ABSTRACT
Antibody single-chain variable fragments (scFvs) are used in a variety of applications, such as for research, diagnosis and therapy. Essential for these applications is the extraordinary specificity, selectivity and affinity of antibody paratopes, which can also be used for efficient protein purification. However, this use is hampered by the high affinity for the protein to be purified because harsh elution conditions, which may impair folding, integrity or viability of the eluted biomaterials, are typically required. In this study, we developed a strategy to obtain structural elements that provide allosteric modulation of the affinities of different antibody scFvs for their antigen. To identify suitable allosteric modules, a complete set of cyclic permutations of calmodulin variants was generated and tested for modulation of the affinity when substituting the linker between VH and VL. Modulation of affinity induced by addition of different calmodulin-binding peptides at physiologic conditions was demonstrated for 5 of 6 tested scFvs of different specificities and antigens ranging from cell surface proteins to haptens. In addition, a variety of different modulator peptides were tested. Different structural solutions were found in respect of the optimal calmodulin permutation, the optimal peptide and the allosteric effect for scFvs binding to different antigen structures. Significantly, effective linker modules were identified for scFvs with both VH-VL and VL-VH architecture. The results suggest that this approach may offer a rapid, paratope-independent strategy to provide allosteric regulation of affinity for many other antibody scFvs.
Introduction
In immunoglobulin single-chain variable fragments (scFvs), the antibody variable domains of the light (VL) and heavy (VH) chain are connected to a single polypeptide. A similar single chain architecture has also been applied for the structurally similar T-cell receptors,Citation1 as well as Fab fragments.Citation2 Both chains are typically connected by an unstructured linker that is flexible and does not show any tendency to interfere with folding of the individual immunoglobulin domains.,Citation3,4 In many cases, these linkers contain assemblies or variations of (Gly4Ser) repeats, the use of which was inspired by the linkers connecting the domains of filamentous bacteriophage minor coat protein III.Citation5 Antibody scFvs are widely used in a variety of applications in research,Citation6 diagnosis and therapy. For example, immunotoxins, which are in clinical testing for cancer therapy, are typically based on an scFv fused to a bacterial toxin to mediate targeted killing.Citation8 Bispecific antibodies comprising 2 different scFv (e.g., bispecific T-cell engagers) can activate and redirect cytotoxic T cells; one of these, blinatumomab (Blincyto®), is approved for the treatment of cancer.Citation9,10 Chimeric antigen receptor (CAR)-T-cell therapy also relies on scFvs specific for malignant cells.Citation11,12
Essential for all of these applications is the extraordinary specificity, selectivity and affinity of antibody paratopes. These properties would also be very useful for the purification of biomaterials, in particular proteins, vaccines or cells. However, the usually very high affinity of antibodies requires harsh elution conditions, which typically impairs folding, integrity or viability of the eluted materials.Citation13 Therefore, antibodies that retain their excellent specificity while being adjustable in respect of their affinity would be advantageous for protein purification,Citation14 cell separation and cell analysis.Citation15 Even the introduction of an affinity-adjustable antibody for therapy may be envisioned, for example as an additional safety mechanism in CAR-T-cell therapy.Citation16 For the latter applications, it is essential that the dissociation of the antibody from the target occurs under physiologic conditions without significant pH or salt concentration changes or the use of denaturing reagents or any other reagents impairing folding, integrity or viability.
Initial studies have already demonstrated the generation of antibodies with changeable affinity. However, most of the used strategies depend on introduction of a second binding site in or very near to the paratope, which directly affects the binding of the antigen to its binding site by steric hindrance or electrostatic repulsion. Fabs that recognized their antigen only in the presence of calcium could be isolated from a human phage library by appropriate selection strategies.Citation17 Single domain VHH antibodies responding to changes in pH were successfully generated by introduction of ionizable histidines in a scanning library approach.Citation18 Despite the fact that histidine “hot-spots” were identified in the paratope of the antibody used in this study, the adaptation of this approach to any antibody with a different paratope structure certainly would require substantial engineering. An approach that works independently from the individual paratope structure would be much more favorable. Along these lines, it has been proposed that a change to the paratope structure, and thus the antigen binding properties, may be induced by an allosteric effect originating in a conformational change within the linker between VH and VL.Citation19 ScFv linker peptides do not contribute to antigen binding and are typically conformationally flexible as they are designed to be compatible with pairing of a large diversity of VH and VL combinations. If exchanged by a structurally defined entity that can undergo induced conformational changes at physiologic conditions, it may be possible to develop a version that can serve as an allosteric regulator for different VH and VL pairs. Such linkers would greatly facilitate the development of affinity-adjustable antibodies, as tedious individual engineering of every paratope with a different specificity could be avoided. In contrast, scFv linkers can be exchanged by a simple cloning step using restriction sites identical, for example in all scFv derived from a phage display library. Initial successful applications of this linker-based approach have been described: the replacement of a (G4S)3-linker with elastin-like polypeptides resulted in scFvs changing their affinities in response to alterations in temperature.Citation20 The introduction of calmodulin and M13 peptide as a linker led to an scFv with calcium-dependent affinity changes.Citation14 However, the described solutions were tested only for one antibody. It has not yet been shown that a particular linker module system can serve as a “switch” in very different scFvs with different paratopes that bind to different classes of antigens.
Calmodulin (CaM) undergoes large conformational changes depending on the presence of calcium and calmodulin-binding peptidesCitation21 (CBP) (). In a calcium- and peptide-unbound form, it adopts a closed conformation (PDB ID: 1CFD).Citation22 The distance between the N- and C-terminus is at its highest in the calcium-bound, open form (PDB ID: 1CLL),Citation23 whereas the termini approach each other when calmodulin binds to a ligand, or a suitable fragment thereof, like peptide M13 (PDB ID: 2BBM).Citation24 Calmodulin and its peptide binding partner M13 have been used as an input domain for the design of various calcium sensors. A Förster resonance energy transfer (FRET)-based system was established by insertion of calmodulin and M13 peptide between 2 different green fluorescent protein-based proteins and calcium-dependent change in FRET was observed.Citation25 Another fluorescent indicator for calcium is based on a circularly permuted green fluorescent protein to which calmodulin and M13 peptide is fused. The fluorescent properties of the fusion protein changed reversibly with the amount of calcium.Citation26 Recently developed switchable enzymes take advantage of proteins that change their conformation in response to a stimulus such as calmodulin. A peptide-dependent TEM-1 β-lactamase was constructed, which showed up to 120-times higher catalytic activity in a calmodulin-binding peptide bound state. This was achieved by insertion of calmodulin into the sequence encoding for the enzyme.Citation21 Furthermore, an allosteric TEM-1 β-lactamase could be generated by insertion of circularly permuted variants of the encoding gene into the E. coli maltose-binding protein gene. A variant was identified that showed a compromised β-lactam hydrolysis activity in absence of maltose, whereas it increased up to 25-fold in the presence of this disaccharide.Citation27,28
Figure 1. Engineering and identification of switchable scFv-CaM variants. (a) Conformation changes of Calmodulin. In a calcium- and peptide-unbound form, calmodulin adopts a closed conformation (PDB ID: 1CFD).Citation22 The distance between the N- and C-terminus is at its highest in the calcium-bound, open form (PDB ID: 1CLL),Citation23 whereas the termini approach each other when binding a calmodulin-binding peptide like M13 (PDB ID: 2BBM).Citation24 (b) Hypothetical mechanism of the Calmodulin-M13 peptide-based switchable scFv-system. Upon conformational change of calmodulin when binding M13 peptide, the paratope conformation of the scFv could be altered by a change in the geometry of VH/VL arrangement.
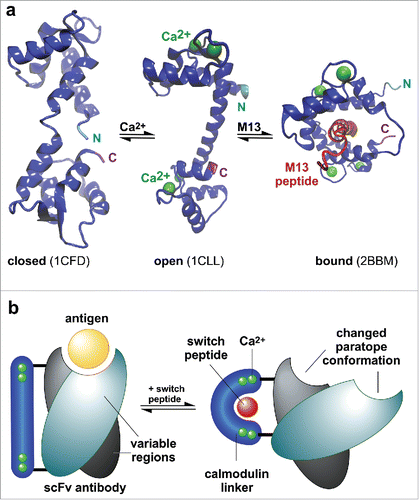
In this study, we developed a strategy to rapidly identify linker-based allosteric actuators to modulate the affinity of different scFvs for their antigen by substitution of the linker between VH and VL with permuted calmodulin variants (). A modulation of affinity induced by addition of different calmodulin-binding peptides was demonstrated for 5 of 6 tested antibody scFvs of different specificities and different architecture, suggesting that our approach may be transferable to many other antibodies.
Results
Identification of switchable anti-lysozyme scFv containing permutated calmodulin-linkers
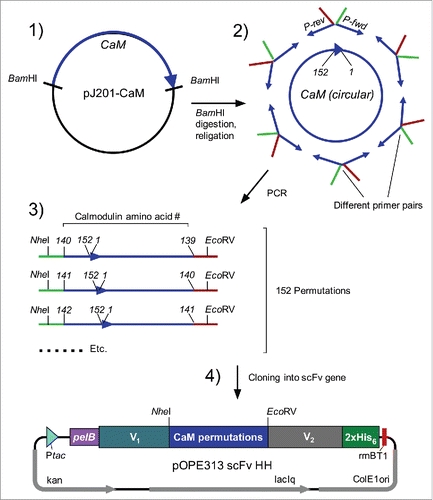
To enable identification of the optimal orientation of the calmodulin molecule between the 2 antigen-binding regions of an antibody that allows modulation of the binding affinity (), a gene library of 152 different variants representing all possible insertion points throughout a circularized calmodulin molecule was generated. First, a cyclic DNA encoding calmodulin was prepared (). This ring served as a template for 152 different sets of oligonucleotide PCR primers. The resulting linear PCR products encode circularized calmodulin with artificially new amino- and carboxy-termini at all other possible amino acid positions. Of the resulting 152 different PCR products, 145 resulted in the production of functional scFvs when used as a linker in the lysozyme binding scFv (D1.3 scFv).
