diversity.
Table 1. Numerical representation of chicken-derived anti-GIPR antibodies.
Chicken antibodies reveal a wide range of affinities to human and murine GIPR
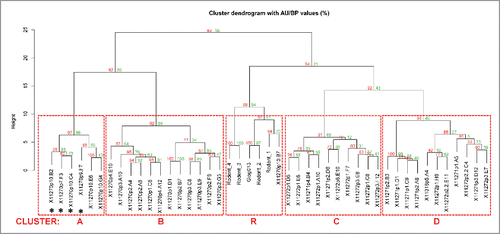
The KD values of GIPR antibodies against human, mouse and rat recombinant antigen (using immobilized extracellular domain protein, see Material & Methods) were determined by surface plasmon resonance (SPR; , Figs. S2 and 3). Affinities against all species antigens were spread over several orders of magnitude (). The chicken-derived antibodies () showed significantly higher affinities, i.e., lower KD values, for human (median 0.7 nM; range 0.009 nM – 212 nM) than for either mouse (median 8.1 nM; range 0.3 nM – 1043 nM) or rat (median 6.7 nM; range 0.2 nM – 3110 nM) antigens. The same trend was present for the mouse and rat hybridoma antibodies (, right panel), which were assessed against human (median 0.3 nM; range 0.02 nM – 3.0 nM) and mouse (median 0.9 nM; range 0.3 nM – 5.2 nM) GIPR antigen. Notably, the antibodies raised from the rodent versus the chicken campaigns differ in their Fc (mouse or rat for rodent, human for chicken) and were thus captured on separate CM5 chips for SPR analysis (see Material & Methods).
Figure 2. Affinity data measured by surface plasmon resonance for anti-GIPR antibodies. (A) summarizes the SPR-derived KD values for anti-GIPR antibodies. The left side shows chicken-derived antibody affinities against human, mouse and rat-GIP receptor extracellular domain (ECD), whereas the right side provides human and mouse GIP receptor ECD affinities for the legacy mouse and rat hybridoma campaigns for comparison. All affinities are shown, irrespective of whether an antibody is antagonistic or not. (B) breaks down the KD values by the antagonistic activity of the chicken-derived antibodies in human or mouse receptor G specific functional cAMP assays. In (C), antibody KD values are grouped by the host chicken, whereas (D) lists chicken-derived antibody affinities based on whether or not they contain a cysteine in the heavy chain CDR3 (left side), as well as by functional activity against human and mouse GIPR and cysteine content (right side). The red lines indicate the population medians. P-values were calculated using Kruskal-Wallis and Mann-Whitney tests. ns - not significant; * p<0.05; ** p<0.01; *** p<0.001; **** p<0.0001.
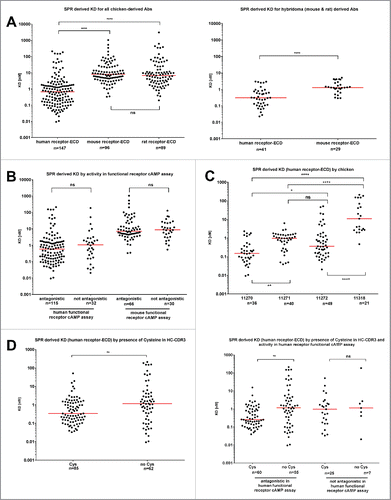
Among the chicken antibody population, antigen affinity was not correlated with functional antagonistic activity in either the human or mouse-GIPR cAMP assay (). The individual host chicken, however, had a strong impact (). Animal 11270 produced antibodies with the lowest KD values against human GIPR antigen (median 0.2 nM; range 0.009 nM – 2.3 nM), whereas animal 11318 yielded antibodies with the highest KD (median 0.9 nM; range 11.1 nM – 212 nM). Antibody populations obtained from animals 11271 (median 1.0 nM; range 0.03 nM – 6.4 nM) and 11272 (median 0.4 nM; range 0.02 nM – 51.2 nM) occupied an intermediate position in affinity and were comparable to each other.
Against the human GIPR antigen, the cysteine containing antibodies displayed a lower median KD (0.3 nM) than the cysteine-free group (1.2 nM) (). However, the antibodies with the lowest overall KD were found in the cysteine-free group, which exhibited a wider spread of measured KDs (range: 0.009 nM – 212 nM) than the cysteine-containing population (range: 0.03 nM – 51.2 nM). When additionally grouping the chicken-derived antibodies by their activity profile (), the median KD of the cysteine containing population was only lower in the antagonistic group, whereas no difference was observed for cysteine-containing vs. cysteine-free antibodies that fail to antagonize GIPR.
The IC50 of the functional antagonistic GIPR cAMP assay is not determined by the presence of cysteines and shows weak correlation to the affinity
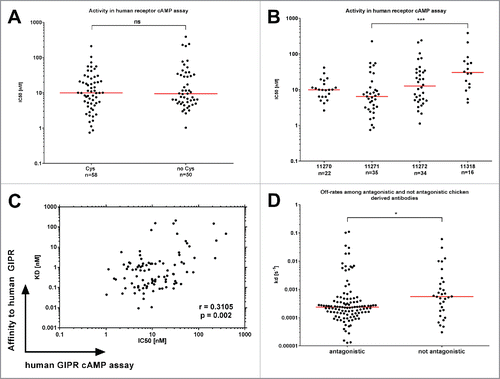
The functional activity of chicken-derived anti-GIPR antibodies was assessed using an αScreen based cAMP in vitro assay (). Briefly, GIPR-expressing cells are stimulated with the endogenous ligand GIP and the resulting cAMP production is quantified. Antagonistic antibodies are characterized by their ability to block the generation of cAMP (for exemplary data, see Fig. S3). The IC50 [nM] of antagonistic antibodies was not affected by the presence (median 10.0 nM; range 0.75 nM – 211.3 nM) or absence (median 9.4 nM; range 1.3 nM – 389.6 nM) of cysteines in the HC-CDR3 (). The host chicken had some effect on the observed strength of functional antagonism (), but the differences are only statistically significant for chicken 11271 (median 6.5 nM) vs. 11318 (median 30.8 nM), and not for other comparisons (medians: 11270: 10.0 nM; 11272: 12.7 nM). There was only a weak correlation between the affinity (KD) and activity (IC50) in the cAMP assay (, correlation coefficient = 0.3105). The median off-rate parameter (kd) among the antagonistic antibodies was significantly lower (2.36E-4 s−1) than among the non-antagonistic fraction (5.53E-4 s−1, ), indicating that longer receptor occupancy of the antibody may benefit antagonism.
Figure 3. Functional activity of chicken-derived anti-GIPR antibodies in cAMP assay. IC50 values for the chicken-derived Abs were determined using an αscreen cAMP assay. (A) illustrates obtained values for functional antibodies broken down by cysteine content, whereas (B) illustrates antibodies raised in different chicken. (C) shows a correlation plot of the KD values vs. the IC50. In (D), the SPR off-rate constants (kd) are compared between antagonistic and not antagonistic chicken-derived antibodies. The red lines indicate the population medians. P-values were calculated using Kruskal-Wallis and Mann-Whitney tests. ns - not significant; * p<0.05; *** p<0.001.
Chicken-derived anti-GIPR antibodies with long heavy chains CDR3 are more likely to contain cysteines and have higher affinities than those with shorter CDR3s
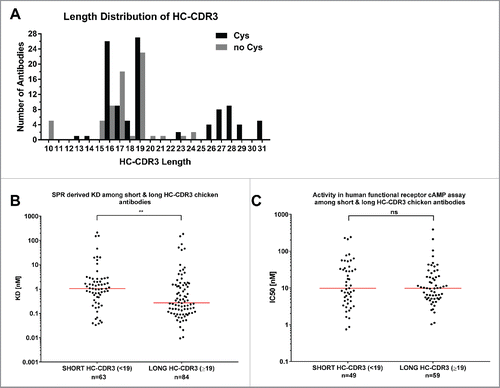
The length of heavy chain CDR3 sequences in our anti-GIPR chicken antibody cohort ranged from 10 to 31 (). As described previously by Wu et al,Citation20 our cysteine-containing antibodies had significantly (p = 0.0016) longer CDR3s (median 19, range 13 – 31) than the cysteine-free antibodies (median 17, range 10 – 24). When binning all antibodies into a short (less than 19 amino acids) and a long (equal to or more than 19 amino acids) heavy chain CDR3 group (), the median affinity to the human GIPR-ECD was lower in the long group (0.27 nM vs 1.04 nM, p = 0.0020). Notably, this distinction was not maintained when looking at the IC50 as a measure of antagonism (),
Figure 4. HC-CDR3 length of anti-GIPR chicken antibodies and effect on affinity and function. The histogram (A) lists the HC-CDR3 length distribution of cysteine-containing (black bars) vs. cysteine-free (gray bars) chicken-derived anti-GIPR antibodies. (B and C) list the affinities (KD) and antagonistic activities (IC50) among antibodies with short (< 19 amino acids) vs. long (≥ 19 amino acids) CDR3 sequences, respectively. The red lines indicate the population median. P-values were calculated using the Mann-Whitney test. ns - not significant; ** p<0.01.
Biolayer interferometry clustering reveals a larger epitope diversity among the chicken-derived antibodies than antibodies from other sources
Biolayer interferometry-based clustering using the Fortebio Octet HTX platform was used to assess the epitope diversity of 40 chicken-derived antibodies, as well as 4 rodent-raised antibodies and one phage display-derived antibody (Gipg013) described previously in the literature Citation17 (and , Fig. S5, ). Briefly, biotinylated human-GIPR ECD was immobilized on streptavidin biosensors and saturated by binding all anti-GIPR antibodies in a first step. In a second step, all antibodies are tested for their ability to bind the GIPR-antibody1 complex. Additional binding indicates the recognition of a distinct epitope. The clustering result, shown as a 2-dimensional matrix (analysis based on method adapted from Liao-Chan et al Citation21) is represented in . The primary antibodies are shown in columns, the secondary antibodies in rows. Rows are sorted according to their Pearson correlation coefficient so that similar antibodies are located next to each other. The column sorting was adapted to match the row sorting, and thus all self-blocking antibody combinations are located on the diagonal (highlighted in red). The color gradient from blue (0) to white (100) was applied to highlight blocking or additional binding.
Figure 5. Biolayer interferometry-derived epitope clustering for anti-GIPR antibodies. A 2-dimensional matrix of the normalized biolayer interferometry assay data used for epitope clustering is shown. 45 anti-GIPR antibodies were assessed, 40 derived from chicken, 5 from other sources (rodent and phage display). The secondary antibodies are shown as rows, the primary antibodies as columns. Rows were sorted by their Pearson correlation coefficient (penultimate column on the right). Following Pearson row sorting, the columns were sorted to match the rows - hence the self-blocking value for each antibody is found on the diagonal (values marked in red). In addition, the Pearson correlation coefficient for the columns is shown in the bottom row. A color gradient from blue (0) to white (100) was applied to the data to highlight cross-blocking or competition. The last-most column indicates the epitope cluster an antibody was assigned based on the dendrogram shown in .
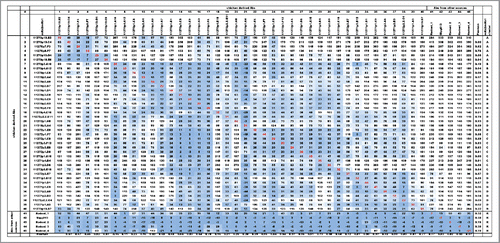
Figure 6. Dendrogram of BLI epitope clustering data. The dendrogram representing clustering of the secondary antibodies was generated in pvclust (see Materials and Methods). The axis on the left (Height) serves as a measure for antibody dissimilarity. Using a height cut-off of 10, 5 antibody clusters (A, B, C, D, R - red dashed boxes emerge). The AU value (%) represents the approximate unbiased p-value computed by pvclust using multiscale bootstrap resampling, whereas the BP value (%) indicates the bootstrap probability. Clusters with high AU values are strongly supported by the data. * denominates non-antagonistic antibodies.
The unsorted matrix was analyzed using pvclust in R.Citation22, 23 Pvclust provides hierarchical clustering of the BLI data (secondary antibodies only,). The ‘Height’ axis provides a measurement for antibody similarity. Using a cut-off of ‘10’, 5 different epitope clusters emerge: A, B, C, D and R. The AU value (%) represents the approximate unbiased p-value computed by pvclust using multiscale bootstrap resampling. Clusters with high AU values are strongly supported by the data. Notably, all 4 rodent-derived antibodies (Rodent_1 – 4) and the phage display antibody Gipg013 cross-compete for binding to the GIPR-ECD and thus fall in the same cluster (R). Hence, they recognize either the same epitope or their epitopes are overlapping in a manner that prevents parallel binding. One chicken-derived antibody (11270p10.B7) can also be found in that cluster. The emergence of the additional clusters (A-D) reveals that there are likely additional epitopes on the GIPR-ECD that could only be accessed via chicken immunization. Notably, the epitope clustering matrix () is not fully symmetric. For example, for chicken-derived Abs 34 (11271p1.D1) and 14 (11270p3.C8) cross-blocking depends on the order in which the antibodies are used. When number 34 is used as the primary and 14 as the secondary antibody, additional binding is observed (normalized binding signal 128). When using 14 first and 34 second, however, cross-blocking (binding signal 25) occurs. These discrepancies may occur when the primary antibody is able to induce conformational changes in the antigen that either enables or blocks binding by a secondary antibody. Elucidating the structural basis for this antibody-GIPR binding behavior will require further research.