Figure 2. Cloning of circularly permutated Calmodulin variants. (1) Excision of gene encoding for calmodulin (CaM) with BamHI from transfer vector pJ201-CaM. (2) Circularisation of linear gene with T4 DNA Ligase and amplification of permutated variants by PCR with appropriate oligonucleotide pairs. (3) Restriction of amplified CaM-variants with NheI and EcoRV and subsequent cloning into target vector (4) encoding for scFv. Abbreviations, P-rev: reverse-oligonucleotide with EcoRV-overhang; P-fwd: forward-oligonucleotide with NheI-overhang. Ptac: tac-promoter; PelB: signal peptide (PhoA was used in case of anti-CD20 scFv); V1/ V2: heavy or light chain variable domain (V1 = VH and V2 = VL in anti-CD4, anti-CD14 and anti-biotin scFv; V1 = VL and V2 = VH in anti-CD20 and anti-FITC scFv); NheI/EcoRV: restriction sites; 2xHis6: histidine-tag; rrnB T1: transcription terminator; ColE1 ori: origin of replication; laqIq: laqIq-promoter; kan: kanamycin resistance; CD: cluster of differentiation; FITC: fluorescein isothiocyanate.
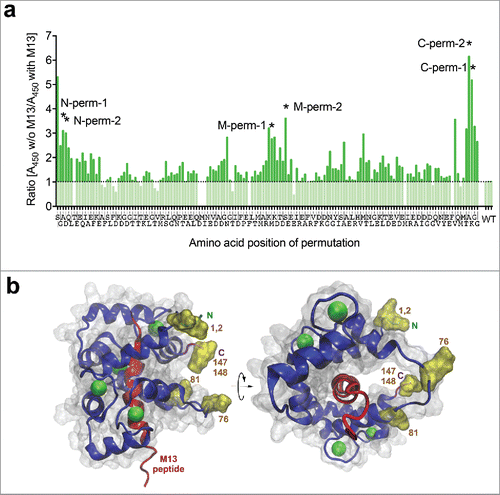
Calmodulin conformation has been shown to change when binding to the calmodulin-binding peptide M13 (residues 577–602 of skeletal muscle myosin light chain kinase).Citation24 To test the influence of M13 peptide on the calmodulin-scFv fusion proteins, all constructs were produced in E. coli using a pOPE vectorCitation29 in microtiter plate format. ScFv-containing periplasmic extracts were analyzed for antigen binding by ELISA in 2 setups: with and without M13 peptide. Five mM Calcium was present in all setups. Nearly all constructs showed a lower binding signal in the presence of M13 peptide (), while no signal changes were observed for the wildtype control ([G4S]3-linker). The largest differences in signal intensities were observed around 3 regions of the calmodulin chain: close to the N-terminus or the C-terminus, and around amino acid 80 (). The 2 fusions from each of these 3 permutation regions with the largest binding difference (named N-perm-1+2, M-perm-1+2, and C-perm-1+2, respectively), as well as the non-permutated CaM variant (i.e., fused by its naive N/C-termini), were used for further analysis.
Figure 3. (a) Identification of switchable anti-lysozyme (D1.3) scFv-CaM-variants by competitive ELISA. The amount of bound D1.3 scFv-variants in 2 different buffer setups (with/without M13 peptide) was compared. The ratio of A450-values obtained for the different setups is displayed in the figure, where a ratio > 1 indicates an M13 dependent signal decrease. Most promising variants are indicated by asterisks and named according to their permutation site (M-perm-1/2: permutated in the middle of the former calmodulin gene; N/C-perm-1/2: permutated N/C-terminally). The respective amino acid of the permutation site is indicated below the corresponding bar. (b) Location of the ligation points of permutated Calmodulins providing the most effective modulation of anti-lysozyme scFv-CaM. Model based on PDB ID: 2BBM. Best insertion points are indicated by yellow labeling of the side chains at the respective positions, further identified by numbers referring to the amino acid position of WT-Calmodulin. N, C indicate N-terminus and carboxy terminus of WT-Calmodulin.
These results confirmed that calmodulin inserted in the linker position between the V domains of antibody D1.3 can induce an M13 peptide-dependent influence on antigen binding.
M13 peptides can induce antigen release from anti-lysozyme scFv D1.3
The calmodulin-mediated affinity change observed in the initial screening was achieved after preincubation with the modulator M13. Next, we designed a release ELISA to test whether M13 peptide binding to the calmodulin linkers can also induce the dissociation of an already established antibody-antigen complex. ScFv-variants were produced in 0.5 L scale and were subsequently purified via affinity chromatography. Expression yields of the different scFv-variants are shown in Table S1. After an initial binding phase of D1.3 scFv variants on antigen in calcium-containing buffer, M13 peptide was added (), with buffer containing calcium alone used for control. In parallel, the same scFvs were analyzed by the preincubation approach described above on the same plate for calibration (). The drop of affinity in the presence of peptide in the release-setup was comparable to the drop observed in the preincubation approach. The highest decrease in signal was observed for the variants with CaM-linkers permutated C-terminally (C-perm-1, C-perm-2) and the M-perm-1 variant.
Figure 4. Analysis of M13 peptide dependent binding behavior of anti-lysozyme scFv-CaM-variants by competitive and release ELISA. The amount of bound D1.3 scFv-variants in 2 different buffer setups (with calcium vs. with calcium and M13) and 2 different ELISA systems (preincubation vs. release) was compared. The ratio of A450-values obtained for the different buffer setups is displayed in the figure, where a ratio > 1 indicates an M13 dependent signal decrease. The mean results with range (indicated by error bars) of 4 independent experiments (n = 4) are shown. Ratios were compared with the corresponding wildtype control via Kruskal-Wallis with Dunn's multiple comparisons test (*p < 0.05; ***p < 0.001). Abbreviations, WT: D1.3 scFv with (G4S)3-linker (wildtype control); CaM linear: D1.3 scFv with linearly cloned calmodulin-linker; w/o: without.
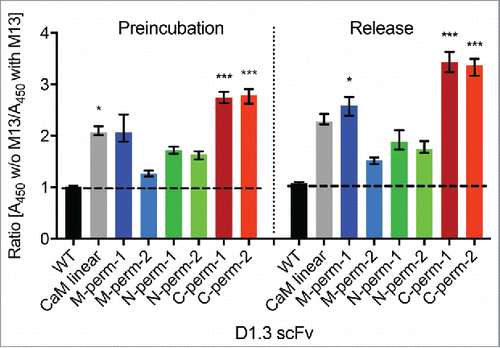
Taken together, these results indicated that a specific release of scFv-CaM-fusion from its antigen was achieved for the D1.3 scFv by adding M13 peptide, indicating a decrease in binding affinity. The use of permutated CaM-variants was advantageous compared with the wildtype (i.e., not permutated) CaM-linker.
M13 peptides modulate antigen binding of anti-lysozyme scFv-CaM-fusions in a concentration-dependent manner
To evaluate whether the affinity modulation of the D1.3 scFv-CaM-variants is specific, the binding of a defined amount of D1.3 scFv (0.1 µM) to its antigen as a function of increasing concentrations of M13 peptide in the presence of calcium was determined. Control titrations were performed in EDTA-containing buffer to assess any calcium-independent effect of M13 peptide. Nearly all analyzed scFv-CaM-variants showed a calcium-dependent decrease in antigen binding with increasing M13 peptide-concentration. At a concentration of 0.1 µM M13 peptide at a molar ratio of 1:1, no further loss of binding signal was observed (). Only the M-perm-2 variant did not show this tendency () and behaved similar to the wildtype control (). The C-terminally permutated () and the M-perm-1 variant () provided the strongest dissociation, corroborating the results shown in . In the EDTA-controls, increasing M13 peptide concentrations did not have an influence on the absorbance measured (). However, overall antigen/scFv-CaM binding was clearly affected by the presence of EDTA, as binding signals obtained in the EDTA-setup were much lower than the corresponding ones with calcium.
Figure 5. Evaluation of the specific M13 peptide dependent decrease of binding signal by titration ELISA. The binding behavior of 0.1 µM D1.3 scFv on lysozyme coated plates as a function of increasing concentrations of M13 peptide was monitored in calcium-containing buffer (empty diamonds) and buffer with EDTA (filled squares). The saturation point (molar ratio 1:1 of M13 peptide:scFv) is indicated by arrows. Absorbance was measured and normalized, where the signal obtained in the calcium-containing buffer without M13 peptide was set to 100%. The mean results with range (indicated by error bars) of 3 independent experiments (n = 3) are shown. Abbreviations, WT: D1.3 scFv with (G4S)3-linker (control) (a); CaM linear: D1.3 scFv with linearly cloned calmodulin-linker (b); M/N/C-perm-1/2: D1.3 scFv with permutated calmodulin-linker (c-h).
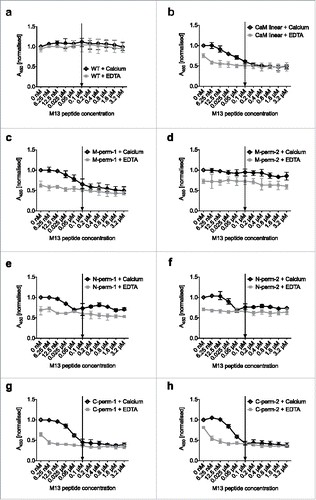
In summary, 6 of 7 tested linker variants provided M13 peptide-dependent antigen binding, with a maximum loss of binding typically obtained at a 1:1 molar ratio of M13 peptide to scFv.