Among the chicken-derived antibodies, the epitope cluster is closely correlated with the heavy chain CDR3 sequence and function
To validate our BLI-based epitope clustering approach (and ), we also analyzed the VH sequences of the 40 chicken-derived antibodies using ClustalW alignment Citation24 and MegAlign (DNAStar) for phylogenetic tree construction (). It became apparent that antibodies that fell into the same BLI-derived epitope cluster are characterized by similar HC-CDR3 sequences. Antibodies with identical HC-CDR3 (denoted as twins) always belong to the same epitope cluster. Notably, each epitope cluster contained both cysteine-containing and cysteine-free antibodies, suggesting that epitope diversity among chicken-derived antibodies is independent of the HC-CDR3 cysteines.
Figure 7. Phylogenetic tree of 40 chicken-derived anti-receptor G antibodies and cluster assignment. The phylogenetic tree (left panel) for the 40 chicken-derived anti-receptor G antibodies used for epitope clustering () was generated in MegAlign using ClustalW alignment of all full length VH sequences. Bootstrap percentage values are shown on each node. The table (right panel) lists antibody IDs, presence of cysteines, antagonistic activity as well as length and amino acid sequence of the HC-CDR3. The symbols in the penultimate column indicate antibodies with identical HC-CDR3s. The last column lists the BLI epitope cluster assignment derived from the dendrogram shown in .

Due to our focus on functionally antagonistic anti-GIPR antibodies, only 4 out of the 40 tested chicken antibodies in the epitope clustering analysis were from the non-antagonistic population. However, all of them fell within the same cluster (cluster A), highlighting a correlation between the epitope and anti-GIPR antibody function.
Discussion
GPCRs, owing to their instability outside of the membrane's lipid bilayer and typically small extracellular domains that can serve as antigens, are considered to be difficult targets for generating antibody therapeutics. We have successfully used chicken immunization to generate a highly diverse set of functional (antagonistic) antibodies against GIPR that cover a broad epitope space. The chicken hosts proved vastly superior in comparison to previously run classic rodent hybridoma campaigns from which we obtained a very small number of antagonistic antibodies.
Chickens readily produce antibodies that are cross reactive to mammalian orthologs. In the case of GIPR, chickens that were immunized and screened (in GEMs) Citation25 exclusively using human GIPR did indeed generate antibodies that were cross-reactive with murine GIPR, albeit at a relatively low frequency (∼5%), which can be considered a “baseline” cross-reactivity rate for this particular target. Protocol adjustments were made that included immunizing with murine GIPR (DNA) as well as screening in GEMs with mGIPR-expressing cells, which combined to significantly enhance the hit rate for human/murine cross-reactive mAbs. It should be noted that, while DNA immunization appeared to be useful in “pushing” the response toward species cross-reactivity, the 2 birds that were immunized exclusively with DNA until the final boost (chickens 11312 and 11318) did not achieve a final titer that was comparable to the other birds that received some protein boosts, and the antibodies recovered were of overall lower affinities. The alternating DNA/protein immunization strategy, however, was quite effective in producing a diverse panel of cross-reactive antibodies.
Mouse/human cross-reactive antibodies are not generally recovered from mice because they represent self-reactive specificities that are typically eliminated from the host animal, so it is somewhat surprising that the few murine- and rat-derived GIPR antibodies that were obtained did in fact cross-react to mGIPR. However, according to our epitope clustering investigations, all rodent-derived antibodies are located within the same cluster (cluster “R”). One chicken-derived antibody was also found in this group. No rodent clones were identified to any of the other 4 non-overlapping epitope clusters that were defined by the panel of chicken antibodies. Many of the antibodies in each of these epitope clusters cross-react with GIPR from all 4 species tested (mouse, rat, cynomolgus, human), and thus can be considered to define “pan-mammalian” epitopes. The absence of such specificities in the rodent panels is likely attributable to the influence of self-tolerance in the host animals. The strong sequence conservation among the rodent and human GIPR-ECD is a challenge in raising diverse sets of antibodies because the human antigen does not elicit a very strong immune response in either mouse or rat, and this limits the epitope space that can be covered by rodent immunization. Chicken immunization, on the other hand, has clearly enabled the generation of antibodies with much larger epitope diversity, which is likely driven by the very low sequence homology of the chicken GIPR ortholog. In general, chicken orthologs have 2–3x more amino acid substitutions than murine orthologs when compared with a given human protein sequence. In some cases, the chicken ortholog is so divergent that it cannot be positively identified, resulting in the chicken being effectively a “knockout” for the gene in question. The ability of chicken immunization to expand the epitope repertoire for other antigens has also recently been described by Abdiche et al.Citation19
It is not clear why such a high frequency of antagonistic antibodies were obtained through the chicken immunization performed in this study, since no special selection was used to enrich for biologic activity. The cell-based GEM screen did bias toward antibodies that recognize native conformation GIPR and were also species cross-reactive, but a bioassay for activity was not used in the GEMs. All cAMP assays were performed after the mAbs were recovered. In the case of GIPR, it may have been serendipitous that the selection strategy we used resulted in a mAb panel that was weighted toward receptor antagonism. While this may not be the case with a different receptor where epitopes associated with antagonism are more rare, it is reasonable to assume that the expanded epitope coverage that is generated through chicken immunization will be generally beneficial in the pursuit of biologically active antibodies to a variety of human targets.