CaM-linker variants provide affinity modulation in other scFv-antibodies
To investigate whether the linkers that modulated the affinity of scFv D1.3 can be used to change affinities of other scFvs, they were cloned into other antibody scFvs providing 5 different specificities, and with different architecture. The specificities were chosen according to their utility in future cell staining and separation applications, as well as their diversity in respect to antigen structures. Three scFvs were directed against different human clusters of differentiation (CD14, CD20, CD4) and 2 against small haptens: biotin and the fluorescent dye fluorescein isothiocyanate (FITC). While the overwhelming number of described scFv have an architecture with the VH at the N-terminus, there are also many functional constructs described with VL-VH arrangement. As it was not obvious from existing data whether a VL-VH or VH-VL arrangement would be better for allowing the allosteric switch by conformation change in the linker connecting the 2 domains, we tried both arrangements. The antigen-binding regions of anti-CD14, anti-CD4 and anti-biotin scFv were assembled as VH-linker-VL, while the anti-CD20 and anti-FITC scFv were arranged as VL-linker-VH constructs.
To identify M13 peptide-dependent scFv-CaM-variants for these different specificities and architectures, human peripheral blood mononuclear cells (PBMC) were stained using E. coli periplasmic extracts and subsequently analyzed by flow cytometry. This strategy allowed for rapid analysis of a large number of variants by a functional assay that already closely resembles a possible final application. To be able to include the anti-hapten scFv-variants in that assay, the cells were prestained with appropriate IgG-conjugates (anti-CD14-biotin or anti-CD14-FITC). Bound scFvs were detected with anti-His-antibodies. Incubation protocols for flow cytometric analysis were comparable to the preincubation ELISA, using buffers with or without M13 peptide. The highest M13 peptide-dependent decrease in fluorescence intensity was obtained for all anti-CD14 scFv-CaM-variants (Fig. S1, first row). The same tendency could be observed for the anti-CD20 scFv-CaM-fusions (Fig. S1, second row), although the difference between the 2 setups was very small. This demonstrates that allosteric modulation can be achieved independently from the order of VH/VL arrangement. In the case of anti-biotin scFvs (Fig. S1, third row), only 2 linker-variants (M-perm-1, M-perm-2) resulted in a visible peptide-dependent decrease of the binding signal, although a very slight decrease was observed for the remaining clones as well (CaM linear, N-perm-1, N-perm-2). Interestingly, the C-terminally permutated variants (C-perm-1, C-perm-2) led to the opposite switching behavior with an increase in fluorescence intensities. The same trend was seen for 3 anti-CD4 scFvs (M-perm-1, C-perm-1, C-perm-2), showing an even higher signal increase than anti-biotin scFv-variants (Fig. S1, fourth row) upon M13 peptide incubation. None of the examined anti-FITC scFvs (Fig. S1, fifth row) showed any modulation of binding activity.
Three of the 5 tested specificities were then produced in 0.5 L scale and subsequently purified. Expression yields of the different scFv-variants are summarized in Table S1. For anti-CD14 scFv and anti-biotin scFv, nearly all scFv-CaM-fusions showed a higher expression rate than the corresponding wildtype. In the case of anti-CD4 scFv, the highest amount of scFv was obtained with the wildtype clone. Therefore, with regard to expression yields, whether the integration of the CaM-linker is beneficial or not depends on the specificity.
The different scFv-variants were then characterized via staining of PBMC with appropriate and comparable scFv titers. For the anti-CD14 scFv-CaM-variants (; ), the same tendency was monitored as in the initial screening with considerable changes in binding signal for all scFv-CaM-fusions. A nearly complete loss of fluorescence signal was observed for the linear variant and the M-perm-1 clone in the staining with crude lysates, which was not the case in the second staining with purified scFvs. This was due to the lower titers of scFv present in the cell extracts, resulting in lower initial binding signals. In case of anti-biotin scFvs (; ), the results from the first screening could be verified with visible changes in binding signals for all scFv-fusions, including the linearly cloned and N-terminally permutated variants. The stainings with purified anti-CD4 scFv-CaM-fusions (; ) mainly supported the results of the first experiment with unpurified extracts. Interestingly, a slight decrease in binding signal could be observed for the M-perm-2 variant, although the M-perm-1 clone behaved in the opposite way with a high increase in fluorescence signal in the presence of M13 peptide. Taking into account the results shown in , the use of C-terminally permutated CaM-variants and those permutated in the middle of the calmodulin encoding gene resulted in the best switchable antibody variants in 3 of the tested specificities. In contrast, the linear variant provided only one highly switchable antibody fragment (anti-CD14 scFv), since the anti-biotin scFv only showed a slight decrease in binding signal and the anti-CD4 scFv showed no change at all.
Figure 6. Identification of switchable scFv-CaM-fusions of different antibodies. (a) Human blood cells (PBMC, peripheral blood mononuclear cells) were stained with purified anti-CD14 scFvs, anti-biotin scFvs or anti-CD4 scFvs containing CaM linkers and subsequently analyzed by flow cytometry. The amount of bound scFv-variants in 2 different setups (dark blue graphs: binding to antigen in presence of calcium only [without M13 peptide]; red graphs: binding to antigen in calcium- and M13 peptide-containing buffer) was compared. Living-PE-positive cells were taken into account. An M13 peptide-dependent change in fluorescence intensity is indicated by orange (signal decrease) and light blue (signal increase) arrows. The results are from a representative experiment (n = 6 [anti-CD14, anti-CD4] or n = 3 [anti-biotin]). (b) Statistical analysis of data shown in (a). The ratio of the median fluorescence intensities (MFI) obtained in the different buffer setups was determined, where a ratio > 1 indicates an M13 dependent change in signal intensity. ScFv-CaM-variants which show an M13 peptide-dependent increase in fluorescence intensity are marked with black diamonds. The mean results with range (indicated by error bars) of 6/3 independent experiments (n = 6 [anti-CD14, anti-CD4] or n = 3 [anti-biotin]) are shown. Ratios were compared with the corresponding wildtype control via Kruskal-Wallis with Dunn's multiple comparisons test (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001). Abbreviations, WT: scFv with (G4S)3-linker (control); CaM linear: scFv with linearly cloned calmodulin-linker; M/N/C-perm-1/2: scFv with permutated calmodulin-linker; w/o: without; CD: cluster of differentiation; PE: phycoerythrin.
![Figure 6. Identification of switchable scFv-CaM-fusions of different antibodies. (a) Human blood cells (PBMC, peripheral blood mononuclear cells) were stained with purified anti-CD14 scFvs, anti-biotin scFvs or anti-CD4 scFvs containing CaM linkers and subsequently analyzed by flow cytometry. The amount of bound scFv-variants in 2 different setups (dark blue graphs: binding to antigen in presence of calcium only [without M13 peptide]; red graphs: binding to antigen in calcium- and M13 peptide-containing buffer) was compared. Living-PE-positive cells were taken into account. An M13 peptide-dependent change in fluorescence intensity is indicated by orange (signal decrease) and light blue (signal increase) arrows. The results are from a representative experiment (n = 6 [anti-CD14, anti-CD4] or n = 3 [anti-biotin]). (b) Statistical analysis of data shown in (a). The ratio of the median fluorescence intensities (MFI) obtained in the different buffer setups was determined, where a ratio > 1 indicates an M13 dependent change in signal intensity. ScFv-CaM-variants which show an M13 peptide-dependent increase in fluorescence intensity are marked with black diamonds. The mean results with range (indicated by error bars) of 6/3 independent experiments (n = 6 [anti-CD14, anti-CD4] or n = 3 [anti-biotin]) are shown. Ratios were compared with the corresponding wildtype control via Kruskal-Wallis with Dunn's multiple comparisons test (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001). Abbreviations, WT: scFv with (G4S)3-linker (control); CaM linear: scFv with linearly cloned calmodulin-linker; M/N/C-perm-1/2: scFv with permutated calmodulin-linker; w/o: without; CD: cluster of differentiation; PE: phycoerythrin.](/cms/asset/d5f4ae81-76c8-42f2-b860-56d73be38596/kmab_a_1277302_f0006_c.gif)
In summary, we have shown that the mechanism of allosteric modulation of binding through a calmodulin-linker/M13 peptide combination under physiologic conditions could be transferred to 4 of the tested 5 antibody specificities. Notably, different linkers could be found to be optimal for different scFv, and even different allosteric effects were observed.
Identification of calmodulin-binding peptides with improved affinity modulating properties
Calmodulin binds to a variety of binding partners. To investigate if other calmodulin-binding peptides or mutants derived from M13 are able to modulate the affinity of the scFv-CaM-fusions, a peptide screening was performed. On the one hand, 38 mutated variants of the M13 peptide were analyzed, including substitution mutants known to have higher affinities for calmodulin,Citation30,31 as well as truncated variants and combinations thereof. In addition, 29 peptides derived from other calmodulin-binding proteins like calcium ATPase,Citation32 spectrinCitation33 and nitric oxidase synthaseCitation34 were analyzed with regard to potential affinity modulating properties. The analysis was performed via competitive staining of PBMC as in the previous experiments. The complete screening (i.e., of the whole peptide libraries) was performed with 4 different linker variants of anti-CD14 scFv (CaM linear, M-perm-2, N-perm-1, C-perm-1) and anti-CD4 scFv (CaM linear, M-perm-1, N-perm-1, C-perm-1). The wildtype of each specificity was used as a control. All 38 mutated variants of the M13 peptide showed affinity modulating properties, whereas 24 of 29 analyzed peptides derived from alternative calmodulin-binding proteins resulted in a change in binding signal in at least one analyzed scFv-CaM-fusion (data not shown). Most of the tested peptides showed an effect comparable or lower than the M13 peptide. We then selected the peptide variants that led to a higher signal change than M13 and analyzed the scFv-CaM-fusions of anti-CD14, anti-biotin and anti-CD4 scFv (Fig. S2+3, ). The sequences of the most effective peptides are summarized in . For anti-CD14 scFv, 3 different variants of the known high affinity mutant of M1330,31 resulted in a greater signal decrease in the clones permutated in the middle of CaM and the linear clone (Fig. S2 + ). CBP-Var8, a derivative of spectrin,Citation33 led to a slightly higher decrease in the C-terminally permutated and the linear variant (Fig. S2 + ). Unexpectedly, a peptide derived from calcium-transporting ATPaseCitation32 (CBP-Var16) resulted in an increase in fluorescence signal in the M-perm-2 and C-perm-2 variants (Fig. S2 + ). The same opposite behavior was monitored for the C-terminally permutated clones of anti-CD4 scFv, where M13 peptide led to an increase in binding signal, while CBP-Var16 resulted in an unexpected decrease in fluorescence intensity (Fig. S3b + ). Two anti-biotin scFv-CaM-fusions (M-perm-1, M-perm-2) showed such an opposite switching behavior in the presence of CBP-Var6, another derivative of calcium-transporting ATPaseCitation32 (Fig. S3a + ). Surprisingly, CBP-Var16 had a further increasing effect on the C-perm-1 clone and showed no opposite switching behavior as in the other scFv-CaM-fusions (Fig. S3a + ). The same was observed for the anti-CD4 scFv M-perm-variants, where CBP-Var6 led to a further increase in fluorescence intensity (Fig. S3b + ). Furthermore, a peptide derived from endothelial nitric oxidase synthaseCitation34 (CBP-Var12) had the same effect on the C-terminally permutated anti-CD4 scFv-CaM-fusions (Fig. S3b + ). Another unexpected decrease in binding signal was observed for the linear anti-CD4 scFv clone, triggered by CBP-Var10, derived from ionotropic glutamate receptor NMDA1,Citation35 although the M13 peptide did not have any influence at all (Fig. S3b + ).
Figure 7. Identification of other calmodulin-binding peptides with affinity modulating properties. Human blood cells (PBMC, peripheral blood mononuclear cells) were stained with purified anti-CD14 scFvs (a), anti-biotin scFv (b) or anti-CD4 scFvs (c) containing CaM-linkers and subsequently analyzed by flow cytometry. Only living (PE-positive) cells were taken into account. The ratio of the median fluorescence intensities (MFI) obtained in 2 different setups (with M13 peptide [M13] or with M13-Var/CBP-Var-peptide [Var]) was determined, where a ratio > 1 indicates an M13-Var/CBP-Var-dependent signal change in comparison to the M13 setup. An opposite switching in binding behavior in comparison to M13 peptide dependent behavior is indicated by a large star above the graph. The mean results with range (indicated by error bars) of 3 independent experiments (n = 3) are shown. Ratios were compared with the corresponding wildtype control via Kruskal-Wallis with Dunn's multiple comparisons test or Mann-Whitney test, respectively (*p < 0.05; **p < 0.01). Abbreviations, WT: scFv with (G4S)3-linker (control); CaM linear: scFv with linearly cloned calmodulin-linker; M/C-perm-1/2: scFv with permutated calmodulin-linker; CD: cluster of differentiation; CBP: calmodulin-binding peptide; Var: variant.
![Figure 7. Identification of other calmodulin-binding peptides with affinity modulating properties. Human blood cells (PBMC, peripheral blood mononuclear cells) were stained with purified anti-CD14 scFvs (a), anti-biotin scFv (b) or anti-CD4 scFvs (c) containing CaM-linkers and subsequently analyzed by flow cytometry. Only living (PE-positive) cells were taken into account. The ratio of the median fluorescence intensities (MFI) obtained in 2 different setups (with M13 peptide [M13] or with M13-Var/CBP-Var-peptide [Var]) was determined, where a ratio > 1 indicates an M13-Var/CBP-Var-dependent signal change in comparison to the M13 setup. An opposite switching in binding behavior in comparison to M13 peptide dependent behavior is indicated by a large star above the graph. The mean results with range (indicated by error bars) of 3 independent experiments (n = 3) are shown. Ratios were compared with the corresponding wildtype control via Kruskal-Wallis with Dunn's multiple comparisons test or Mann-Whitney test, respectively (*p < 0.05; **p < 0.01). Abbreviations, WT: scFv with (G4S)3-linker (control); CaM linear: scFv with linearly cloned calmodulin-linker; M/C-perm-1/2: scFv with permutated calmodulin-linker; CD: cluster of differentiation; CBP: calmodulin-binding peptide; Var: variant.](/cms/asset/40847ace-607a-40cd-9d5c-030bff43d9da/kmab_a_1277302_f0007_c.gif)
Table 1. Calmodulin-binding peptides with affinity modulating properties.
In summary, we have shown that the majority of the tested calmodulin-binding peptides led to a modulation of affinity in the scFv-CaM-fusions. Some candidates were identified that resulted in an even higher decrease or increase of binding signal than the wildtype variant of M13. Furthermore, some peptides resulted in an opposite switching behavior. Taken together, the screening of the described collection of additional modulation peptides increased the number of solutions for allosteric modulation.
Affinity changes are not a result of scFv dimerization/multimerization
To exclude that the M13 peptide-dependent decrease in binding affinity was a result of dimer or multimer formation, a phenomenon well described to be related to changes in linker length of scFvs,Citation36,37,38 an investigation by size-exclusion chromatography (SEC) was performed for the anti-CD14 scFv M-perm-2 variant. Anti-CD14 scFv-WT was used as a control. First, monomeric scFvs were purified by preparative SEC (Fig. S4a+b). Staining of PBMC with the monomeric fractions indicated that the tendency of switching ability was the same as before the purification (Fig. S4c). Then, the potential M13-dependent change of conformation was analyzed by analytical SEC. ScFv was mixed with calcium (setup A) or calcium and M13 peptide (setup B) and was subsequently separated by SEC. The M-perm-2 variant showed, along with the wildtype control, nearly the same elution profile under the 2 tested conditions (). The single peaks were identified as monomeric scFvs, for both CaM-variant and wildtype control, by comparison of the respective retention times with an analyzed molecular weight standard.
Figure 8. Size-exclusion chromatography of anti-CD14 scFv M-perm-2. The conformation of anti-CD14 scFv-WT (a,c) and anti-CD14 scFv M-perm-2 (b,d) in calcium-containing buffer (a,b) and calcium- and M13 peptide-containing buffer (c,d) was determined by SEC. Molar mass calibration was done with a gel filtration standard (filled squares) containing thyroglobulin (670 kDa), bovine γ-globulin (158 kDa), chicken ovalbumin (44 kDa), equine myoglobin (17 kDa) and vitamin B-12 (1.35 kDa).
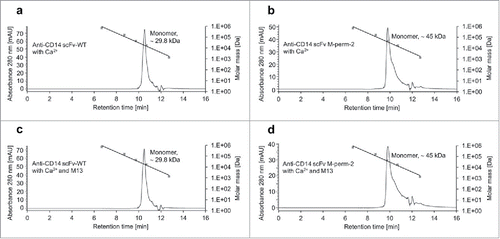
Taken together, these results indicated that the M13-dependent change of binding affinity of anti-CD14 scFv M-perm-2 is not a result of a change of the monomer/dimer ratio.
Determination of dissociation constants by equilibrium binding titration
Previous experiments showed that most scFv-variants with CaM-linkers lead to fluorescence signal intensities that are comparable to the corresponding wildtype clones when used for staining of PBMC in calcium-containing buffer at similar molar concentrations (). It seemed that the affinities of scFvs are not significantly altered by exchange of the GlySer linkers with CaM-linker variants without peptides. To investigate this in more detail, dissociation constants (KD) for anti-CD14 scFv M-perm-2 and the corresponding wildtype control were determined by equilibrium binding titration.Citation39 The titration was performed by staining of PBMC as in the previous experiments, but only in calcium-containing buffer. The incubation of the scFvs was performed at 24°C for 45 min to allow equilibrium binding. For the wildtype, a KD of 284.6 ± 20.96 nM was determined, whereas the KD of the M-perm-2 variant was 427.2 ± 22.27 nM and therefore increased by a factor of 1.5 in comparison to the wildtype ( and ). Thus, the initial binding affinity of the anti-CD14 scFv was only very slightly decreased by integration of the CaM-linker.
Figure 9. Determination of dissociation constants (KD) by equilibrium binding titration. Equilibrium binding constants (KD) for anti-CD14 scFv-WT and anti-CD14 scFv M-perm-2 were determined by staining of PBMC at 24°C with titers in the range of 1 nM to 60 µM, without (blue) and with (red) M13 peptide. Shown is the normalized mean fluorescence gained at the corresponding concentration of 3 titrations (n = 3) of independent experiments. Living-FITC-positive cells were taken into account. KD was determined with a nonlinear regression model using GraphPad Prism 7.
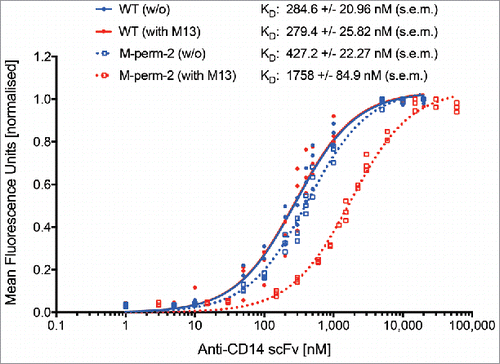
Next, binding affinities were determined in M13 peptide-containing buffer. While the wildtype KD (279.4 ± 25.82 nM) did not differ significantly from the KD determined in peptide-free buffer, the M-perm-2 variant showed an M13 peptide-dependent decrease in affinity by a factor of 4.1 (1758 ± 84.9 nM) ().