Additional cysteines and disulfide bonds, a particular feature of chicken-derived antibodies used to stabilize long heavy chain CDR3 loops,Citation20 may be considered liabilities for biopharmaceutical development. The immunogenic potential of these structures in humans is unknown at present, and as such may pose a challenge for humanization, although examples of humanizing these types of antibodies have been recently published.Citation26, 27 Replacing the cysteines via mutagenesis may have an unpredictable effect on the antibodies' affinity, binding mode or may even disrupt antigen recognition altogether. Further, additional cysteines can result in non-classical disulfide bond formation or result in increased proportions of free sulfhydryl groups, which may cause the emergence of difficult to control antibody subpopulations during manufacturing, hamper formulation development or promote antibody aggregation.Citation28-31 However, we have no direct evidence of our non-canonical cysteine-containing clones being problematic in terms of either expression or aggregation when prepared at research scale; we have not evaluated stability differences between Cys and non-Cys clones. Certainly many camelid VHH antibodies that are being developed for therapeutic use contain non-canonical cysteine residues that presumably are necessary for both activity and stability.Citation32, 33
Whether the development risk of such clones is real or perceived, the avoidance of antibodies with these potential liabilities may be important to allow a smooth drug development process given the current state of knowledge. For our GIPR program, this goal was straightforward to achieve. While the majority (60%) of our anti-GIPR antibodies contained cysteines in the heavy chain CDR3, we found comparable fractions of antagonism among both cysteine-containing and cysteine-free antibodies. The median KDs for the cysteine-containing antibodies was lower than for the cysteine-free population, as was the case for antibodies with heavy chain CDR3s containing 19 or more amino acids, which indicates that longer CDR3 loops enable optimal binding to the GIPR-ECD. Nevertheless, the overall lowest KD values were found among antibodies in the cysteine-free fraction and the presence of cysteines had no discernible effect on the observed IC50 in the functional GIPR cAMP assay. Additionally, each identified epitope cluster contained at least one cysteine-free antibody, suggesting that the presence of longer CDR3s and disulfide-bonds is not an absolute necessity to obtain epitope diversity in chicken-derived antibodies. We conclude that screening a sufficiently high number of antibodies raised in chicken enables the selection of cysteine-free antibodies with desired functional profiles if this feature is desirable for a particular antibody development project. Furthermore, recent advances in the development of a genetically engineered chicken that produces human sequence antibodies may provide a viable alternative to take advantage of chicken host immune recognition without introducing potential sequence liabilities.Citation34-36
Our epitope clustering experiments revealed several interesting points. Cluster assignment obtained from the biolayer interferometry studies was well supported by the antibody sequence data, in particular the heavy chain CDR3 sequence. Our finding that chicken-derived antibodies containing similar or identical heavy chain CDR3 sequences fall in the same BLI clusters, and therefore recognize the same epitope, underlines that this part of the antibody structure, which possesses the highest sequence diversity, serves as the key determinant for specificity and selectivity.Citation37 Notably, we also saw a relationship between functional antagonism and epitope cluster. The 4 non-antagonistic antibodies clustered together, revealing that there are specific functional (allowing antibody-mediated antagonism when bound) and non-functional epitopes on the GIPR-ECD. However, 2 functional, i.e., antagonistic, antibodies, also fell within this cluster. Both share a unique HC-CDR3 sequence distinct from the non-functional antibodies in the cluster. This suggests that the detailed structure of the antibody paratope, i.e., the overall combined architecture of CDRs and frameworks, has an effect on antagonistic functionality when bound to a particular epitope of the GIPR-ECD. Detailed investigations into the antibody-receptor protein-protein interactions will be required to elucidate the underlying principles of this behavior.
Finally, the epitope clustering results appeared to be asymmetric for several antibodies, meaning that whether or not an antibody pair competes for the binding of the GIPR-ECD is dependent on which antibody was used first. Such asymmetries in epitope binning analyses have been reported in several studies in the literature, across different types of epitope mapping methodologies.Citation21, 38-42 Liao Chan et al Citation21 have attributed these observations to the possibility that the first epitope-paratope interaction blocks the second one due to steric, allosteric or electrostatic effects. It is conceivable that an antibody binding the GIPR-ECD induces conformational changes, in the same manner that the endogenous ligand GIP rearranges the secondary structure of the ECD upon binding the receptor.Citation9, 43-46 Such a subtle structural change may either enable or prevent the binding of a second antibody and help explain the observed clustering asymmetry.
In conclusion, we recommend considering chickens as alternative hosts in antibody generation campaigns, particularly when aiming to access difficult antigenic targets, such as GPCRs, where high levels of rodent-human sequence conservation may limit the antibody yield from classic mouse or rat hybridoma approaches. In our specific case, which aimed at raising functionally antagonist antibodies against the human GIP receptor, chicken immunization resulted in a much larger number of antibodies overall, a higher fraction of antagonistic antibodies and greater epitope diversity than rodent hybridoma technology.
Materials and methods
Antigen generation
Two types of GIPR-ECD fusion proteins were produced for use in this study. Human, mouse and rat-GIPR-ECD rabbit (rb) Fc fusion proteins were generated for biophysical assays, and human GIPR-ECD human Fc fusion protein (hGIPR-Fc) was used for immunization purposes. In both cases, expression vectors based on pTT5 were constructed with sequences coding for the receptor extracellular domains fused to sequences for the Fc domains. Proteins were produced by transient expression in HEK293-E6 cells and purified by standard Protein A chromatography. In addition, the rb-Fc fusion proteins contained a thrombin cleavage site between the ECD and Fc to facilitate removal of the Fc domain. Sequences are shown in Supplement 1.
Chicken immunization
A total of 5 female white leghorn chickens were used for the program, all starting immunization at 8–9 weeks of age. Animals were immunized with hGIPR-Fc as purified protein, or full-length mouse or human GIPR cDNA, or with an alternating regimen of both DNA and protein (). Two animals were immunized with DNA and followed with a final boost either of CHO cells expressing human GIPR (chicken 11312), or hGIPR-Fc (chicken 11318). For the remaining animals, initial boosts with 200 µg protein were mixed with an equal volume of Imject Freund's complete adjuvant (VWR, PI77140) and administered intramuscularly. All subsequent boosts with 100 µg protein were mixed with an equal volume of Imject Freund's incomplete adjuvant (VWR, PI77145) and administered intramuscularly. DNA immunizations were performed in accordance with the Bio-Rad GeneGun protocol (Bio-Rad; Hercules, CA, USA). Briefly, gold particles were coated with 4 µg plasmid DNA encoding a CMV-based expression cassette containing either full-length human GIPR, or full length murine GIPR, and administered intradermally using the GeneGun at 400 PSI.
Table 2. Chicken immunization regimes.
Once GIPR-specific titer plateaued in the plasma, chickens were killed, spleens were removed and a single cell suspension prepared and cells were cryopreserved for single B cell cloning.