Discussion
In this study, we showed that the permutated calmodulin-derived linkers between the 2 variable domains of scFv of different immunoglobulins provided modulation of the antigen binding of 5 of 6 scFvs with different specificities and different V domain arrangements. This confirms our initial design hypothesis that conformation changes induced in the linker are able to influence antigen binding by an allosteric mechanism, suggesting that this is achieved by a structural change induced in the paratope structure, like, for example, the change of VH to VL angle. Affinity modulation could successfully be achieved for an anti-lysozyme antibody, known to bind to its antigen by a large rather flat interaction surface,Citation40 and 4 other antibodies, including a hapten binding scFv, where a small, groove like paratope structure can be assumed. This indicates that the allosteric mechanism seems to be quite robust and compatible with a variety of different V domains.
The allosteric switch module was designed to be easily introduced into any typical antibody scFv as a substitute for the usual linker, including scFv with different VH/VL arrangements. Our results suggest that screening of a limited number of linkers with different permuted CaM-variants (in particular M-perm-1+2, C-perm-1+2) can be combined with a small set of affinity-modulating calmodulin-binding peptides to identify functional and the most suitable combinations for a given scFv specificity. Of course, for other scFvs to be modulated in the future, the optimal calmodulin permutation point may well be different from the M-perm-1/2 and C-perm-1/2 variants preferentially used in this study, but this can easily be tested with the method used for D1.3 scFv. Only one of 6 tested scFv-specificities did not result in switchable scFv-CaM-variants. This anti-FITC-clone 4M5.3 is an scFv with an extraordinary strong monovalent antigen-binding affinity of 270 fM. The bound hapten is buried in a deep, hydrophobic pocket. Citation41,42 The failure to find a solution for this antibody therefore is still compatible with our hypothesis of allosteric changes because we may not have been able to recognize an affinity shift in the range observed for the other scFvs with the assay conditions used. Further, allosteric changes mediated by slight changes of VH/VL orientation may effect a larger, rather flat antigen binding surface stronger than a small hydrophobic pit.
The switching effect (i.e., the extent and direction of affinity change) of certain peptides differed among CaM-variants and scFv-specificities. This may be explained by the diverse structures of the scFvs. Different antibodies are known to have quite different VH/VL geometry by nature, with packing angles observed to vary between −31.0° and −60.8°. Citation43 Therefore, we expected that the conformational change induced by the CaM-linkers must be different for different scFvs to enable distortion of that particular paratope to result in a decrease of affinity. The same hypothesis may also explain a stabilization of the paratope by a CaM-linker, which has a much more rigid structure after M13 peptide binding than the unstructured GlySer linker it replaces. The natural degree of freedom of VH/VL movement in an scFv that results from the lack of constant regions may be limited by a CaM-linker, leading to an increased affinity, which was indeed found in a few cases as well. However, whether this simple hypothesis is sufficient to explain the results in detail could only be answered by determining high resolution molecular structures. Nevertheless, by a standardized screening of combinations of different CaM-linkers and calmodulin-binding peptides, different solutions for the different individual antibodies were found, except for an extremely high affinity antibody with a very small hydrophobic antigen binding pocket. Interestingly, CBP-Var6 and CBP-Var16, peptides derived from calcium-transporting ATPase,Citation32 also showed an opposite switching behavior in comparison to the effect observed for peptide M13. This may be attributed to their different binding mode to CaM. Previous studies showed that these peptides mainly interact with the C-terminal lobe of CaM,Citation44 whereas peptide M13 interacts with both domains of Calmodulin.Citation24 Consequently, the conformational change induced by binding of CBP-Var6 or CBP-Var16 may differ from the change induced by M13 peptide, thus offering an explanation for the observed opposite switching behavior.
Previous approaches and strategies for the generation of antibodies that allow regulation of their affinity typically relied on the establishment of phage librariesCitation17 or amino acid scanning libraries.Citation18 These library approaches have the disadvantage that each scFv candidate has to undergo the whole process of mutagenesis, selection and functional screening individually, as there is no common molecular mechanism. Any structural solution identified with these methods always was strongly dependent on the sequence of that particular paratope. Many paratopes may not be amenable at all to incorporation of the desired switch structure. In contrast, a VH/VL linker-based system can be tested independently from individual paratope structures, and therefore promises to provide a much quicker approach, since the introduction of the allosteric module only requires a one-step cloning to an existing set of mutant linkers.
None of the described previously linker-based approaches has yet been shown to be transferable to a variety of different antibodies. Further, the described solutions have other disadvantages. For example, the elastin-based setup described by Megeed et al.Citation20 is based on the temperature-dependent conformational changes of elastin-like polypeptides. A significant modulation of antibody affinity was only reached at rather high temperatures (55°C), which comes close to the denaturation temperature of many V domains and would render this approach useless for many applications, such as cell separation. A system that is functional at physiologic temperatures would be more preferable. In the calmodulin-based setup described by Kobatake et al.,Citation14 the binding affinity of the antibody increased with addition of calcium. Hence, a release of an already bound antibody might be possible by addition of chelating agents such as EDTA. These chelating agents may be disadvantageous for some applications or downstream processing. Furthermore, their use in therapeutic applications would by hampered by the calcium levels existing in vivo. The use of a peptide-dependent switch working at constant and physiologic calcium concentrations, as described in this publication, would avoid this problem.
In the setup described by Kobatake et al.,Citation14 native calmodulin was used as a linker. In our study, it could be shown that permuted variants provided switching even in cases where the native CaM did not work. Consequently, based on our data and the above observations, we assume that no scFv linker will be capable of switching every possible VH/VL or VL/VH combination. Therefore, the presented strategy is based on the test of CaM-linkers in various (if necessary all possible) orientations combined with the additional possibility to use different binding modes for modulation. We conclude that this combinatorial screening approach could allow identification of functional and affinity switchable scFv-variants for a large variety of other specificities, even without a priori knowledge about a certain paratope structure.
A potential use of a switchable antibody fragment is in high specificity affinity chromatography for protein purification. Most current antibody-based purification techniques require harsh elution conditions due to the high affinity of antibodies, which typically impair the folding or integrity of the eluted target proteins.Citation13 The mild elution of bound materials by addition of peptides under physiologic buffer conditions using affinity-switchable antibodies may allow the development of novel and attractive alternatives. Moreover, affinity modulated antibodies could be used for reversible cell staining and cell separation.Citation15 Present reversible systems for this task usually use chemical coupling of functional moieties like biotin or derivatives to the antibody.Citation45 Chemical modification of an antibody is often laborious, stoichiometrically not well defined and may be accompanied with loss of function. In contrast to that, our calmodulin-linker scFvs could be expressed in a host of choice and may be used directly after purification without further modifications. Even a therapeutic use of switchable scFvs may be envisioned, for example as an additional safety mechanism in CAR-T-cell therapy.Citation16 In that case, it would be essential that the peptide used for affinity-modulation of the scFv does not interfere with the patients' native calmodulin molecules. A possible solution may be the selection of peptides not binding to wildtype calmodulin, for example by peptide phage display.Citation46
For effective and efficient employment of the scFv-CaM-fusions in protein purification and cell separation, further improvements of the herein described system would be required. As a next step, additional engineering of the linker can be expected to further improve individual modulation strength, for example by adding or removing several amino acids from the CaM linker or mutating the calmodulin encoding gene for the generation of a calmodulin high affinity mutant for calcium or certain peptides.Citation47 Moreover, the selection and screening of new calmodulin-binding peptides in a library approach may lead to improved combinations of permuted CaM variants and corresponding binding peptides. This could be achieved by creation of peptide libraries and screening for the desired characteristics by phage displayCitation46 or very-high-density peptide arrays.Citation48 Another approach to enhance the modulation may be the replacement of calcium with other ions known to bind calmodulin and induce activation of binding partners.Citation49,50 Finally, the details of the structural changes, despite the fact that they may be difficult to investigate given the known problems with scFv crystallization, will be very interesting and help to understand antibody-antigen interactions.
In summary, we designed a library of switch modules consisting of a small set of permuted CaM-linkers and a manageable amount of calmodulin-binding peptides. By functional screening of combinations of these sets, it should be possible to identify functional and affinity switchable scFv-variants of different architectures and with a large variety of specificities.
Materials and methods
Cloning of permutated calmodulin variants
The gene encoding for human calmodulin (GenBank: AAD45181.1) was optimized for recombinant expression in E. coli, flanking sequences encoding for BamHI recognition sites were added and the construct was synthesized by DNA2.0 (Menlo Park, USA). The plasmid bearing the gene of interest was transformed and amplified in E. coli NEB® 5-α (New England Biolabs) as indicated in the manufacturer's manual and purified by using the NucleoBond® Xtra Maxi Plasmid Purification Kit (Macherey-Nagel). The gene encoding for calmodulin was excised from the plasmid by restriction with BamHI (all enzymes were obtained from New England Biolabs, if not indicated otherwise) and gel extraction of the fragment by using the NucleoSpin® Gel and PCR clean-up Kit (Macherey-Nagel). Thereafter, the fragment was circularised by ligation with T4 DNA ligase (o/n, 16°C). The reaction was stopped (65°C, 10 min) and circular forms were isolated by gel extraction. The circularized fragment was used as template for the generation of permutated variants by PCR. 152 different oligonucleotide pairs (Metabion) were designed for the amplification of all possible permutated variants of calmodulin. Oligonucleotides contained overlapping sequences encoding for NheI (forward; overlapping sequence [before restriction]: GGAGGTGCTAGC-) and EcoRV (reverse; overlapping sequence [before restriction]: CCTCCAGATATC-) recognition sites to facilitate cloning into the target vector pOPE313 D1.3 scFv HH. The resulting linear PCR products encode circularized calmodulin with artificially new amino- and carboxy-termini at all other possible amino acid positions and were inserted between the V domains, completely substituting the region encoding the conventional linker peptide. PCR was performed with Phusion® High-Fidelity DNA Polymerase as indicated in the manufacturer's manual. The size of the products was verified by gel electrophoresis and the remaining reaction mixture was desalted by using the NucleoSpin® Gel and PCR clean-up Kit, followed by restriction with NheI und EcoRV. The reaction was stopped (80°C, 20 min) and directly used for ligation into the equally treated vector backbone, a pOPE vectorCitation29 containing the scFv genes. ScFvs used for the generation of switchable scFv-CaM-fusions were derived from the following clones: anti-lysozyme (D1.3)Citation51 anti-CD14 (TÜK4),Citation52 anti-CD4 (Q425),Citation53 anti-biotin (Bio-3 18E7.2; Miltenyi Biotec), anti-FITC (4M5.3),Citation41 anti-CD20 (LT20).Citation54 Partial blunt-end ligation was performed with T4 DNA ligase (o/n, 16°C). Reaction was stopped (65°C, 10 min) and directly transformed into the expression strain E. coli W3110 by heat-shock using standard protocols.Citation55 Positive clones were identified by colony PCR using REDTaq® DNA Polymerase (Sigma-Aldrich) and sequencing (GATC Biotech, Cologne, Germany).
Protein expression
E. coli W3110 cells harbouring the desired pOPE construct were grown overnight at 37°C and 250 rpm in 30 mL 2× YP-GK-medium (2× YP-medium [16 g L−1 soy peptone, 10 g L−1 yeast extract, 5 g L−1 NaCl, pH 7.0] containing 100 mM glucose and 50 µg/mL kanamycin). The next day, 500 mL fresh medium was inoculated to an OD600 of 0.1 and shaken in 2 L shake flasks (37°C, 250 rpm) until an OD600 of 1.0 was reached. Protein expression was induced with a final concentration of 0.2 mM IPTG and cultures were further incubated at 25°C for 4 h. Bacteria were harvested by centrifugation (4,000 g, 20 min, 4°C) and the bacterial pellet was directly processed or stored at −20°C.
Purification of recombinant scFvs
The bacterial pellet was resuspended in 10 mL TE buffer per g pellet (100 mM Tris, 10 mM EDTA; pH 9.0 or pH 7.4, depending on the isoelectric point of the scFv-fusion) and incubated overnight at 37°C at 250 rpm. The next day, Benzonase® Nuclease (final concentration: 1 U/mL; Merck), MgCl2 (20 mM) and Halt™ Protease Inhibitor Cocktail (Thermo Fisher Scientific) were added. The mixture was incubated for 1 h at 37°C and 250 rpm. Afterwards, the protein containing supernatant was separated from the cell debris by centrifugation (5,000 g, 20 min, RT) and prepared for purification by addition of 11× dilution buffer (110 mM Tris, 550 mM NaCl, 55 mM imidazole; pH 9.0 or pH 7.4). Purification was done according to the manufacturer's manual with 250 µL Nickel Sepharose™ 6 resin (GE Healthcare). Target protein containing fractions were pooled and dialyzed 2 times at 4°C against 200 volumes of 1× tris-buffered saline (TBS) (50 mM Tris, 150 mM NaCl; pH 8.0) for 2 h and finally overnight against 500 buffer volumes. Protein concentration was determined with the Pierce™ Coomassie Protein Assay Kit (Thermo Fisher Scientific) according to the manufacturer's instructions.
Protein expression in MTP-format
Cells harboring the desired construct were grown overnight at 37°C and 1,000 rpm in 96-well polypropylene U-bottom plates (Greiner Bio-One) in 180 µL 2× YP-GK-medium per well. The next day, 170 µL fresh medium was inoculated with 5 µL overnight culture and incubated at 1,000 rpm for 6 h at 30°C. Protein expression was induced with a final concentration of 0.2 mM IPTG and cultures were incubated overnight at 25°C. Bacteria were harvested by centrifugation (4,000 g, 20 min, 4°C). For periplasmic extraction of target protein, the pellets were resuspended in 100 µL TE buffer per well and shaken for 2 h at 37°C and 1,000 rpm. The protein containing supernatant was separated from the cells by centrifugation (4,000 g, 20 min, RT) and directly used for ELISA or staining of PBMC.
Competitive ELISA
100 ng of lysozyme was coated to 96-well Nunc MaxiSorp® ELISA plates (Thermo Fisher Scientific) in 1× TBS (pH 8.0) overnight at 4°C. The next day, plates were washed 3 times with 1× TBST (1× TBS + 0.05% [v/v] Tween®20; pH 8.0) and afterwards blocked with 1× B-TBS (1× TBS + 1% [w/v] bovine serum albumin; pH 8.0) for at least 1 h at RT. Crude lysates of D1.3 scFv-variants from microtiter plate expression were diluted 1:10 in 1× B-TBS/5 mM CaCl2 without (setup A) or with 1 µM M13 peptide (Anaspec) (setup B). Purified scFvs were also diluted in the mentioned buffers to appropriate concentrations (0.1 µM). The diluted scFvs were preincubated in 96-well polypropylene plates for 1 h at RT and afterwards 100 µL of the protein solution was transferred to the blocked and washed ELISA plates. After incubation at RT for 1.5 h, plates were washed again and horseradish peroxidase (HRP)-conjugated anti-His-antibody (1:10,000 diluted in 1× B-TBS, 100 µL per well; clone: GG11–6F4.3.2; Miltenyi Biotec; Cat. #130–092–785) was added. After another washing step, visualization of bound antibody-complexes was performed by addition of 100 µL TMB (3,3,5,5′-tetramethylbenzidine) substrate (Seramun Diagnostica) per well. The reaction was stopped with 100 µL 0.5 M H2SO4 and absorbance (450 nm) was measured with a Versamax® ELISA microplate reader (Molecular Devices, Sunnyvale, USA).
Release ELISA
The release ELISA differed from the competitive ELISA only in the preincubation step and an additional release step was performed. Purified scFvs for both setups were diluted to appropriate concentrations (0.1 µM) in 1× B-TBS/5 mM CaCl2 and directly transferred to the blocked ELISA plates. After initial binding of the scFvs (1.5 h, RT), plates were washed and different release-buffers were added. For the control (setup A), wells were filled with 100 µL 1× B-TBS/5 mM CaCl2, whereas 1× B-TBS/5 mM CaCl2/1 µM M13 peptide was added in setup B. After incubation for 1 h at RT, plates were treated comparable to the competitive ELISA.
Titration ELISA
The titration ELISA differed from the competitive ELISA only in the buffer composition used for preincubation. From column 11 to 2, M13 peptide concentration was sequentially diluted (dilution factor: 1:2) in 1× TBS/5 mM CaCl2 (highest concentration in column 11: 3.2 µM; lowest concentration in column 2: 6.25 nM; control in column 1: 0 nM) or 1× B-TBS/5 mM EDTA.
Staining of PBMC with scFvs
All antibodies, staining reagents and cytometers used for flow cytometry applications were from Miltenyi Biotec (Bergisch Gladbach, Germany). First stainings with crude lysates were performed in 1.5 mL microtubes. For stainings with anti-biotin scFv-variants and anti-FITC scFv-variants, PBMC were prestained with appropriate IgG-conjugates. For anti-biotin scFv-screenings, 1×106 PBMC were incubated on ice for 10 min in 110 µL 1× B-TBS (1× TBS + 0.5% [w/v] bovine serum albumin)/5 mM CaCl2 (pH 7.4) containing anti-CD14-biotin (dilution: 1:11; clone: TÜK4; Cat. #130–098–380), whereas for analysis of anti-FITC scFv-variants anti-CD14-FITC (dilution: 1:11; clone: TÜK4; Cat #130–080–701) was used. The reaction was stopped by addition of 1 mL buffer and centrifugation at 300 g for 5 min at 4°C. The supernatant was removed completely and the pellet was stored on ice and resuspended in buffer immediately before the next staining step.
Next, 20 µL EDTA-containing periplasmic extracts were mixed with an excess of CaCl2 (20 mM), followed by incubation for 5 min at RT and addition of 20 µM M13 peptide (setup B) or an equal amount of 1× B-TBS/5 mM CaCl2 (pH 7.4/8.0) (setup A). After preincubation (45 min, RT), the extracts were chilled on ice for 5 min and 1×106 PBMC in 80 µL 1× B-TBS/5 mM CaCl2 were added. The mixture was incubated on ice for 20 min and the reaction was stopped by addition of 1 mL buffer and centrifugation at 300 g for 5 min at 4°C. The cells were resuspended in 110 µL 1× B-TBS/5 mM CaCl2 containing anti-His-PE (phycoerythrin) (dilution: 1:11; clone: GG11–8F3.5.1; Cat. #130–092–691) or anti-His-APC (allophycocyanin) (dilution: 1:11; clone: GG11–8F3.5.1; in-house conjugate), incubated on ice for 10 min, followed by a further washing step. The pellet was stored on ice and resuspended in 1 mL buffer containing propidium iodide (final concentration: 1 µg/mL) immediately before measurement. The analysis was performed on a MACSQuant® Analyzer 10 with corresponding software (MACSQuantify™) in single tube mode. A total of 10,000 events were collected for each sample.