Polyclonal immune responses
Plasma was collected bi-weekly during the immunization to determine titer. High binding ELISA plates were coated with 2 ug/ml of hGIPR-Fc or purified Fc in phosphate-buffered saline (PBS) overnight at 4°C. Plates were blocked with 3% dry milk in PBS + 0.05% Tween-20 (PBSM) for 1 hr at room temperature. Plates were washed with PBS+0.05% Tween-20 (PBST) and 50 ul of diluted plasma was added. Plasma was diluted with PBSM starting at 1:100 followed by 7, 5-fold dilutions down the ELISA plate. Plasma was incubated for 2 hours at room temperature then washed off with PBST. One hundred microliters of rabbit anti-chicken IgY HRP (Sigma, A9046) diluted 1:5000 with PBSM was added and incubated for 1 hour at room temperature. Plates were washed with PBST and developed with 50 ul of TMB and stopped with 50 ul 1N HCl. ELISA plates were read at 450 nm using the BioTek Synergy H1 Hybrid reader (Biotek; Vinooski, VT, USA).
Monoclonal antibody generation
Screening single B cells using the GEM assay
We used a single lymphocyte screening and recovery method, the GEM assay,Citation25, 47 to isolate antigen-specific monoclonal antibodies from the GIPR-immunized chickens. The GEM assay involves placing single antibody-secreting lymphocytes in proximity with reporters (which can be cells or beads). The secreted antibody diffuses locally within the GEM and has the opportunity to bind to the reporters. Bound antibody can be detected either directly through the use of a secondary antibody, or by eliciting a response in the reporter that generates a visual signal. Each GEM may contain multiple types of reporters that can be differentiated from each other based on color.
In this study, GEMs were prepared with both beads and cells. When beads were used, GIPR-Fc or Fc was coated onto white or blue beads, respectively. This approach allowed for the immediate elimination of any clones binding the Fc portion of the immunogen. CHO cells expressing GIPR were also used in the GEMs, with target specificity controlled for by the inclusion of parental CHO cells labeled with an alternative dye. Even though stable CHO lines were available expressing each of 4 species of GIPR (human, cynomolgus, rat, mouse), we generally opted for use of the CHO cell line expressing murine GIPR because we considered it more likely to identify pan-species cross reactive antibodies in an animal that was immunized with the human GIPR.
Expression and initial characterization of recombinant antibodies
Selected GEMs were isolated and antibody genes amplified through RT-PCR and cloned into the mammalian expression vector pF5a (Promega, C9401) in scFv-Fc format (with Fc derived from human IgG1 sequence). Plasmids containing recombinant scFv-Fc from the GEM harvests were transiently transfected into HEK293 cells and clonal supernatants were harvested. Supernatants were tested for specificity and species cross-reactivity on parental CHO, and human and murine GIPR-expressing CHO cells using flow cytometry. All clones that bound both hGIPR and mGIPR CHO cells were sequenced (n = 694), and unique clones (n = 462) were re-transfected to generate material for further testing. Concentrations of the clones in the 2 ml supernatant were determined and binding to hGIPR-Fc confirmed in ELISA format. A more detailed flow cytometry analysis of species cross-reactivity was performed using parental, human, murine, rat and cynomolgus GIPR expressing CHO cell. All clones were tested at a concentration of 5 ug/ml and compared with a positive control antibody.
Fluorescence-activated cell sorting methodology
hGIPR, mGIPR and parental CHO cells were lifted from culture flasks using StemPro Accutase (Invitrogen, A1110501). Cells were counted and re-suspended at 2 million cells per milliliter in FACS buffer (PBS + 1% BSA + 0.1% NaN3) and 50 ul was put into each well of a 96-well U bottom plate. Supernatants were collected from 96-well transfections of HEK293 cells and centrifuged to remove debris. 50 ul of supernatant was added to each of the 3 cell lines, hGIPR mGIPR, and parental and incubated for 1 hour at 4°C. Cells were washed 2 times with FACS buffer and re-suspended in 100 ul of 5 ug/ml anti-human IgG Alexa 488 (Jackson ImmunoResearch, 109-546-098). Cells were incubated for 45 min at 4°C and washed 2 times with FACS buffer. Cells were re-suspended in 150 ul of FACS buffer and fluorescence data was gathered using the Attune acoustic focusing cytometer (Thermo Fisher Scientific; Waltham, MA, USA).
EC50 determination for binding strength to cellular hGIPR
Human GIPR and parental CHO cells were lifted from culture flasks using StemPro Accutase (Invitrogen, A1110501). Cells were counted and re-suspended at 2 million cells per milliliter in FACS buffer (PBS + 1% BSA + 0.1% NaN3) and 50 µl was put into each well of a 96-well U bottom plate. Supernatants were collected from 6 well transfections of HEK293 cells and centrifuged to remove debris. Antibodies, diluted to 10 µg/ml followed by 7, 3-fold dilutions, were added to each of the cell lines, hGIPR and parental and incubated for 1 hour at 4°C. Cells were washed 2 times with FACS buffer and re-suspended in 100 µl of 5 µg/ml anti-human IgG Alexa 488 (Jackson ImmunoResearch, 109-546-098). Cells were incubated for 45 min at 4°C and washed 2 times with FACS buffer. Cells were re-suspended in 150 µl of FACS buffer and fluorescence data was gathered using the Attune acoustic focusing cytometer and the EC50 was determined using Prism v6 (Graphpad software). All antibodies were compared with a control antibody at the same concentrations.
Antibodies from previous immunization campaign
GIPR antibodies were also previously raised in classic rodent immunization and hybridoma campaigns (both rat and mouse immunizations). Animals were immunized using GIPR-ECD-Fc fusion protein, cell lines over-expressing GIPR or GIPR-specific cDNA. Overall yields of antibodies binding to and antagonizing GIPR were low (), prompting the switch to chicken immunization. For comparative purposes, screening data for a selection of rodent hybridoma supernatants is shown in .
Table 3. Rodent immunization campaigns.
In the epitope clustering analysis shown in , a total of 5 anti-GIPR recombinant antagonist antibodies were included to compare them to the chicken panel. Four (Rodent_1–4) were generated at Boehringer Ingelheim using hybridoma technology. Rodent_1 is derived from a mouse, whereas Rodent_2–4 are products of rat immunization. The antibody Gipg013 is the result of a phage display campaign described by Ravn et al.Citation17 All recombinant antibodies comprise the V-regions fused to human IgG1 constant regions and were expressed by using the transient CHO-3E7 system Citation48 and purified by routine Protein A chromatography. The biophysical characteristics (SPR KD & IC50 in cAMP assay) of those recombinant anti-GIPR antibodies are listed in .
Table 4. SPR affinity and cAMP activity of anti-GIPR reference antibodies from rodent immunization and phage display.