Stainings with purified scFvs were performed in 96-well polypropylene plates. Prestainings of PBMC were performed as described above if necessary. Purified scFvs were diluted to appropriate concentrations in 50 µL 1× B-TBS/5 mM CaCl2 (pH 7.4/8.0) per well. Similar molar concentrations were used for the CaM-linker variants and the corresponding wildtype controls. For competitive screenings, peptide (M13 peptide [Anaspec], M13-variants library and CBP (calmodulin-binding peptide) library [Genscript]) was added in molar excess in a total volume of 5 µL per well, whereas the control stainings were supplied with 5 µL 1× B-TBS/5 mM CaCl2. Stainings and analysis were performed as described above with 2×105 cells per sample. Cells were washed by addition of 175 µL buffer and centrifugation at 300 g for 10 min at 4°C.
Equilibrium binding titration
One×106 PBMC were mixed with different amounts of purified anti-CD14 scFv-variants (1 nM to 60 µM) in a total volume of 100 µL 1x B-TBS/240 µM CaCl2 (pH 8.0) (exception: 1 mL in case of 1 nM) or 100 µL 1x B-TBS/240 µM CaCl2 (pH 8.0) containing appropriate amounts of M13 peptide. Samples were incubated at 24°C for 45 min. The reaction was stopped by addition of 1 mL buffer and centrifugation at 300 g for 5 min at 4°C. Cells were resuspended in 110 µL 1x B-TBS/240 µM CaCl2 containing anti-His-FITC (dilution: 1:11; clone: GG11–8F3.5.1; Cat. #130–092–675), incubated on ice for 10 min to prevent significant dissociation of bound scFv, followed by a further washing step. The pellet was stored on ice and resuspended in 0.9 mL buffer containing propidium iodide immediately before measurement. Mean fluorescence units were normalized and used for calculation of KD-values with GraphPad Prism 7 (GraphPad Software, USA) using a nonlinear regression model (one-site specific binding).
Size-exclusion chromatography
Preparative SEC for purification of monomeric scFvs was done with an ÄKTA purifier system (GE Healthcare). A maximal sample volume of 900 µL (> 700 µg/mL) was injected onto a self-packed column (Superdex™ 200 Prep Grade; GE Healthcare) at a flow rate of 1 mL/min. For analytical SEC, 20 – 50 µg scFv was mixed with an 8-fold molar excess of calcium (setup A) or an 8-fold molar excess of calcium and a 1.5-fold molar excess of peptide M13 (setup B) and incubated at RT for 45 min. Afterwards, the samples were injected onto a Yarra SEC-3000 column (Phenomenex) using the Agilent 1100 HPLC-system (Agilent Technologies) at a constant flow rate of 0.35 mL/min. For calibration issues, a gel filtration standard (1.35 kDa – 670 kDa) (Bio-Rad) was analyzed.
Statistical analysis
Statistical analyses of peptide-dependent signal changes were performed with GraphPad Prism 7 (GraphPad Software, USA). Measured absorbances (A450) or median fluorescence intensities (MFI) obtained in different buffer setups were compared by determination of the signal ratios. Higher values were always divided by lower values. Ratios obtained for different scFv-CaM-fusions were compared with the corresponding wildtype control (WT with [G4S]3-linker) via nonparametric Kruskal-Wallis with Dunn's multiple comparisons post-hoc test (≥3 groups) or via Mann-Whitney test (2 groups), respectively. The α value was always set to 0.05.
Disclosure of potential conflicts of interest
The authors have filed a patent application based on this work.
Supplementary_Table_1_and_Supplementary_Figures_1-4.docx
Download MS Word (3 MB)Acknowledgments
We thank Anna Bergmann and Torsten Schardt (Miltenyi Biotec) for excellent technical assistance in size-exclusion chromatography. We are grateful to Doris Meier (TU Braunschweig) for excellent technical assistance in initial experiments. We would like to thank Dagmar Häußler and Ian Johnston (Miltenyi Biotec) for reading the manuscript.
References
- Aggen DH, Chervin AS, Schmitt TM, Engels B, Stone JD, Richman SA, Piepenbrink KH, Baker BM, Greenberg PD, Schreiber H, et al. Single-Chain VαVβ T Cell Receptors Function Without Mispairing With Endogenous TCR Chains. Gene Ther 2012; 19:365-74; PMID: 21753797; http://dx.doi.org/10.1038/gt.2011.104
- Hust M, Jostock T, Menzel C, Voedisch B, Mohr A, Brenneis M, Kirsch MI, Meier D, Dübel S. Single chain Fab (scFab) fragment. BMC Biotechnol 2007; 7:14; PMID: 17346344; http://dx.doi.org/10.1186/1472-6750-7-14
- Huston JS, Levinson D, Mudgett-Hunter M, Tai MS, Novotny J, Margolies MN, Ridge RJ, Bruccoleri RE, Haber E, Crea R. Protein engineering of antibody binding sites: Recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc Natl Acad Sci 1988; 85:5879-83; PMID:3045807; http://dx.doi.org/10.1073/pnas.85.16.5879
- Bird RE, Hardman KD, Jacobson JW, Johnson S, Kaufman BM, Lee SM, Lee T, Pope SH, Riordan GS, Whitlow M. Single-Chain Antigen-Binding Proteins. Science 1988; 242:423-6; PMID: 3140379; http://dx.doi.org/10.1126/science.3140379
- van Wezenbeek P, Schoenmakers JGG. Nucleotide sequence of the genes III, VI and I of bacteriophage M13. Nucleic Acids Res 1979; 6:2799-818; PMID:379830; http://dx.doi.org/10.1093/nar/6.8.2799
- Dübel S, Stoevesandt O, Taussig MJ, Hust M. Generating recombinant antibodies to the complete human proteome. Trends Biotechnol 2010; 28:333-9; PMID:20538360; http://dx.doi.org/10.1016/j.tibtech.2010.05.001
- Rajput R, Sharma G, Rawat V, Gautam A, Kumar B, Pattnaik B, Pradhan HK, Khanna M. Diagnostic potential of recombinant scFv antibodies generated against hemagglutinin protein of influenza A virus. Front Immunol 2015; 6:440; PMID: 26388868; http://dx.doi.org/10.3389/fimmu.2015.00440
- Chaudhary VK, Queen C, Junghans RP, Waldmann TA, FitzGerald DJ, Pastan I. A recombinant immunotoxin consisting of two antibody variable domains fused to Pseudomonas exotoxin. Nature 1989; 339:394-7; PMID: 2498664; http://dx.doi.org/10.1038/339394a0
- Mack M, Riethmüller G, Kufer P. A small bispecific antibody construct expressed as a functional single-chain molecule with high tumor cell cytotoxicity. Proc Natl Acad Sci 1995; 92:7021-5; PMID:7624362; http://dx.doi.org/10.1073/pnas.92.15.7021
- Wolf E, Hofmeister R, Kufer P, Schlereth B, Baeuerle PA. BiTEs: bispecific antibody constructs with unique anti-tumor activity. Drug Discov Today 2005; 10:1237-44; PMID: 16213416; http://dx.doi.org/10.1016/S1359-6446(05)03554-3
- Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric Antigen Receptor-Modified T Cells in Chronic Lymphoid Leukemia. N Engl J Med 2011; 365:725-33; PMID: 21830940; http://dx.doi.org/10.1056/NEJMoa1103849
- Brentjens RJ, Davila ML, Riviere I, Park J, Wang X, Cowell LG, Bartido S, Stefanski J, Taylor C, Olszewska M, et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci Transl Med 2013; 5:177ra38; PMID: 23515080; http://dx.doi.org/10.1126/scitranslmed.3005930
- Moser AC, Hage DS. Immunoaffinity chromatography: an introduction to applications and recent developments. Bioanalysis 2010; 2:769-90; PMID: 20640220; http://dx.doi.org/10.4155/bio.10.31
- Kobatake E, Kosaku C, Hanzawa S, Mie M. Construction of affinity changeable antibody in response to Ca2+. Biotechnol Lett 2012; 34:1019-23; PMID: 22350334; http://dx.doi.org/10.1007/s10529-012-0881-z
- Miltenyi S, Müller W, Weichel W, Radbruch A. High Gradient Magnetic Cell Separation with MACS. Cytometry 1990; 11:231-8; PMID: 1690625; http://dx.doi.org/10.1002/cyto.990110203
- Shi H, Liu L, Wang Z. Improving the efficacy and safety of engineered T cell therapy for cancer. Cancer Lett 2013; 328:191-7; PMID: 23022475; http://dx.doi.org/10.1016/j.canlet.2012.09.015
- Hironiwa N, Ishii S, Kadono S, Iwayanagi Y, Mimoto F, Habu K, Igawa T, Hattori K. Calcium-dependent antigen binding as a novel modality for antibody recycling by endosomal antigen dissociation. MAbs 2016; 8:65-73; PMID: 26496237; http://dx.doi.org/10.1080/19420862.2015.1110660
- Murtaugh ML, Fanning SW, Sharma TM, Terry AM, Horn JR. A Combinatorial Histidine Scanning Library Approach to Engineer Highly pH-Dependent Protein Switches. Protein Sci 2011; 20:1619-31; PMID: 21766385; http://dx.doi.org/10.1002/pro.696
- Dübel S. Fv constructs with an influenceable affinity for a substance that is to be linked. US Patent 10/344,447. 2005;
- Megeed Z, Winters RM, Yarmush ML. Modulation of Single-Chain Antibody Affinity with Temperature-Responsive Elastin-Like Polypeptide Linkers. Biomacromolecules 2006; 7:999-1004; PMID: 16602713; http://dx.doi.org/10.1021/bm0507002
- Meister GE, Joshi NS. An Engineered Calmodulin-Based Allosteric Switch for Peptide Biosensing. ChemBioChem 2013; 14:1460-7; PMID: 23825049; http://dx.doi.org/10.1002/cbic.201300168
- Kuboniwa H, Tjandra N, Grzesiek S, Ren H, Klee CB, Bax A. Solution structure of calcium-free calmodulin. Nat Struct Biol 1995; 2:768-76; PMID: 7552748; http://dx.doi.org/10.1038/nsb0995-768
- Chattopadhyaya R, Meador WE, Means AR, Quiocho FA. Calmodulin Structure Refined at 1.7 Å Resolution. J Mol Biol 1992; 228:1177-92; PMID: 1474585; http://dx.doi.org/10.1016/0022-2836(92)90324-D
- Ikura M, Clore G, Gronenborn A, Zhu G, Klee C, Bax A. Solution Structure of a Calmodulin-Target Peptide Complex by Multidimensional NMR. Science 1992; 256:632-8; PMID: 1585175; http://dx.doi.org/10.1126/science.1585175
- Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 1997; 388:882-7; PMID: 9278050; http://dx.doi.org/10.1038/42264
- Nagai T, Sawano A, Park ES, Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2+. Proc Natl Acad Sci 2001; 98:3197-202; PMID: 11248055; http://dx.doi.org/10.1073/pnas.051636098
- Guntas G, Mitchell SF, Ostermeier M. A Molecular Switch Created by In Vitro Recombination of Nonhomologous Genes. Chem Biol 2004; 11:1483-7; PMID: 15555998; http://dx.doi.org/10.1016/j.chembiol.2004.08.020
- Guntas G, Ostermeier M. Creation of an Allosteric Enzyme by Domain Insertion. J Mol Biol 2004; 336:263-73; PMID: 14741221; http://dx.doi.org/10.1016/j.jmb.2003.12.016
- Dübel S, Breitling F, Klewinghaus I, Little M. Regulated Secretion and Purification of Recombinant Antibodies in E. coli. Cell Biophys 1992; 21:69-79; PMID:1285332; http://dx.doi.org/10.1007/BF02789479
- Montigiani S, Neri G, Neri P, Neri D. Alanine Substitutions in Calmodulin-binding Peptides Result in Unexpected Affinity Enhancement. J Mol Biol 1996; 258:6-13; PMID: 8613992; http://dx.doi.org/10.1006/jmbi.1996.0229
- Hultschig C, Hecht H-J, Frank R. Systematic Delineation of a Calmodulin Peptide Interaction. J Mol Biol 2004; 343:559-68; PMID: 15465045; http://dx.doi.org/10.1016/j.jmb.2004.08.012
- James P, Maeda M, Fischer R, Verma AK, Krebs J, Penniston JT, Carafoli E. Identification and Primary Structure of a Calmodulin Binding Domain of the Ca2+ Pump of Human Erythrocytes. J Biol Chem 1988; 263:2905-10; PMID: 2963820
- Leto T, Pleasic S, Forget B, Benz EJJ, Marchesi V. Characterization of the Calmodulin-binding Site of Nonerythroid α-Spectrin. Recombinant protein and model peptide studies. J Biol Chem 1989; 264:5826-30; PMID:2647727
- Matsubara M, Titani K, Taniguchi H. Interaction of Calmodulin-Binding Domain Peptides of Nitric Oxide Synthase with Membrane Phospholipids: Regulation by Protein Phosphorylation and Ca2+ -Calmodulin †. Biochemistry 1996; 35:14651-8; PMID: 8931564; http://dx.doi.org/10.1021/bi9613988
- Ehlers MD, Zhang S, Bernhardt JP, Huganir RL. Inactivation of NMDA Receptors by Direct Interaction of Calmodulin with the NR1 Subunit. Cell 1996; 84:745-55; PMID: 8625412; http://dx.doi.org/10.1016/S0092-8674(00)81052-1
- Holliger P, Prospero T, Winter G. Diabodies: Small bivalent and bispecific antibody fragments. Proc Natl Acad Sci U S A 1993; 90:6444-8; PMID: 8341653; http://dx.doi.org/10.1073/pnas.90.14.6444
- Arndt KM, Müller KM, Plückthun A. Factors Influencing the Dimer to Monomer Transition of an Antibody Single-Chain Fv Fragment†. Biochemistry 1998; 37:12918-26; PMID: 9737871; http://dx.doi.org/10.1021/bi9810407
- Schmiedl A, Zimmermann J, Scherberich JE, Fischer P, Dübel S. Recombinant variants of antibody 138H11 against human gamma-glutamyltransferase for targeting renal cell carcinoma. Hum Antibodies 2006; 15:81-94; PMID: 17065739
- Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Isolating and engineering human antibodies using yeast surface display. Nat Protoc 2006; 1:755-68; PMID: 17406305; http://dx.doi.org/10.1038/nprot.2006.94
- Amit AG, Mariuzza RA, Phillips SE, Poljak RJ. Three-Dimensional Structure of an Antigen-Antibody Complex at 2.8 Å Resolution. Science 1986; 233:747-53; PMID: 2426778; http://dx.doi.org/10.1126/science.2426778
- Boder ET, Midelfort KS, Wittrup KD. Directed evolution of antibody fragments with monovalent femtomolar antigen-binding affinity. Proc Natl Acad Sci U S A 2000; 97:10701-5; PMID: 10984501; http://dx.doi.org/10.1073/pnas.170297297
- Midelfort KS, Hernandez HH, Lippow SM, Tidor B, Drennan CL, Wittrup KD. Substantial Energetic Improvement with Minimal Structural Perturbation in a High Affinity Mutant Antibody. J Mol Biol 2004; 343:685-701; PMID: 15465055; http://dx.doi.org/10.1016/j.jmb.2004.08.019
- Abhinandan KR, Martin ACR. Analysis and prediction of VH/VL packing in antibodies. Protein Eng Des Sel 2010; 23:689-97; PMID: 20591902; http://dx.doi.org/10.1093/protein/gzq043
- Elshorst B, Hennig M, Försterling H, Diener A, Maurer M, Schulte P, Schwalbe H, Griesinger C, Krebs J, Schmid H, et al. NMR Solution Structure of a Complex of Calmodulin with a Binding Peptide of the Ca 2+ Pump †, ‡. Biochemistry 1999; 38:12320-32; PMID: 10493800; http://dx.doi.org/10.1021/bi9908235
- Dose C, Brieden J. Release system for cell-antibody-substrate conjugates containing a polyethylene glycol spacer unit. European Patent 12189516.3. 2014.
- Choi JY, Lee SH, Park CY, Heo W Do, Kim JC, Kim MC, Chung WS, Moon BC, Cheong YH, Kim CY, et al. Identification of Calmodulin Isoform-specific Binding Peptides from a Phage-displayed Random 22-mer Peptide Library. J Biol Chem 2002; 277:21630-8; PMID: 11901148; http://dx.doi.org/10.1074/jbc.M110803200
- Yosef E, Politi R, Choi MH, Shifman JM. Computational Design of Calmodulin Mutants with up to 900-Fold Increase in Binding Specificity. J Mol Biol 2009; 385:1470-80; PMID:18845160; http://dx.doi.org/10.1016/j.jmb.2008.09.053
- Loeffler FF, Foertsch TC, Popov R, Mattes DS, Schlageter M, Sedlmayr M, Ridder B, Dang F-X, von Bojničić-Kninski C, Weber LK, et al. High-flexibility combinatorial peptide synthesis with laser-based transfer of monomers in solid matrix material. Nat Commun 2016; 7:11844; PMID: 27296868; http://dx.doi.org/10.1038/ncomms11844
- Habermann E, Crowell K, Janicki P. Lead and Other Metals can Substitute for Ca2+ in Calmodulin. Arch Toxicol 1983; 54:61-70; PMID: 6314931; http://dx.doi.org/10.1007/BF00277816
- Chao SH, Suzuki Y, Zysk JR, Cheung WY. Activation of Calmodulin by Various Metal Cations as a Function of Ionic Radius. Mol Pharmacol 1984; 26:75-82; PMID: 6087119
- Mariuzza RA, Jankovic DL, Boulot G, Amit AG, Saludjian P, Le Guern A, Mazié JC, Poljak RJ. Preliminary Crystallographic Study of the Complex Between the Fab Fragment of a Monoclonal Anti-lysozyme Antibody and its Antigen. J Mol Biol 1983; 170:1055-8; PMID: 6644814; http://dx.doi.org/10.1016/S0022-2836(83)80206-X
- Goyert SM, Ferrero E. Biochemical analysis of myeloid antigens and cDNA expression of gp55 (CD14). In: McMichael AJ, editor. Leucocyte Typing III: White Cell Differentation Antigens. New York (USA): Oxford University Press; 1987. page 613-8.
- Healey D, Dianda L, Moore JP, McDougal JS, Moore MJ, Estess P, Buck D, Kwong PD, Beverley PC, Sattentau QJ. Novel Anti-CD4 Monoclonal Antibodies Separate Human Immunodeficiency Virus Infection and Fusion of CD4+ Cells from Virus Binding. J Exp Med 1990; 172:1233-42; PMID: 1698911; http://dx.doi.org/10.1084/jem.172.4.1233
- Polyak MJ, Deans JP. CD20 Workshop Panel report. In: Mason D, Andre P, Bensussan A, Buckley C, Civin C, Clark E, De Haas M, Goyert S, Hadam M, Hart D, et al., editors. Leucocyte Typing VII. New York (USA): Oxford University Press; 2002. page 93-101.
- Sambrook J, Russel DW. Molecular Cloning: A Laboratory Manual. 3rd ed. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2001.