Surface plasmon resonance (biacore) methodology
All affinity measurements were performed and analyzed using a Biacore T200 (GE Healthcare; Chicago, IL, USA) using a single cycle kinetics protocol. Briefly, test antibodies were captured on CM5 chips (GE Healthcare, BR100012) using the human (for antibodies with human Fc) or mouse (for antibodies with mouse or rat Fc) antibody capture kits (GE Healthcare, BR100838 & BR100839). Human, mouse or rat-GIPR-ECD (thrombin cleaved from respective GIPR-ECD-rb-Fc antigen) were subsequently flown over the sensor surface at 5 different concentrations (0.125 µg/ml, 0.25 µg/ml, 0.5 µg/ml, 1 µg/ml and 2 µg/ml). The resulting curve was fitted in the Biacore T200 Evaluation software version 2.0 using a 1:1 fit to obtain KD values. A selection of curves can be found in the Fig. S2, the isoaffinity plot for all chicken antibodies binding to human GIPR-ECD is shown in supplement 3.
Functional GIPR cAMP assay
The antagonistic activity of antibodies was tested using the αScreen Functional cAMP assay kit (Perkin-Elmer, 6760625R) according to the manufacturer's instructions. A CHO cell line over-expressing human GIPR was used. Briefly, 10000 cells/50 µl/well were seeded in 384-well plates and incubated for 24 h at 37°C and 5% CO2. Then, cells were transferred into 100 µl assay buffer supplemented with serial dilutions of the antibodies and kept for 15 mins at 26°C. Subsequently, 100 pM of the receptor agonist GIP were added with a further incubation step of 30 min at 26°C. In parallel, a cAMP standard curve was prepared according to the manufacturer's instructions. 10 µl of the lysis/detection buffer, anti-cAMP acceptor beads, streptavidin donor beads and biotinylated cAMP mixture were added to each well and incubated for 2 h at RT in the dark. A serial dilution of the GIPR agonist human GIP (Sigma, G2269) was used as a positive control. Plates were measured using an Envision reader (Perkin-Elmer; Waltham, MA, USA). cAMP concentrations were interpolated from the standard curves and obtained values were plotted against the antibody concentrations. Each sample was measured in duplicate. A curve (4 parametric logistic dose response model) was fitted to obtain IC50 values. Exemplary data can be found in the supplement 4.
Epitope clustering analysis
Prior to epitope clustering, the human GIPR-ECD-rb-Fc fusion protein was biotinylated using the EZ-Link Sulfo NHS Biotin kit (Thermo Fisher Scientific, 21217) according to manufacturer's instructions. Measurements were performed using a Fortebio Octet HTX (Pall; Port Washington, NY, USA) high throughput biolayer interferometry system and streptavidin-coated biosensors (Pall, 18–5020) on which the antigen was immobilized. Antibodies were binned in-tandem - sensors were incubated with the first antibody, followed by a baseline and incubation with the second antibody. An antibody not binding to the antigen was used as a control. Antibodies were diluted to 25 µg/ml in PBS before performing the assay. Biosensors were regenerated in 10 mM glycine pH 3. Further details about the assay setup can be found in Fig. S5. The Octet Data Analysis software version 8 was used to process the data and create a matrix. The obtained nM shifts were normalized by dividing them by the value obtained using only the secondary antibody and multiplied by 100. The highest self-binding signal is used to judge the threshold for competition or additional binding. Primary antibodies are arranged in columns, secondary antibodies in columns. This data matrix was then analyzed using a method adapted from Liao-Chan et al.Citation21 Briefly, in Excel, the rows were sorted using the PEARSON function in a way that neighboring antibodies had the highest correlation coefficient. In addition, the unsorted matrix, with secondary antibody signals transposed into columns, were clustered using pvclust Citation22 in R version 6.1.3.Citation23 Prior to clustering, the matrix was normalized using the ‘scale’ function in R. In pvclust, clustering was performed using correlation as a distance measure and the Ward function as the clustering method. Specifically, the following command was used: result <- pvclust(scaled_data, method.hclust = ”ward.D2”, method.dist = ”correlation”, nboot = 10000)
Sequence analysis
VH sequences of the chicken-derived antibodies were aligned using MegAlign (DNAStar; Madison, WI, USA) and the ClustalW method.Citation24 MegAlign was also used to construct the phylogenetic tree shown in .
Data visualization and statistical analyses
Figures were created and using Prism v6 (Graphpad Software Inc.; La Jolla, CA, USA), as were statistical analyses. P-value calculations were performed using the non-paired, non-parametric Mann-Whitney (2 group comparisons) and Kruskal-Wallis (multi-group comparisons) tests.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental_Data.pdf
Download PDF (1.1 MB)Acknowledgments
The authors wish to thank Julie Ritchie for supporting antigen production and immunization. We are further grateful to Tobias Litzenburger, Nikolai Roosz and Michael Ritter for producing the GIPR-ECDs for biophysical analysis as well as Biacore and data analysis support. Our thanks goes to Yvonne Roth for performing cAMP functional assays and to Simon Plyte and Clive Long for invaluable scientific discussion and support.
Funding
This work was funded by Boehringer Ingelheim.
References
- Baggio LL, Drucker DJ. Biology of incretins: GLP-1 and GIP. Gastroenterology 2007; 132:2131-57; PMID:17498508; http://dx.doi.org/10.1053/j.gastro.2007.03.054
- McIntosh CHS, Widenmaier S, Kim S-J. Glucose-dependent insulinotropic polypeptide (Gastric Inhibitory Polypeptide; GIP). Vitam Horm 2009; 80:409-71; PMID:19251046; http://dx.doi.org/10.1016/S0083-6729(08)00615-8
- Christensen M, Vedtofte L, Holst JJ, Vilsbøll T, Knop FK. Glucose-dependent insulinotropic polypeptide: a bifunctional glucose-dependent regulator of glucagon and insulin secretion in humans. Diabetes 2011; 60:3103-9; PMID:21984584; http://dx.doi.org/10.2337/db11-0979
- Irwin N, Gault V, Flatt PR. Therapeutic potential of the original incretin hormone glucose-dependent insulinotropic polypeptide: diabetes, obesity, osteoporosis and Alzheimer's disease? Expert Opin Investig Drugs 2010; 19:1039-48; PMID:20698813; http://dx.doi.org/10.1517/13543784.2010.513381
- Flatt PR. Dorothy Hodgkin Lecture 2008. Gastric inhibitory polypeptide (GIP) revisited: a new therapeutic target for obesity-diabetes? Diabet Med 2008; 25:759-64.; PMID: 18513308; http://dx.doi.org/10.1111/j.1464-5491.2008.02455.x
- Meier JJ, Nauck MA, Schmidt WE, Gallwitz B. Gastric inhibitory polypeptide: the neglected incretin revisited. Regul Pept 2002; 107:1-13; PMID:12137960; http://dx.doi.org/10.1016/S0167-0115(02)00039-3
- Nauck MA, Baller B, Meier JJ. Gastric inhibitory polypeptide and glucagon-like peptide-1 in the pathogenesis of type 2 diabetes. Diabetes 2004; 53(Suppl 3):S190-6; PMID:15561910; http://dx.doi.org/10.2337/diabetes.53.suppl_3.S190
- Usdin TB, Mezey E, Button DC, Brownstein MJ, Bonner TI. Gastric inhibitory polypeptide receptor, a member of the secretin-vasoactive intestinal peptide receptor family, is widely distributed in peripheral organs and the brain. Endocrinology 1993; 133:2861-70; PMID:8243312; http://dx.doi.org/10.1210/endo.133.6.8243312
- Cordomí A, Ismail S, Matsoukas M-T, Escrieut C, Gherardi M-J, Pardo L, Fourmy D. Functional elements of the gastric inhibitory polypeptide receptor: Comparison between secretin- and rhodopsin-like G protein-coupled receptors. Biochem Pharmacol 2015; 96:237-46; PMID:26043830; http://dx.doi.org/10.1016/j.bcp.2015.05.015
- Miller LJ, Dong M, Harikumar KG, Gao F. Structural basis of natural ligand binding and activation of the Class II G-protein-coupled secretin receptor. Biochem Soc Trans 2007; 35:709-12; PMID:17635130; http://dx.doi.org/10.1042/BST0350709
- Hollenstein K, de Graaf C, Bortolato A, Wang M-W, Marshall FH, Stevens RC. Insights into the structure of class B GPCRs. Trends Pharmacol Sci 2014; 35:12-22; PMID:24359917; http://dx.doi.org/10.1016/j.tips.2013.11.001
- Miyawaki K, Yamada Y, Ban N, Ihara Y, Tsukiyama K, Zhou H, Fujimoto S, Oku A, Tsuda K, Toyokuni S, et al. Inhibition of gastric inhibitory polypeptide signaling prevents obesity. Nat Med 2002; 8:738-42; PMID:12068290; http://dx.doi.org/10.1038/nm727
- Yamada C, Yamada Y, Tsukiyama K, Yamada K, Yamane S, Harada N, Miyawaki K, Seino Y, Inagaki N. Genetic inactivation of GIP signaling reverses aging-associated insulin resistance through body composition changes. Biochem Biophys Res Commun 2007; 364:175-80; PMID:17937928; http://dx.doi.org/10.1016/j.bbrc.2007.09.128
- Pathak V, Gault VA, Flatt PR, Irwin N. Antagonism of gastric inhibitory polypeptide (GIP) by palmitoylation of GIP analogues with N- and C-terminal modifications improves obesity and metabolic control in high fat fed mice. Mol Cell Endocrinol 2015; 401:120-9; PMID:25449420; http://dx.doi.org/10.1016/j.mce.2014.10.025
- Gault VA, McClean PL, Cassidy RS, Irwin N, Flatt PR. Chemical gastric inhibitory polypeptide receptor antagonism protects against obesity, insulin resistance, glucose intolerance and associated disturbances in mice fed high-fat and cafeteria diets. Diabetologia 2007; 50:1752-62; PMID:17558485; http://dx.doi.org/10.1007/s00125-007-0710-4
- McClean PL, Irwin N, Cassidy RS, Holst JJ, Gault VA, Flatt PR. GIP receptor antagonism reverses obesity, insulin resistance, and associated metabolic disturbances induced in mice by prolonged consumption of high-fat diet. Am J Physiol Endocrinol Metab 2007; 293:E1746-55; PMID: 17848629; http://dx.doi.org/10.1152/ajpendo.00460.2007
- Ravn P, Madhurantakam C, Kunze S, Matthews E, Priest C, O'Brien S, Collinson A, Papworth M, Fritsch-Fredin M, Jermutus L, et al. Structural and pharmacological characterization of novel potent and selective monoclonal antibody antagonists of glucose-dependent insulinotropic polypeptide receptor. J Biol Chem 2013; 288:19760-72; PMID: 23689510; http://dx.doi.org/10.1074/jbc.M112.426288
- Hutchings CJ, Koglin M, Marshall FH. Therapeutic antibodies directed at G protein-coupled receptors. mAbs 2010; 2:594-606; PMID: 20864805; http://dx.doi.org/10.4161/mabs.2.6.13420
- Abdiche YN, Harriman R, Deng X, Yeung YA, Miles A, Morishige W, Boustany L, Zhu L, Izquierdo SM, Harriman W. Assessing kinetic and epitopic diversity across orthogonal monoclonal antibody generation platforms. mAbs 2016; 8:264-77; PMID: 26652308; http://dx.doi.org/10.1080/19420862.2015.1118596
- Wu L, Oficjalska K, Lambert M, Fennell BJ, Darmanin-Sheehan A, Shúilleabháin DN, et al. Fundamental characteristics of the immunoglobulin VH repertoire of chickens in comparison with those of humans, mice, and camelids. J Immunol 2012; 188:322-33; PMID: 22131336; http://dx.doi.org/10.4049/jimmunol.1102466
- Liao-Chan S, Zachwieja J, Gomez S, Duey D, Lippincott J, Theunissen J-W. Monoclonal antibody binding-site diversity assessment with a cell-based clustering assay. J Immunol Methods 2014; 405:1-14; PMID: 24380699; http://dx.doi.org/10.1016/j.jim.2013.12.007
- Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 2006; 22:1540-2; PMID: 16595560; http://dx.doi.org/10.1093/bioinformatics/btl117
- Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria, 2015.
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007; 23:2947-8; PMID: 17846036; http://dx.doi.org/10.1093/bioinformatics/btm404
- Mettler Izquierdo S, Varela S, Park M, Collarini EJ, Lu D, Pramanick S, Rucker J, Lopalco L, Etches R, Harriman W. High-efficiency antibody discovery achieved with multiplexed microscopy. Microscopy (Oxford, England) 2016; 65:341-52; PMID:27107009; http://dx.doi.org/10.1093/jmicro/dfw014
- Baek DS, Kim YS. Humanization of a phosphothreonine peptide-specific chicken antibody by combinatorial library optimization of the phosphoepitope-binding motif. Biochem Biophys Res Commun 2015; 463:414-20; PMID: 26036575; http://dx.doi.org/10.1016/j.bbrc.2015.05.086
- Townsend S, Fennell BJ, Apgar JR, Lambert M, McDonnell B, Grant J, Wade J, Franklin E, Foy N, Ní Shúilleabháin D, et al. Augmented binary substitution: Single-pass CDR germ-lining and stabilization of therapeutic antibodies. Proc Natl Acad Sci U S A 2015; 112:15354-9; PMID: 26621728; http://dx.doi.org/10.1073/pnas.1510944112
- Liu H, May K. Disulfide bond structures of IgG molecules. mAbs 2012; 4:17-23; PMID: 22327427; http://dx.doi.org/10.4161/mabs.4.1.18347
- Liu H, Chumsae C, Gaza-Bulseco G, Hurkmans K, Radziejewski CH. Ranking the susceptibility of disulfide bonds in human IgG1 antibodies by reduction, differential alkylation, and LC-MS analysis. Anal Chem 2010; 82:5219-26; PMID: 20491447; http://dx.doi.org/10.1021/ac100575n
- Brych SR, Gokarn YR, Hultgen H, Stevenson RJ, Rajan R, Matsumura M. Characterization of antibody aggregation: role of buried, unpaired cysteines in particle formation. J Pharm Sci 2010; 99:764-81; PMID: 19691118; http://dx.doi.org/10.1002/jps.21868
- Hutterer KM, Hong RW, Lull J, Zhao X, Wang T, Pei R, Le ME, Borisov O, Piper R, Liu YD, et al. Monoclonal antibody disulfide reduction during manufacturing: Untangling process effects from product effects. mAbs 2013; 5:608-13; PMID: 23751615; http://dx.doi.org/10.4161/mabs.24725
- Van Roy M, Ververken C, Beirnaert E, Hoefman S, Kolkman J, Vierboom M, Breedveld E, 't Hart B, Poelmans S, Bontinck L, et al. The preclinical pharmacology of the high affinity anti-IL-6R Nanobody(R) ALX-0061 supports its clinical development in rheumatoid arthritis. Arthritis Res Ther 2015; 17:135; PMID:25994180; http://dx.doi.org/10.1186/s13075-015-0651-0
- Van Heeke G, Allosery K, De Brabandere V, De Smedt T, Detalle L, de Fougerolles A. Nanobodies(R) as inhaled biotherapeutics for lung diseases. Pharmacology & Therapeutics 2017; 169:47-56; PMID:27373507; http://dx.doi.org/10.1016/j.pharmthera.2016.06.012
- Schusser B, Collarini EJ, Yi H, Izquierdo SM, Fesler J, Pedersen D, Klasing KC, Kaspers B, Harriman WD, Van de Lavoir MC, et al. Immunoglobulin knockout chickens via efficient homologous recombination in primordial germ cells. Proc Natl Acad Sci USA 2013; 110:20170-5; PMID: 24282302; http://dx.doi.org/10.1073/pnas.1317106110
- Leighton PA, Schusser B, Yi H, Glanville J, Harriman W. A diverse repertoire of human immunoglobulin variable genes in a chicken B cell line is generated by both gene conversion and somatic hypermutation. Front Immunol 2015; 6; PMID: 25852694; http://dx.doi.org/10.3389/fimmu.2015.00126
- Schusser B, Yi H, Collarini EJ, Izquierdo SM, Harriman WD, Etches RJ, Leighton PA. Harnessing gene conversion in chicken B cells to create a human antibody sequence repertoire. PLoS One 2013; 8:e80108; PMID: 24278246; http://dx.doi.org/10.1371/journal.pone.0080108
- Xu JL, Davis MM. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity 2000; 13:37-45; PMID: 10933393; http://dx.doi.org/10.1016/S1074-7613(00)00006-6
- Abdiche YN, Miles A, Eckman J, Foletti D, Van Blarcom TJ, Yeung YA, Pons J, Rajpal A. High-throughput epitope binning assays on label-free array-based biosensors can yield exquisite epitope discrimination that facilitates the selection of monoclonal antibodies with functional activity. PLoS One 2014; 9:e92451; PMID: 24651868; http://dx.doi.org/10.1371/journal.pone.0092451
- Abdiche YN, Malashock DS, Pinkerton A, Pons J. Exploring blocking assays using Octet, ProteOn, and Biacore biosensors. Anal Biochem 2009; 386:172-80; PMID: 19111520; http://dx.doi.org/10.1016/j.ab.2008.11.038
- Abdiche YN, Lindquist KC, Stone DM, Rajpal A, Pons J. Label-free epitope binning assays of monoclonal antibodies enable the identification of antigen heterogeneity. J Immunol Methods 2012; 382:101-16; PMID: 22609372; http://dx.doi.org/10.1016/j.jim.2012.05.010
- Nagata S, Numata Y, Onda M, Ise T, Hahn Y, Lee B, Pastan I. Rapid grouping of monoclonal antibodies based on their topographical epitopes by a label-free competitive immunoassay. J Immunol Methods 2004; 292:141-55; PMID: 15350519; http://dx.doi.org/10.1016/j.jim.2004.06.009
- Miller PL, Wolfert RL, Diedrich G. Epitope binning of murine monoclonal antibodies by a multiplexed pairing assay. J Immunol Methods 2011; 365:118-25; PMID: 21223970; http://dx.doi.org/10.1016/j.jim.2010.12.021
- Venneti KC, Malthouse JPG, O'Harte FPM, Hewage CM. Conformational, receptor interaction and alanine scan studies of glucose-dependent insulinotropic polypeptide. Biochim Biophys Acta 2011; 1814:882-8; PMID: 21539943; http://dx.doi.org/10.1016/j.bbapap.2011.04.002
- Underwood CR, Parthier C, Reedtz-Runge S. Structural basis for ligand recognition of incretin receptors. Vitam Horm 2010; 84:251-78; PMID:21094903; http://dx.doi.org/10.1016/B978-0-12-381517-0.00009-6
- Tikhele SH, Pissurlenkar RRS, Srivastava S, Saran A, Coutinho EC. Mapping interactions of gastric inhibitory polypeptide with GIPR N-terminus using NMR and molecular dynamics simulations. J Pept Sci 2010; 16:383-91; PMID:20607844; http://dx.doi.org/10.1002/psc.1250
- Pal K, Melcher K, Xu HE. Structure and mechanism for recognition of peptide hormones by Class B G-protein-coupled receptors. Acta Pharmacol Sin 2012; 33:300-11; PMID: 22266723; http://dx.doi.org/10.1038/aps.2011.170
- Harriman WD. Gel microdrop composition and method of using the same. Crystal Bioscience Inc., 2013.
- Durocher Y, Loignon M. Process, Vectors and Engineered Cell Lines for Enhanced Large-Scale Transfection. 2009.
